Abstract
The structure and function of microorganisms that live in and on us, the human microbiota, are a tremendous resource. Microbiota may help to explain individual variability in health outcomes and be a source of new biomarkers for environmental exposures and of novel prognostic and diagnostic indicators. The increase in availability of low-cost, high-throughput techniques makes it relatively straightforward to include microbiota assessments in epidemiologic studies. With the recent joint publications of the findings of the Human Microbiome Consortium and related studies, the consequent surge of interest in microbiome research, and remarkable media attention, the time is ripe for epidemiologists to contribute their expertise to and translate results of microbiota research for population health.
Keywords: Human Microbiome Project, microbiome, microbiota
The first phase of the Human Microbiome Project (HMP) was recently completed; the results of this comprehensive analysis of microbial life existing in and on the healthy human body appeared in June 2012 issues of the journals Nature (1–4), Science (5–14), and PLoS (15–24). These studies demonstrated that 1) the composition and function of human microbiota contribute to metabolic, regulatory, and morphogenetic networks in the human host, making humans a “superorganism” that relies on human and extrahuman genomes for functioning (25); 2) the composition of microbiota is essentially unique to the individual; and 3) the functions of microbiota are conserved across individuals. This strongly suggests that characterizing human microbiota structure and function in well-designed epidemiologic studies will give new insight into disease etiology, help to explain variability in host susceptibility, and be a source for novel diagnostic and prognostic markers.
On the human body, there are 10 bacterial cells for every human cell and an estimated 10 viral particles for every bacterial cell (26). Microbes are not limited to the skin, mouth, and gastrointestinal tract. Instead, every body surface that has a portal to the external environment is colonized by microbes, including the lung (27). These communities of bacteria, archaea, viruses, and fungi interact directly and indirectly with the host, shaping the immune system, developing tissues, and resisting or enhancing invasion by pathogens. The gut microbiota are as metabolically active as our livers, making them effectively another organ (28). Gut microbiota modulate lipid metabolism and glucose homeostasis, absorb dietary fats and lipid-soluble vitamins, and detoxify xenobiotics among other functions. Gut microbiota can activate (or inactivate) drugs, create a toxic by-product from a drug or food, or alter host drug metabolism by changing host gene expression (13). There are hints that the composition of gut microbiota can mediate the effectiveness of oral vaccines (29) and modify risk of chronic diseases such as asthma (30).
All microbiota are known to resist pathogen invasion, modulate pH and other functions, and respond rapidly to changes in their environment. Therefore, changes in the types or relative amounts of bacteria present or changes in their functional output might be used as markers of exposure to environmental hazards or of disease risk. In a rat model, the composition of intestinal microbiota changes following radiation, making it potentially a more sensitive marker of biological dose (31). Gut microbial profiles are associated with obesity (32) and oral microbial profiles with pancreatic cancer (33), although whether these are risk markers or risk factors remains to be seen.
Similarly, as we understand the essential functions performed by microbiota, we might either introduce microbes to perform functions (probiotics) or add nutrients to enhance existing functions (prebiotics). It is possible to change microbiota to treat disease: Total fecal transplants have proven successful for treating intractable antibiotic-associated diarrhea (34). The bacteria found on health-care workers' hands might be modified to enhance resistance to colonization by important hospital pathogens (35), urinary catheters might be inoculated with prebiotics to reduce risk of colonization by harmful bacteria, or eczema might be treated indirectly by modifying gut microbiota. Drugs or vaccines might be administered with prebiotics or probiotics to enhance their effects or minimize adverse effects.
