Abstract
The mRNA 3′-untranslated region (3′-UTR) modulates message stability, transport, intracellular location and translation. We have discovered a novel nuclear poly(A) polymerase termed Star-PAP (nuclear speckle targeted PIPKIα regulated-poly(A) polymerase) that couples with the transcriptional machinery and is regulated by the phosphoinositide lipid messenger phosphatidylinositol-4,5-bisphosphate (PI4,5P2), the central lipid in phosphoinositide signaling. PI4,5P2 is generated primarily by type I phosphatidylinositol phosphate kinases (PIPKI). Phosphoinositides are present in the nucleus including at nuclear speckles compartments separate from known membrane structures. PIPKs regulate cellular functions by interacting with PI4,5P2 effectors where PIPKs generate PI4,5P2 that then modulates the activity of the associated effectors. Nuclear PIPKIα interacts with and regulates Star-PAP, and PI4,5P2 specifically activates Star-PAP in a gene- and signaling-dependent manner. Importantly, other select signaling molecules integrated into the Star-PAP complex seem to regulate Star-PAP activities and processivities toward RNA substrates, and unique sequence elements around the Star-PAP binding sites within the 3′-UTR of target genes contribute to Star-PAP specificity for processing. Therefore, Star-PAP and its regulatory molecules form a signaling nexus at the 3′-end of target mRNAs to control the expression of select group of genes including the ones involved in stress responses.
Introduction
Stress responses are critical biological processes involving intricate cellular signaling cascades and protein synthesis, turnover/redistribution and changes in gene expression required for organismic survival. Under a specific stress condition, only select genes are transcribed and processed via signaling activation of transcription factors and mRNA processing machineries. While signaling regulations of gene expression at the 5′-end of messages have been extensively studied, the hunt for signaling relays at the 3′-end that are equally essential as those at the 5′-end during processing of the transcripts has just begun.
Nascent mRNAs are tailed at the 3′-end by a stretch of poly-adenosine monophosphates called poly(A) tail. This occurs after 3′-end cleavage of the messages at a site situated ~20 nt downstream of the poly(A) signal (AAUAAA). These processes at the 3′-end of a transcript require high fidelity of the corresponding sequence elements within the mRNA and well-tuned coordination of the processing factors that often bind the message. Non-permissive alterations in this highly ordered process could result in malfunctioning of the 3′-end processing machineries and generation of unnecessary transcripts that could impact the survival of cells and organisms.
Studies in human diseases such as genetic disorders and cancers have detected mutations in the cis-regulatory elements of mRNAs, changes in the levels of the associated regulatory molecules and even the variations in the expression and activities of poly(A) polymerase. In addition, utilization of alternative 3′-UTR sequences has been implicated in embryonic development. These collectively highlight the abundance of the messages that the 3′-end of mRNAs carries, and thus regulation strategies targeting the tails of these mRNAs represent a novel venue for the intervention of human diseases.
The nuclear speckle targeted poly(A) polymerase (PAP), Star-PAP, was identified as a stress signal-regulated mRNA 3′-end processing enzyme involved in cleavage and polyadenylation of target pre-mRNAs. Interestingly, Star-PAP recruits common mRNA 3′-end processing factors that are also components of the canonical PAP complex and recruit additional enzymes to form and activate the Star-PAP complexes. Star-PAP is regulated downstream of stress signals and differentially couples with specific activating molecules (transactivating factors) for the processing of select target mRNAs. These mRNAs have well defined sequence elements at the 3′-end matching with Star-PAP binding specificity. Therefore, Star-PAP is not only a biosensor for the transduction of stress signals within cell nucleus but also an effector for the fate of target mRNAs. Here the roles of Star-PAP in mRNA processing and stress response are discussed.
Star-PAP enzymatic activities and activation for target RNA processing
Star-PAP is a non-canonical PAP (Mellman et al, 2008), a novel member of the DNA Pol β superfamily of terminal nucleotidyl transferases (Martin et al, 2008; Schmidt & Norbury, 2010; Stevenson & Norbury, 2006). It controls the 3′-end processing of select genes involved in various cellular processes such as oxidative stress response and apoptosis (Li et al, 2012; Mellman et al, 2008). In vitro, Star-PAP can add poly(A) tail to short oligonucleotides showing its polyadenylation property, yet it also has uridylation activity (Mellman et al, 2008; Trippe et al, 2006). Nevertheless, at physiological concentrations of ATP, adenylation activity competes over uridylation activity suggesting the primary role of Star-PAP as a PAP in cells (Mellman et al, 2008). In addition, Star-PAP was shown to be involved in both cleavage and polyadenylation of its target heme oxygenase 1 (HO-1) pre-mRNA (Laishram & Anderson, 2010; Mellman et al, 2008). Thus, Star-PAP activity is discussed in the context of in vitro non-specific polyadenyation, specific cleavage, and target mRNA polyadenylation.