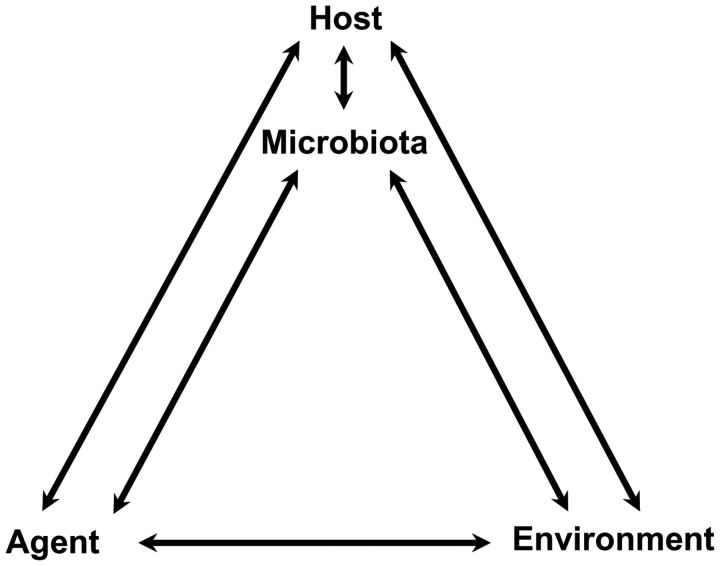
The potential of using microbiota structure or function as part of a diagnosis or of manipulating microbiota to treat or prevent disease is evident, but much work remains to realize this potential. Mechanistic studies are surely needed (36), but to translate these findings to clinical or public health interventions requires epidemiology. Epidemiologists are uniquely positioned to characterize the complex microbiota among large human populations and to help understand the direct and indirect impacts on disease. Are there measures of microbiota structure or function that correspond to health or disease? Are these measures risk markers, risk factors, or modifiers of either? How amenable are they to intervention? What is the variability of various measures of microbiota by person, place, and time and how do these change by host, agent, and environment (Figure 1)? As yet, however, the terms “microbiota” (the collection of microorganisms from a defined environment) and “microbiome” (the collection of genes harbored by microbiota) rarely appear in the epidemiologic literature. A search for articles on “microbiota, human and disease” using PubMed shows the number of papers published on this topic since 2000 rising essentially exponentially (Figure 2). Only 13 of these appeared in the American Journal of Epidemiology, Epidemiology, Annals of Epidemiology, International Journal of Epidemiology, and the Journal of the National Cancer Institute (searched on July 12, 2012).
Figure 1.
Microbiota can modify the effects of agent and environment on the host and indicate host changes in response to agent and environmental exposures.
Figure 2.
Frequency of papers found on PubMed search for “microbiota, human and disease,” on July 12, 2012.
Microbiota assessments are easily added to existing epidemiologic studies. Sample collection, storage, and consequent analyte extraction are generally very straightforward, although special preservatives must be used if samples are to be tested for transcripts. Basically, any sample from a human body site can be tested (Figure 3). The amount of material needed to assess microbiota structure or function is small and, with proper storage that minimizes analyte degradation, even previously collected specimens can be tested. For example, DNA has been extracted and screened from Gram's stain slides that had been stored in a slide box at room temperature for several years (37). Similarly, we conducted a metabolomic screen on frozen saliva collected for another purpose (B.F., unpublished data). However there are some important challenges in incorporating measures of microbiota in epidemiologic studies.
Figure 3.
Human microbiome sampling examples.
One challenge relates to the nature of what is being measured. Microbiota are living organisms so, unlike other genetic data but similar to many exposures assessed in epidemiology, microbiota are dynamic. Much like blood pressure is known to fluctuate but “healthy” falls within a certain range, microbiota change. How rapidly human microbiota respond to changes in the health status of the host or to environmental exposures is unknown and probably varies by microbiota location. Therefore, determining the range of normal variation over various time periods by site is essential for determining an appropriate sampling scheme for epidemiologic applications. Second, variation in measurement reflects not just true biological variance but variance from technical issues with analyte extraction method, amplification and sequencing primer sets, sequencing platform, and so on. Currently, the technology is under rapid development; reproducibility must be accounted for as each new technology is implemented (38, 39).
Once obtained, samples can be tested for the type and relative abundance of bacteria, virus, and fungi present (metagenomes); the functional potential of these microorganisms (metatranscriptomics); the actual functions being performed (metaproteomics); and what cellular by-products were produced (metabolomes) (Web Figure 1 available at http://aje.oxfordjournals.org/). Currently, the primary method for characterizing microbiota uses metagenomics, which we briefly describe. For more in-depth discussions of these techniques, including metagenomics, the reader is referred to several excellent recent reviews (40–43). For metagenomics, the most common technique uses polymerase chain reaction followed by sequencing of a segment of the ribosomal gene that is present in all cells (16S RNA for bacteria and 18S RNA for fungi). The advantage of using segments of the ribosomal gene is that there are several databases available to which the sequences can be mapped. This makes it possible to determine which phylum, genus, or species is present, but the resolution is dependent on the segment(s) amplified. It is also possible to sequence all DNA present by using a comprehensive shotgun metagenomic approach, whereby DNA from the collective genome of microorganisms is sheared and sequenced in small fragments. This approach enables detection of known genes and thus determination of the functional potential. In either case, sample preparation upon DNA extraction is required, involving reduction of sample contaminants (e.g., removal of host DNA), amplification, attachment of barcodes, and so on. For whole-genome metagenomic sequencing, further processing involves fragmentation of DNA and construction of paired-end libraries. Several sequencing platforms exist based on different technologies (e.g., capillary, pyrosequencing, integrated semiconductors), each bringing its own level of expected throughput, bias, and error rate. For example, the read length generated by the Roche 454 platforms (Roche Diagnostics Corporation, Indianapolis, Indiana) is a lengthy 400–700 base pairs, yet their error rate, mostly because of insertions and deletions, is approximately 1%. Conversely, the read length generated by the Illumina HiSeq platforms (Illumina, Inc., San Diego, California) is around 200–300 base pairs, and their error rate, mostly because of substitutions, is just over 0.1% (44).