Non-specific polyadenylation
Star-PAP has PAP activity and can add long (A)-tail toward nonspecific substrates such as generic A15 poly(A) RNA oligo primer or A45 oligo primer of the sequence (UAGGGA5)A15, and also RNA L1 substrate (Mellman et al, 2008). Intriguingly, while the polyadenylation activity of recombinant Star-PAP was stimulated >10 fold by phosphatidylinositol 4,5-biphosphate (PI4,5P2), Star-PAP purified from cells required priming with oxidative stress (tBHQ) or DNA damage signal (etoposide treatment) for PI4,5P2 stimulation of its polyadenylation activity (Li et al, 2012; Mellman et al, 2008). The stimulation bears two facets. First, treatment of cells with the agonists causes an increase in PAP activity of Star-PAP for the initiation resulting in the formation of enhanced very short (A)-chain (at least 1 adenosine addition). Second, Star-PAP isolated from tBHQ or etoposide treated cells shows a dramatic increase in the processivity of the polymerase in the presence of PI4,5P2, resulting in the formation of increased longer (A)-chain. Both initiation and processivity of Star-PAP activity are regulated by protein kinases such as casein kinase I (CKI) and protein kinase C delta (PKCδ), and signaling events like DNA damage and oxidative stress (Gonzales et al, 2008; Laishram et al, 2011; Li et al, 2012; Mellman et al, 2008).
Characterization of Star-PAP non-specific polyadenylation activity using PAP assays at different time points illustrated a high processivity of Star-PAP. Star-PAP extended the A45-primer to the maximum (A)-tail length (>800 nucleotides) in less than 5 minutes (Li et al, 2012; Mellman et al, 2008). The amount of (A)-tailed substrates increased linearly with time in both the initiation and the processivity of Star-PAP activity (Li et al, 2012). Various factors limit the poly(A) tail length to 200-300 adenosine residues within the cell (Mandel et al, 2008; Zhao et al, 1999). Purified Star-PAP, like other PAPs, can add several hundred long (A)-tail to the oligo substrates or non-specific templates in vitro (Kyriakopoulou et al, 2001; Martin & Keller, 1996; Mellman et al, 2008; Nagaike et al, 2005; Raabe et al, 1991; Zhelkovsky et al, 1995). This is due to the high processivity of the enzyme and absence of poly(A) tail length control mechanisms in vitro.
Specific cleavage of target pre-mRNA
Microarray analysis demonstrated that Star-PAP regulates oxidative stress-responsive genes such as HO-1 and NQO-1 (Mellman et al, 2008). Oxidative stress agonist tBHQ stimulates HO-1 expression in a Star-PAP-dependent manner (Laishram & Anderson, 2010; Mellman et al, 2008). Measurement of mRNA cleavage using a pair of primers across the cleavage site showed an increased accumulation of uncleaved HO-1 transcripts upon Star-PAP knockdown. The non Star-PAP targets such as GCLC and GAPDH, however, were not affected, indicating the involvement of Star-PAP in mRNA 3′-end processing of specific genes (Mellman et al, 2008). This was supported by the measurement of the poy(A) tail of HO-1 mRNA using 3′-RACE assay, where knockdown of Star-PAP resulted in loss of both the oxidative induced and uninduced 3′-end tail formation of HO-1 mRNA even in the presence of oxidative stress (Laishram & Anderson, 2010). Further, primer extension by reverse transcription using a 5′-end radio-labeled primer specific to HO-1 UTR downstream of cleavage site corroborated the observations of the Star-PAP requirement for the cleavage and polyadenylation of HO-1 pre-mRNA (Laishram & Anderson, 2010).
Although cleavage and polyadenylation reactions are tightly coupled in vivo, the two processes can be uncoupled in vitro using chain terminating ATP molecules (3′-dATP) (Humphrey et al, 1987; Takagaki et al, 1989). In vitro cleavage reactions using an in vitro transcribed HO-1 UTR RNA fragment encompassing all cis-acting elements required for cleavage and polyadenylation and active HeLa nuclear extracts confirmed the requirement of Star-PAP in HO-1 pre-mRNA cleavage (Laishram & Anderson, 2010). Hela nuclear extract prepared from Star-PAP knockdown cells failed to cleave HO-1 UTR RNA. Recombinant Star-PAP but not PAPα was able to rescue the cleavage defect. There was a dose dependent increase in the cleavage of HO-1 pre-mRNA with increasing concentrations of Star-PAP in the rescue experiment, confirming the role of Star-PAP in the cleavage. Interestingly, oxidative stress treatment of the cells stimulated the cleavage reaction and PI4,5P2 addition did not affect the cleavage of HO-1 pre-mRNA, suggesting that the phosphatidylinositol-4-phosphate 5-kinase I alpha (PIPKIα) activities are not involved in this process for this gene (Laishram & Anderson, 2010).
Specific polyadenylation of target pre-mRNA
Polyadenylation is always coupled with the preceding 3′-end cleavage in vivo (Colgan & Manley, 1997; Proudfoot, 2004; Zhao et al, 1999). Various previous experiments demonstrated the role of Star-PAP in the 3′-end formation of its target mRNA. Therefore, the polyadenylation activity of Star-PAP toward target pre-mRNA was tested using the HO-1 UTR RNA in an in vitro coupled cleavage and polyadenylation assay (Laishram & Anderson, 2010). The HeLa nuclear extract showed specific polyadenylation after cleavage of the HO-1 RNA substrate. Knockdown of Star-PAP abolished both cleavage and polyadenylation of the RNA. In addition, pre-cleaved HO-1 mRNA remained unpolyadenylated with the nuclear extract from Star-PAP knockdown cells (Laishram & Anderson, 2010). Unlike the non-specific polyadenylation, the addition of poly(A) tail to the HO-1 UTR RNA by Star-PAP was limited to ~250 adenosine residues. Thus, the reconstitution experiment reiterated that Star-PAP is indeed the PAP responsible for polyadenylation of target HO-1 mRNA. The polaydeylation activity was as in the case of non-specific polyadenylation, enhanced several fold by the addition of PI4,5P2. However, tBHQ treatment did not impact the specific polyadenylation. As noted before, tBHQ treatment but not PI4,5P2 addition stimulated the cleavage reaction (Mellman et al, 2008). Together, this supports a hypothesis where oxidative stress-induced cleavage reaction of HO-1 pre-mRNA is stimulated by PI4,5P2-mediated enhancement of Star-PAP polyadenylation. It suggests an interconnected pathway of phosphoinositide and oxidative stress signal regulating Star-PAP-mediated cleavage and polyadenylation of its target pre-mRNAs.