Analysis of the resulting sequences involves filtering reads to remove artifacts introduced by polymerase chain reaction and sequencing errors, sequence assembly (if applicable), sequence alignment, taxonomic classification, functional assignment of reads and identification of metabolic pathways (if applicable), and use of phylogenetic and/or taxonomic binning approaches for microbial community characterization. Several analytical tools, mostly open source and each with its advantages and disadvantages, have been and continue to be developed to address the challenges of handling very large, multidimensional data (45). Although demanding the novice user to overcome a learning curve in terms of ecological theory and familiarity with command line interface, most include an integrated pipeline for comprehensive analysis that is not insurmountable to the first-time user. However, if preferred, most large research institutions, especially those having sequencing core centers, are now starting to provide bioinformatics support; this partnership should be fostered.
The resulting data structure (e.g., relative abundance of taxa per sample) and some of the analytical software may be unfamiliar, but to epidemiologists—who are used to large data sets that integrate multiple different types of measures—this should be a tractable problem. As in genetic studies, there are multiple variables; analytical techniques include ordination techniques (e.g., principal component analysis), cluster analysis, factor analysis, regression-based techniques, and analysis-of-variance–based techniques comparing microbial community matrices to one another. As a starting point, readers are directed to the Human Microbiome Project website, that includes the software, online resources, and standard operating protocols used as part of the Human Microbiome Project (http://www.hmpdacc.org/tools_protocols/tools_protocols.php).
Adding microbiota assessments to ongoing epidemiologic studies where samples are already collected only marginally increases the costs of sample processing. For metagenomics and metatranscriptomics, the costs of DNA and RNA extraction and preparation for sequencing are approximately $100 per sample, but as high-throughput procedures are implemented, these costs are falling rapidly. Once samples are prepared, the cost of determining microbiota structure by sequencing the 16S ribosomal RNA gene has been reported to be as low as $11 per sample (46). Sequencing service agreements can vary, with some facilities charging for repeat runs, sequence quality assessment, deconvoluting, eliminating bad sequences, and constructing high-quality reads. Untargeted metabolomic screens are significantly more expensive (approximately $200–$400 per sample), but once pathways of interest are identified, the costs for targeted assays are at least an order of magnitude lower. These tests are available at most major research institutions, and increasingly services are offered by private companies. There are also costs associated with learning how to manage and analyze these types of data. It is possible, however, to partner with academic institutions where this work is ongoing or to obtain training at the short courses and workshops that are increasingly available.
The first phase of the HMP was just a first step; increasingly, Human Microbiome Project researchers are turning their attention to the role of microbiota in health and disease. Epidemiologists should also, as the ability to characterize the genes and functions of our microbiota, our “supergenome,” has tremendous potential to help resolve outstanding issues in population health. Knowledge of microbiota functions might help to resolve outstanding questions about individual variability in susceptibility to environmental and infectious exposures. Differences in microbiota by place may further explain geographical variation in disease incidence. Considering how changes in medical practice, such as use of antibiotics, impact the microbiota might help to explain changes in disease incidence and prevalence over time. It is not enough, however, for epidemiologists to characterize human microbiota in health and disease. Epidemiologists need to rethink their paradigm of the role of microbes. Much of epidemiology (and public health) was built upon methods developed to identify infectious causes of disease. As we have outlined here, our microbiota are ourselves: Their genes are also our genes, and their functionings are our functionings. Without microbes, our bodies cannot develop or function properly (47). Just as some cancers result from normal cell function gone awry, some diseases result from our microbiota gone awry. Might microbiota explain some of the variability of susceptibility to disease, tolerance to environmental hazards, and response to treatment? Thinking of microbes as self provides another lever to pry open the proverbial black box.
ACKNOWLEDGMENTS
Author affiliation: Center for Molecular and Clinical Epidemiology of Infectious Diseases, Department of Epidemiology, University of Michigan School of Public Health, Ann Arbor, Michigan (Betsy Foxman, Mariana Rosenthal).
The authors contributed equally to the work.
This work was supported by the Interdisciplinary Training Program in Infectious Diseases (T32 AI049816 (M.R.) and R01 DE014899 (B.F.)).
We thank Dr. Sara Adar for her helpful comments on an earlier draft of this manuscript and Dr. Claudia Rosenthal for her illustrations.
Conflict of interest: none declared.
REFERENCES
- 1.Human Microbiome Project Consortium. A framework for human microbiome research. Nature. 2012;486(7402):215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Relman DA. Microbiology: learning about who we are. Nature. 2012;486(7402):194–195. doi: 10.1038/486194a. [DOI] [PubMed] [Google Scholar]
- 4.Knight R, Jansson J, Field D, et al. Unlocking the potential of metagenomics through replicated experimental design. Nat Biotechnol. 2012;30(6):513–520. doi: 10.1038/nbt.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mueller K, Ash C, Pennisi E, et al. The gut microbiota. Introduction. Science. 2012;336(6086):1245. doi: 10.1126/science.336.6086.1245. [DOI] [PubMed] [Google Scholar]
- 6.Costello EK, Stagaman K, Dethlefsen L, et al. The application of ecological theory toward an understanding of the human microbiome. Science. 2012;336(6086):1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336(6086):1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balter M. Taking stock of the human microbiome and disease. Science. 2012;336(6086):1246–1247. doi: 10.1126/science.336.6086.1246. [DOI] [PubMed] [Google Scholar]
- 9.Hvistendahl M. Pigs as stand-ins for microbiome studies. Science. 2012;336(6086):1250. doi: 10.1126/science.336.6086.1250. [DOI] [PubMed] [Google Scholar]
- 10.Nicholson JK, Holmes E, Kinross J, et al. Host-gut microbiota metabolic interactions. Science. 2012;336(6086):1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 11.Hood L. Tackling the microbiome. Science. 2012;336(6086):1209. doi: 10.1126/science.1225475. [DOI] [PubMed] [Google Scholar]
- 12.Hvistendahl M. My microbiome and me. Science. 2012;336(6086):1248–1250. doi: 10.1126/science.336.6086.1248. [DOI] [PubMed] [Google Scholar]
- 13.Haiser HJ, Turnbaugh PJ. Is it time for a metagenomic basis of therapeutics? Science. 2012;336(6086):1253–1255. doi: 10.1126/science.1224396. [DOI] [PubMed] [Google Scholar]
- 14.Kamada N, Kim YG, Sham HP, et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science. 2012;336(6086):1325–1329. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papa E, Docktor M, Smillie C, et al. Non-invasive mapping of the gastrointestinal microbiota identifies children with inflammatory bowel disease. PLoS One. 2012;7(6):e39242. doi: 10.1371/journal.pone.0039242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flores R, Shi J, Gail MH, et al. Association of fecal microbial diversity and taxonomy with selected enzymatic functions. PLoS One. 2012;7(6):e39745. doi: 10.1371/journal.pone.0039745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jumpstart Consortium Human Microbiome Project Data Generation Working Group. Evaluation of 16S rDNA-based community profiling for human microbiome research. PLoS One. 2012;7(6):e39315. doi: 10.1371/journal.pone.0039315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aagaard K, Riehle K, Ma J, et al. A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy. PLoS One. 2012;7(6):e36466. doi: 10.1371/journal.pone.0036466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wylie KM, Truty RM, Sharpton TJ, et al. Novel bacterial taxa in the human microbiome. PLoS One. 2012;7(6):e35294. doi: 10.1371/journal.pone.0035294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li K, Bihan M, Yooseph S, et al. Analyses of the microbial diversity across the human microbiome. PLoS One. 2012;7(6):e32118. doi: 10.1371/journal.pone.0032118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cantarel BL, Lombard V, Henrissat B. Complex carbohydrate utilization by the healthy human microbiome. PLoS One. 2012;7(6):e28742. doi: 10.1371/journal.pone.0028742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu B, Faller LL, Klitgord N, et al. Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PLoS One. 2012;7(6):e37919. doi: 10.1371/journal.pone.0037919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Licciardi PV, Toh ZQ, Dunne E, et al. Protecting against pneumococcal disease: critical interactions between probiotics and the airway microbiome. PLoS Pathog. 