Mechanism of Star-PAP-mediated cleavage and polyadenylation of target pre-mRNA
Star-PAP regulates the cleavage and polyadenylation of its target HO-1 pre-mRNA. In addition to unique regulatory kinases such as PIPKIα and CKIα, Star-PAP associates with all the canonical 3′-end processing factors including CPSF (160, 73, 100, 30 and hFip1 subunits), symplekin, CstF and hFip1 (Laishram & Anderson, 2010; Mellman et al, 2008) but is devoid of detectable PAPα and vice versa, indicating the distinct nature of the Star-PAP and canonical PAPα complexes (Mellman et al, 2008). GST-pull down experiments demonstrated that Star-PAP directly interacts with CPSF 160 and CPSF 73, which are key factors for cleavage reaction. Star-PAP also associates with target pre-mRNAs such as HO-1 and BIK at 3′-UTR. PAPα on the other hand does not associate with the Star-PAP targets such as HO-1 or BIK UTR RNA (Laishram & Anderson, 2010; Li et al, 2012). These results indicate that Star-PAP and PAPα exist in two distinct 3′-end processing complexes each having a distinct niche of target genes (Fig. 1).
Fig.1.
Mechanism of Star-PAP mediated mRNA 3′-end processing. Stress signals activate specific protein kinases and other factors, which are targeted to the Star-PAP complex and induce Star-PAP activation. In turn, other regulatory proteins including the CPSF subunits required for the 3′-end processing could be recruited to the complex. Star-PAP binds a GC-rich RNA sequence upstream of the poly(A) signal. The U/GU deficient DSE is suboptimal for CstF 64 binding, which in combination with Star-PAP binding, excludes PAPα recruitment and the formation of PAPα-based processing complex.
Star-PAP binds target pre-mRNA UTR
Since Star-PAP has two RNA binding motifs: zinc finger (ZF) and RNA recognition motif (RRM), and associates with target RNAs in the cell, it was hypothesized that Star-PAP directly interacts with its target mRNAs. Electrophoretic mobility shift assays (EMSA) using radio-labeled HO-1 or GAPDH UTR RNA fragments revealed that Star-PAP indeed binds its target HO-1 RNA directly and specifically (Laishram & Anderson, 2010). The binding exhibited around ~10 nM of half maximal binding which is consistent with the general binding properties of most RNA binding proteins and transcription factors (Chen & Varani, 2005; Lee & Schedl, 2006; Lunde et al, 2007; Ptashne & Gann, 1997). Deletion of either ZF or RRM resulted in a reduction in the affinity of binding. However, loss of binding was observed only when both domains were deleted. While full length Star-PAP showed half maximal binding of ~10 nM, individual deletions showed half maximal binding of 20-30 nM. Interestingly, a peptide comprised of both ZF and RRM domains of Star-PAP bound with ~5 fold higher affinity to HO-1 RNA with a half maximal binding of ~2 nM confirming that Star-PAP RNA binding requires both ZF and RRM domains and suggesting that the full length Star-PAP partially folds over the ZF and RRM domains (Laishram & Anderson, 2010). The involvement of two domains for RNA recognition, however, complicates the determination of actual binding sequence. Therefore, the actual binding sequence might involve the combination of multiple nucleotide elements interacting with ZF or RRM or both. The multiple contacts between Star-PAP and target pre-mRNA would in turn enhance both the specificity and flexibility for targeting specific RNA sequences.
Since Star-PAP was found to directly bind HO-1 RNA, the binding region and the required sequence for Star-PAP interaction with HO-1 RNA was further investigated by RNA footprinting (Laishram & Anderson, 2010). Both RNase T1 and RNase S showed a similar protection of a region around ~60 nt footprint on the RNA. It extended from ~108 to ~50 nucleotide upstream of cleavage site followed by weak protection extended up to ~45 nt downstream (Laishram & Anderson, 2010). Based on this observation, deletion analysis of the protected region also resulted in the loss of Star-PAP binding and failure of cleavage by HeLa nuclear extract (Laishram & Anderson, 2010). This further confirmed that Star-PAP binds a region extending from ~108 to ~50 nucleotides upstream of the cleavage site.
Star-PAP recruits CPSF factors to promote specific cleavage
RNA immunoprecipitation (RIP) analysis identified association of the CPSF subunits (160, 100, 73, and 30) with HO-1 pre-mRNA in a Star-PAP dependent manner (Laishram & Anderson, 2010), suggesting a role of Star-PAP in the assembly of the CPSF complex onto target RNAs. This was corroborated by EMSA experiments with HO-1 UTR RNA and the immunoprecipitated CPSF 160 complex from the cells in the presence or absence of Star-PAP knockdown and HO-1 UTR RNA (Laishram & Anderson, 2010). Similar experiments with AAUAAA containing UTR RNA and nuclear extract showed a mobility-shifted complex of the 3′-end processing factors assembled on the RNA (Humphrey et al, 1987). For the Star-PAP target HO-1 RNA, a distinct 3′-end processing complex formed in nuclear extract, which was not observed upon Star-PAP knockdown (Laishram & Anderson, 2010). This demonstrated that Star-PAP is required for the stable assembly of the cleavage complex on HO-1 RNA.
Star-PAP specific recruitment of CPSF was further defined using recombinant proteins. Recombinant CPSF 160 exhibited weak binding to the HO-1 UTR RNA, which in the presence of recombinant Star-PAP formed a stronger ternary complex of CPSF 160, HO-1 RNA and Star-PAP (Laishram & Anderson, 2010). In a dose dependent EMSA experiment, CPSF 160 bound to the HO-1 RNA with a half maximal binding of ~60 nM. However, in the presence of Star-PAP, the binding affinity was increased with a half maximal binding of ~20 nM, suggesting that Star-PAP recruits CPSF 160 to assemble a cleavage complex. This was confirmed by an in vitro reconstitution of cleavage assay using limited number of 3′-end processing factors (Laishram & Anderson, 2010). To date, no successful reconstitution of the in vitro cleavage reaction with recombinant cleavage factors was reported, likely due to the requirement of a complex set of proteins for the cleavage reaction in vivo (Shi et al, 2009). Yet, for Star-PAP, specific cleavage of HO-1 RNA in vitro was reconstituted with Star-PAP along with CPSF 160 and 73 subunits (Laishram & Anderson, 2010). While CPSF 73 alone exhibited non-specific cleavage, it specifically cleaved HO-1 RNA at the cleavage site in the presence of Star-PAP and CPSF 160 (Laishram & Anderson, 2010; Mandel et al, 2006). Specific cleavage was weak indicating that additional cleavage factors or regulation is required to obtain the optimal cleavage in vivo (Humphrey et al, 1987; Mandel et al, 2008; Takagaki et al, 1989). Nevertheless, this represents the first reconstitution of a specific cleavage reaction in vitro with recombinant and a limited number of 3′-end processing factors (Laishram & Anderson, 2010; Marzluff, 2010). This reconstitution suggests that the failure to reconstitute the cleavage reaction in vitro in the previous studies would probably relate to the inability to assemble a strong complex on the 3′-UTR RNA substrate to recruit the CPSF 73. In case of Star-PAP, the association of Star-PAP with CPSF 160 and the HO-1 RNA was sufficiently strong to recruit and position CPSF 73 at the correct cleavage site.
Model of Star-PAP-mediated 3′-end processing
Star-PAP binds a GC rich region upstream of the poly(A) signal of its target pre-mRNA and also directly interacts with CPSF 160 and 73 subunits (Laishram & Anderson, 2010; Li et al, 2012). CPSF 160 feebly interacts with the poly(A) signal on the target RNA, and its cooperation with other cleavage factors in the absence of Star-PAP is not stable enough to assemble a processing complex on the RNA. Star-PAP direct binding to the RNA and an additional interaction with CPSF 160 then help recruit CPSF complex to the Poly(A) signal. The direct interaction with CPSF 160 and Star-PAP will recruit CPSF 73 to the complex and direct it to the correct cleavage site (Fig. 1). The interaction-effected recruitment of Star-PAP and CPSF on cleavage site provides the cue for remaining cleavage factors to be engaged at the site of assembly. Cleavage ensues with the action of CPSF 73 followed by subsequent polyadenylation. This mechanism is in contrast to the PAPα-mediated 3′-end processing where PAPα does not have RNA specificity nor binds RNA (Martin & Keller, 1996; Wahle, 1991; Zhao et al, 1999). Instead, PAPα is recruited by CPSF complex (Keller et al, 1991). CPSF 160 associates with the poly(A) signal and co-operates with CstF, which binds to GU/U-rich DSE and CF Im that recognizes UGUA sequence on the pre-mRNA to assemble a stable complex. This complex by virtue of CPSF interaction with PAPα, recruits PAPα to the cleavage site for polyadenylation (Brown & Gilmartin, 2003; Gilmartin & Nevins, 1991; Keller et al, 1991; Murthy & Manley, 1995). Thus, Star-PAP is the first example of a PAP that recruits CPSF subunits to promote efficient cleavage of an mRNA and represents a novel mechanism for cleavage and polyadenylation of pre-mRNAs.
It is not clear why Star-PAP target mRNAs are exclusively regulated by Star-PAP and not by PAPα. One obvious reason for the specificity is Star-PAP binding to its target pre-mRNA upstream of the poly(A) signal (Laishram & Anderson, 2010). Star-PAP footprint on HO-1 and BIK pre-mRNAs and RNA sequence analysis of Star-PAP target genes indicated a GC rich sequence ~40-60 nucleotides upstream of the cleavage site (Laishram & Anderson, 2010; Li et al, 2012). Yet, it does not answer why PAPα cannot process Star-PAP target genes while there are intact poly(A) signals and cleavage sites. Bioinformatics analysis showed that there appears to be U/GU-depleted downstream sequence element (DSE) present in the Star-PAP target genes (Li et al, 2012). Moreover, the Star-PAP target BIK UTR RNA does not have an optimal DSE, which is critical for CstF binding (Li et al, 2012). Therefore, one possible reason for PAPα exclusion from the Star-PAP target genes could be that the low U/GU sequence (suboptimal DSE) downstream of cleavage site renders the pre-mRNA inaccessible to CstF and thus prevents the recruitment of PAPα (Fig.1).