2012;8(6):e1002652. doi: 10.1371/journal.ppat.1002652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abubucker S, Segata N, Goll J, et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput Biol. 2012;8(6):e1002358. doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murdoch TB, Detsky AS. Time to recognize our fellow travellers. J Gen Intern Med. 2012;27(12):1704–1706. doi: 10.1007/s11606-012-2105-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clemente JC, Ursell LK, Parfrey LW, et al. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148(6):1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beck JM, Young VB, Huffnagle GB. The microbiome of the lung. Transl Res. 2012;160(4):258–266. doi: 10.1016/j.trsl.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bocci V. The neglected organ: bacterial flora has a crucial immunostimulatory role. Perspect Biol Med. 1992;35(2):251–260. doi: 10.1353/pbm.1992.0004. [DOI] [PubMed] [Google Scholar]
- 29.Björkstén B. Diverse microbial exposure—consequences for vaccine development. Vaccine. 2012;30(29):4336–4340. doi: 10.1016/j.vaccine.2011.10.074. [DOI] [PubMed] [Google Scholar]
- 30.Blaser MJ. Equilibria of humans and our indigenous microbiota affecting asthma. Proc Am Thorac Soc. 2012;9(2):69–71. doi: 10.1513/pats.201108-048MS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lam V, Moulder JE, Salzman NH, et al. Intestinal microbiota as novel biomarkers of prior radiation exposure. Radiat Res. 2012;177(5):573–583. doi: 10.1667/rr2691.1. [DOI] [PubMed] [Google Scholar]
- 32.Ley RE, Turnbaugh PJ, Klein S, et al. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444(7122):1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 33.Farrell JJ, Zhang L, Zhou H, et al. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut. 2012;61(4):582–588. doi: 10.1136/gutjnl-2011-300784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landy J, Al-Hassi HO, McLaughlin SD, et al. Review article: faecal transplantation therapy for gastrointestinal disease. Aliment Pharmacol Ther. 2011;34(4):409–415. doi: 10.1111/j.1365-2036.2011.04737.x. [DOI] [PubMed] [Google Scholar]
- 35.Rosenthal M, Goldberg D, Aiello A, et al. Skin microbiota: microbial community structure and its potential association with health and disease. Infect Genet Evol. 2011;11(5):839–848. doi: 10.1016/j.meegid.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gordon JI. Honor thy gut symbionts redux. Science. 2012;336(6086):1251–1253. doi: 10.1126/science.1224686. [DOI] [PubMed] [Google Scholar]
- 37.Srinivasan U, Ponnaluri S, Villareal L, et al. Gram stains: a resource for retrospective analysis of bacterial pathogens in clinical studies. PLoS One. 2012;7(10):e42898. doi: 10.1371/journal.pone.0042898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Claesson MJ, Wang Q, O'Sullivan O, et al. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 2010;38(22):e200. doi: 10.1093/nar/gkq873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuan S, Cohen DB, Ravel J, et al. Evaluation of methods for the extraction and purification of DNA from the human microbiome. PLoS One. 2012;7(3):e33865. doi: 10.1371/journal.pone.0033865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuczynski J, Lauber CL, Walters WA, et al. Experimental and analytical tools for studying the human microbiome. Nat Rev Genet. 2011;13(1):47–58. doi: 10.1038/nrg3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13(4):260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamady M, Knight R. Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Res. 2009;19(7):1141–1152. doi: 10.1101/gr.085464.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang JY, Karr JR, Watrous JD, et al. Integrating ‘-omics’ and natural product discovery platforms to investigate metabolic exchange in microbiomes. Curr Op Chem Biol. 2011;15(1):79–87. doi: 10.1016/j.cbpa.2010.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scholz MB, Lo CC, Chain PSG. Next generation sequencing and bioinformatic bottlenecks: the current state of metagenomic data analysis. Curr Opin Biotechnol. 2012;23(1):9–15. doi: 10.1016/j.copbio.2011.11.013. [DOI] [PubMed] [Google Scholar]
- 45.Gonzales A, Knight R. Advancing analytical algorithms and pipelines for billions of microbial sequences. Curr Opin Biotechnol. 2012;23(1):64–71. doi: 10.1016/j.copbio.2011.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caporaso JG, Lauber CL, Costello EK, et al. Moving pictures of the human microbiome. Genome Biol. 2011;12(5):R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449(7164):811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]