Regulation of Star-PAP activity by protein kinases and stress signaling
The observations that Star-PAP activities are regulated and Star-PAP targets select mRNA sequences for processing suggest that Star-PAP is a sensor at the 3′-end of a transcript under processing. To date, several signaling molecules have been identified as direct Star-PAP activators and two major stress-responsive pathways regulating Star-PAP activities defined.
PIPKIα and PI4,5P2 regulation of Star-PAP activity
Star-PAP was identified in a yeast two-hybrid screen using the PIPKIα C-terminal nuclear speckle-targeting region as bait (Mellman et al, 2008). Interestingly, while readily detectable in the Star-PAP complex, PIPKIα was excluded from the PAPα complex, and its messenger product PI4,5P2 that markedly triggered Star-PAP processivity failed to stimulate PAPα polyadenylation activities (Mellman et al, 2008), indicating a PAP-specific manner of PIPKIα and PI4,5P2 regulation. This also highlights the notion that PIPKIα exerts its function by targeting PI4,5P2 effector proteins. PIPKs interact with receptor targets and spatially and temporally generate PI4,5P2, which associates with the target proteins via unique phosphoinositide binding domains to activate the targets (Heck et al, 2007).
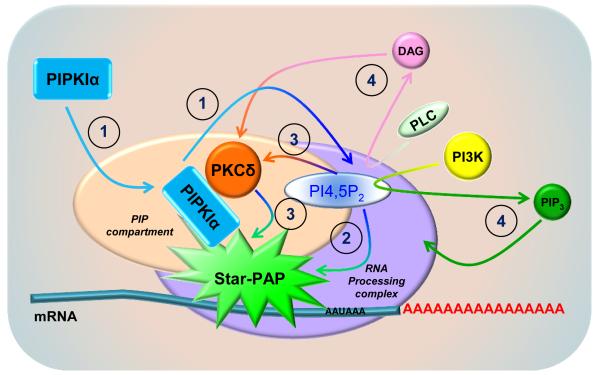
PIPKIα activation of Star-PAP might be achieved through four possible ways (Fig. 2). First, activation of the locally targeted PIPKIα within a phosphatidylinositol phosphate (PIP)-containing compartment in the vicinity of pre-mRNA templates generates PI4,5P2, which further reinforced the formation and configuration of the Star-PAP processing complex. This facilitates the recruitment of other protein kinases such as PKCδ required for Star-PAP activation (Li et al, 2012). Second, the locally generated PI4,5P2 directly activates Star-PAP. This is supported by the evidence that PI4,5P2 stimulates the RNA processing activities of both recombinant Star-PAP in vitro and isolated Star-PAP from cells (Mellman et al, 2008). Third, PI4,5P2, acting as a direct messenger, binds and activates other kinases such as PKCδ within the Star-PAP complex that further activates Star-PAP (Li et al, 2012). Fourth, PI4,5P2 is further catalytically processed by nuclear PLCs or PI3K to generate second messengers that switch on additional pathways inducing Star-PAP activation although this might not apply to all Star-PAP targets since under certain conditions diacylglycerol (DAG) failed to mediate PIPKIα- and PKCδ-regulated Star-PAP activities (Li et al, 2012). The existence of a nuclear phosphoinositide signaling network independent of the cytosolic counterpart and separated from the nuclear membrane structures has been well established (Barlow et al, 2009; Cocco et al, 1987; Cocco et al, 1988; Payrastre et al, 1992). Interestingly, PIPKIα, PI4,5P2 and PI3K all assemble at nuclear speckles where Star-PAP and other mRNA processing factors dwell (Boronenkov et al, 1998; Didichenko & Thelen, 2001; Osborne et al, 2001), implicating potential PI3K/Akt signaling switches at this compartment.
Fig. 2.
PIPKIα and PI4,5P2 regulation of Star-PAP activation. PIPKIα activation and PI4,5P2 generation could activate Star-PAP through four potential mechanisms under different conditions. ① Activation of the locally targeted PIPKIα within a PIP-containing compartment generates PI4,5P2, which in turn participates the conformational changes of the Star-PAP complex and facilitates the recruitment of PKCδ and other activating enzymes. ② The locally generated PI4,5P2 stimulates Star-PAP activation together with other co-factors. ③ PI4,5P2 directly activates PKCδ, which phosphorylates and enhanced Star-PAP activities required for mRNA processing. ④ The nuclear PLCs or PI3K processes PI4,5P2 and generates second messengers that are in involved in additional signaling events for Star-PAP activation.
CKI-mediated regulation of Star-PAP activity
The CKI family of Ser/Thr-specific protein kinases requires “priming” phosphorylation by other kinases or by CKI itself (Flotow & Roach, 1991; Gross & Anderson, 1998). The isoform CKIα is a PI4,5P2-sensitive kinase co-localized with Star-PAP at nuclear speckles (Gonzales et al, 2008; Gross et al, 1999). Star-PAP is phosphorylated at the proline rich region (PRR) in the catalytic domain by CKI isoforms α and ε and this is critical for Star-PAP activity (Laishram et al, 2011). The PRR contains several Ser and Thr residues, which are putative sites of CKI phosphorylation. It is therefore likely that Star-PAP is phosphorylated at multiple sites within the PRR region. This is consistent with the observation that none of the individual mutations of the Ser or Thr residues in the PRR abolished the in vitro kinase activity of CKI toward Star-PAP (Gonzales et al, 2008; Laishram et al, 2011). Oxidative stress treatment resulted in the induction of Star-PAP phosphorylation, signifying that stress signaling pathways work in concert with phosphorylation of Star-PAP (Laishram et al, 2011).
Since CKI activity is required for the expression of some of the Star-PAP target genes, the role of phosphorylation on the Star-PAP polyadenylation activity was studied using affinity purified Star-PAP (Gonzales et al, 2008; Laishram et al, 2011). Treatment of the Star-PAP complex with λ-phosphatase resulted in the inhibition of Star-PAP polyadenylation activity, demonstrating that phosphorylation is critical for Star-PAP activity. Both the initiation and processivity of poly(A) tail addition activities were affected by the dephosphorylation of Star-PAP. Dephosphorylated Star-PAP purified from cells exhibited only minimal activity irrespective of stimulation by PI4,5P2 and/or oxidative stress (Laishram et al, 2011). Concomitantly, loss of CKI activity resulted in the loss of Star-PAP polyadenylation activity. Conversely, only when both CKIα and CKIε isoforms were knocked down, was Star-PAP polyadenylation activity abolished, suggesting that these CKI isoforms are redundant in the cell (Laishram et al, 2011). In addition, the target gene such as HO-1 expression was affected by the knockdown of both α and ε isoforms, and single CKI isoform knockdown did not significantly affect the expression. Moreover, both of the isoforms were shown to regulate 3′-end processing of Star-PAP target HO-mRNA (Laishram et al, 2011). Thus, one of the two isoforms may serve as the major CKI that regulates Star-PAP phosphorylation and HO-1 expression, and the other isoform could act as a substitute kinase to regulate HO-1 3′-end processing and could act more specifically in other pathways regulating Star-PAP.
PCKδ regulation of Star-PAP activity
PKCδ belongs to the novel PKC subfamily, whose members are commonly regulated by diacylglycerol, a second messenger generated by phospholipase C hydrolysis of PI4,5P2. Yet, some PKCs directly interact with and are regulated by PI4,5P2 (Chauhan & Brockerhoff, 1988; Huang & Huang, 1991; Lee & Bell, 1991). PKCδ is a key enzyme in apoptosis pathways and was shown to function within nucleus downstream of DNA damage (Humphries et al, 2006; Steinberg, 2004). Though, its nuclear targets and mechanisms of action remain elusive.
The full length PKCδ, but not the cleaved form, was shown to be targeted to nuclear speckles where it associates with the Star-PAP complex. Further, PKCδ is required for Star-PAP processing of the transcript encoding the pro-apoptotic gene BIK (Li et al, 2012). PKCδ is recruited to the Star-PAP complex by binding to PIPKIα and can directly phosphorylate Star-PAP and potentiate Star-PAP polyadenylation activity (Li et al, 2012). PI4,5P2 dose-dependently increased PKCδ phosphorylation of its substrates. Remarkably, PKCδ phosphorylation of Star-PAP in the presence of PIPKIα required PI4,5P2, demonstrating that PIPKIα activation and PI4,5P2 generation are prerequisites for PKCδ activation and subsequent modulation of Star-PAP. PKCδ is key in the activation pathway since PI4,5P2-potentiated and DNA damage-induced Star-PAP stimulation of polyadenylation activity were abolished by loss of PKCδ (Li et al, 2012). In this signaling cascade, PI4,5P2 but not diacylglycerol acted as a direct activator of PKCδ that then activates Star-PAP and possibly other downstream events. Therefore it would be of interest to know what defines the difference in the PKCδ activation mechanism in the Star-PAP complex, and how the local pool of PI4,5P2 is metabolized after the activation.
Stress signaling regulation of Star-PAP and control of stress-responsive gene expression
The 3′-UTR is important for mRNA stability, localization, export and translation. Our data indicate that multiple signal transduction pathways occur at the 3′-end of RNA transcripts. This is an emerging field of study and represents a “hotspot” for the regulation of mRNA processing and gene expression. For example, the MAPKs have been shown to phosphorylate the inhibitory RNA-binding proteins FBP2/3 and TTP at the 3′-UTR and promote mRNA stability and expression (Danckwardt et al, 2011; Hitti et al, 2006). Genotoxic stress could induce dissociation of the mRNA decay-promoting AUF1 and the translational suppressor TIAR from GADD45α, a DNA damage-inducible gene, thereby increase its expression required for DNA repair and genomic stability (Lal et al, 2006).
RNA polymerase II (Pol II) is the core of the transcriptional machinery and its phosphorylation status changes during signaling and downstream of stress responses. Laminar shear stress-induced Pol II phosphorylation at carboxy-terminal domain (CTD) serine 2 increased Pol II binding to the endothelial nitric oxide synthase (eNOS) gene and enhanced its expression (Moore et al, 2010). Of note, while serine 2 phosphorylated in the CTD of RNA Pol II is involved in 3′-end processing of certain mRNAs, it fails to act for the others (Fujita et al, 2008; Gomes et al, 2006). Interestingly, Pol II is able to carry certain transactivating factors from the 5′ to the 3′ of the gene and the 3′-UTR RNA is important for the factors to interact with the transcript. When yeast experiencing osmotic stress, the stress-activated protein kinase Hog1 is recruited to Pol II during elongation of the mRNA of the stress-responsive gene STL1 and this requires the 3′-UTR region of the gene even in the presence of the wild type promoter (Proft et al, 2006), suggesting that the 3′-UTR has signal sensing regions that are independent of the mRNA 5′-end.
It is becoming evident that polyadenylation of mRNA by PAP is a signal-regulated event. Hyperphosphorylation of PAP by the maturation/mitosis-promoting factor, MPF, inhibits PAP activity during cell cycle (Colgan et al, 1996). In addition, different cleavage sites seem to be used for the generation of specific transcripts during stress response and embryogenesis (Berger & Meselson, 1994; Hall-Pogar et al, 2005; Ji et al, 2009). Therefore, exploring the regulation of the 3′-end processing will shed light on the control of gene expression and response of cells under stress conditions.
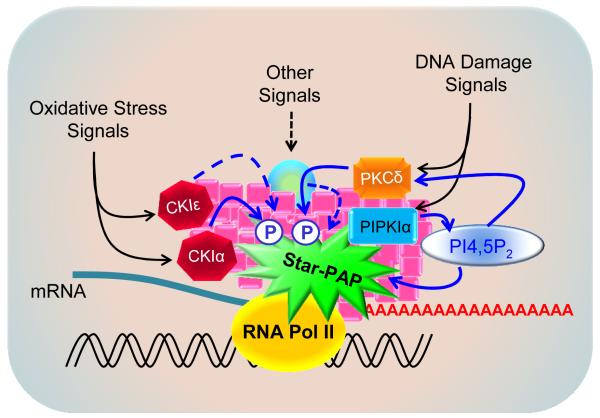
As a nuclear PAP, Star-PAP functions at the 3′-end of newly transcribed mRNAs by integrating upstream stress signals and binding to specific sequence elements within the 3′-UTR of transcripts. Star-PAP activity and processivity are tightly regulated by signaling molecules such as PIPKIα, PI4,5P2, CKIα/ε and PKCδ. Two distinct stress signaling pathways, the oxidative and the DNA damage cascades, have been defined for the regulation of Star-PAP control of stress-responsive gene expression (Fig. 3).
Fig. 3.
Signaling regulation of Star-PAP mediated 3′-end processing. Star-PAP-mediated mRNA 3′-end cleavage and polyadenylation precede transcription termination. Star-PAP activities were stimulated by various signaling molecules in response to different upstream signals as exemplified by the oxidative and DNA damage signaling cascades, where the protein kinases CKIα/ε and PKCδ phosphorylate and activate Star-PAP in the presence of locally generated PI4,5P2.
Oxidative stress and DNA damage both stimulated the Star-PAP cleavage and polyadenylation of RNA substrates and both are required for Star-PAP polymerase activity (Li et al, 2012; Mellman et al, 2008). Treatment of cells with oxidative or DNA damage agonists primed Star-PAP for the PI4,5P2 stimulation of Star-PAP activity. DNA damage stimulated both the initiation (short A-tail) and the priming of the PI4,5P2-stimulated processivity (long A-tail formation) of Star-PAP polyadenylation activity. Remarkably, knockdown of PKCδ abolished etoposide-induced Star-PAP activity as well as the initiation (short tail) and processivity (long tail) of polyadenylation by Star-PAP suggesting that PKCδ is required for Star-PAP activation in this DNA damage pathway (Li et al, 2012).
The expression and 3′-end processing of Star-PAP target BIK also require PKCδ downstream of DNA damage stimulation. PKCδ knockdown blocked etoposide priming for PIP2 stimulation of Star-PAP polyadenylation activity, but did not affect the oxidative stress stimulated Star-PAP polyadenylation activity. This demonstrates that the DNA damage and PKCδ-mediated Star-PAP regulatory pathways are distinct from that of the oxidative stress modulated Star-PAP pathway (Laishram & Anderson, 2010; Li et al, 2012) (Fig. 3). In addition, oxidative stress did not affect the expression of PKCδ-regulated BIK mRNA processing, and the DNA damage pathway did not significantly alter the oxidative stress-regulated HO-1 expression suggesting a signal-mediated differential regulation of Star-PAP. PKCδ knockdown also did not affect the HO-1 expression (Li et al, 2012). These observations indicate that distinct signaling pathways and complexes regulate Star-PAP control of 3′-end processing of specific target mRNAs (Fig. 3).
Perspective
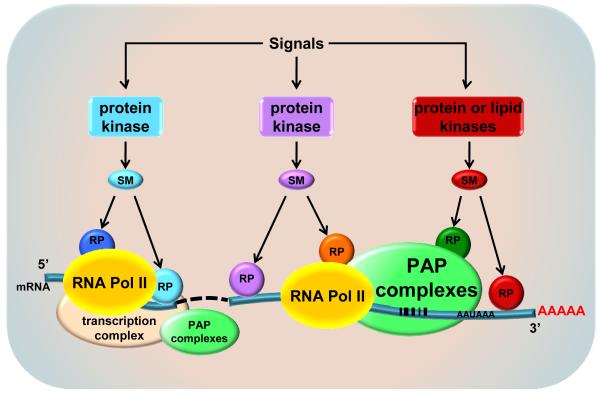
The mRNA 3′-end processing machinery has a different composition compared to the 5′-end transcription initiation complex. However, key components of the 3′-end processing machinery including CPSF 73 (Glover-Cutter et al, 2008), PAPα and Star-PAP (unpublished) are detectable at the 5′-end of genes, presumably through association with RNA Pol II. As the transcriptional complex progresses from the 5′-end to the 3′-end, RNA Pol II associates with some 3′-end processing components. Yet, PAPα and Star-PAP are always in separate complexes, indicating that the distinct PAPs differentially integrate into the RNA Pol II complexes during transcription elongation of specific gene transcripts. Along the gene, the PAPα- or Star-PAP-RNA Pol II complexes must recruit specific signaling molecules, kinases or other transactivators that modulate Star-PAP or canonical PAP specificity (Fig. 4). The complexes of RNA Pol II with the 3′-end processing machinery and kinase transactivators are gene specific and modulate the PAPα and Star-PAP specificity toward specific pre-mRNAs and polyadenylation sites. As the RNA Pol II complexes progress down the gene and generate transcript, Star-PAP specifically binds the RNA at the 3′-end and recruits processing factors that cleave the RNA followed by Star-PAP polyadenylation of the message.
Fig. 4.
Interactive model of signaling regulation of mRNA 3′-end processing – a big picture. The mRNA 3′-end carries unique sequence elements for specific PAP-containing processing complex. Additionally, the processing is regulated by signaling pathways involving multiple levels of modulation by protein kinases, other signaling molecules/second messengers (SM), and regulatory proteins (RP). The interactions between these signaling cascades and between the mRNA 3′- and 5′-ends as well as the connective transcriptional processes form a dynamic nexus for the control of mRNA processing and gene expression.
Star-PAP differentially mediates target gene expression downstream of oxidative and DNA damage pathways, indicating that unique transactivators are incorporated into the 3′-end processing complexes. In addition, for Star-PAP targets, there appears to be unique sequence elements within the 3′-UTR RNAs. As not all genes are Star-PAP targets, the Star-PAP and canonical PAP targets appear to have distinct sequence elements around the 3′-end cleavage sites (Laishram & Anderson, 2010; Li et al, 2012). This suggests that within a single gene that has multiple poly(A) signals, some sites may be targeted by Star-PAP and others by canonical PAPs. There are three canonical PAPs in human, PAPα, PAPβ and PAPγ. While PAPβ is a testis-specific cytoplasmic PAP, PAPα and PAPγ are expressed in nucleus of most cells. It is not clear how these PAPs define their target messages and how they conspire in processing a single transcript with multiple poly(A) signals.
Phosphorylation of PAPs is a critical way of regulating the activities of the enzyme required for RNA processing. Star-PAP phosphorylation by CKIα/ε and PKCδ and canonical PAP phosphorylation by MPF exemplify this regulation. Besides the common domains that the canonical PAPs have, Star-PAP has additional domains that contain putative phosphorylation sites by other kinases. This feature allows the flexibility of Star-PAP regulation by different signaling pathways and integration of select proteins for activation, suggesting that certain genes are only regulated by Star-PAP downstream of specific signals.
Star-PAP is regulated by the lipid messenger PI4,5P2 that is generated by PIPKIα and possibly other PIP kinases. The organization of PI4,5P2 in the nucleus that regulates Star-PAP is not known but has significant implications for spatial regulation of Star-PAP-controlled gene expression.
Acknowledgements
We would like to acknowledge the National Institutes of Health (NIH), the American Heart Association (AHA) and the Wellcome Trust-India Alliance for funding.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barlow CA, Laishram RS, Anderson RA. Nuclear phosphoinositides: a signaling enigma wrapped in a compartmental conundrum. Trends Cell Biol. 2009 doi: 10.1016/j.tcb.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL, Meselson M. Production and cleavage of Drosophila hsp70 transcripts extending beyond the polyadenylation site. Nucleic Acids Res. 1994;22(15):3218–3225. doi: 10.1093/nar/22.15.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boronenkov IV, Loijens JC, Umeda M, Anderson RA. Phosphoinositide signaling pathways in nuclei are associated with nuclear speckles containing pre-mRNA processing factors. Mol Biol Cell. 1998;9(12):3547–3560. doi: 10.1091/mbc.9.12.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KM, Gilmartin GM. A mechanism for the regulation of pre-mRNA 3′ processing by human cleavage factor Im. Mol Cell. 2003;12(6):1467–1476. doi: 10.1016/s1097-2765(03)00453-2. [DOI] [PubMed] [Google Scholar]
- Chauhan VP, Brockerhoff H. Phosphatidylinositol-4,5-bisphosphate may antecede diacylglycerol as activator of protein kinase C. Biochem Biophys Res Commun. 1988;155(1):18–23. doi: 10.1016/s0006-291x(88)81043-x. [DOI] [PubMed] [Google Scholar]
- Chen Y, Varani G. Protein families and RNA recognition. FEBS J. 2005;272(9):2088–2097. doi: 10.1111/j.1742-4658.2005.04650.x. [DOI] [PubMed] [Google Scholar]
- Cocco L, Gilmour RS, Ognibene A, Letcher AJ, Manzoli FA, Irvine RF. Synthesis of polyphosphoinositides in nuclei of Friend cells. Evidence for polyphosphoinositide metabolism inside the nucleus which changes with cell differentiation. Biochem J. 1987;248(3):765–770. doi: 10.1042/bj2480765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocco L, Martelli AM, Gilmour RS, Ognibene A, Manzoli FA, Irvine RF. Rapid changes in phospholipid metabolism in the nuclei of Swiss 3T3 cells induced by treatment of the cells with insulin-like growth factor I. Biochem Biophys Res Commun. 1988;154(3):1266–1272. doi: 10.1016/0006-291x(88)90276-8. [DOI] [PubMed] [Google Scholar]
- Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11(21):2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- Colgan DF, Murthy KG, Prives C, Manley JL. Cell-cycle related regulation of poly(A) polymerase by phosphorylation. Nature. 1996;384(6606):282–285. doi: 10.1038/384282a0. [DOI] [PubMed] [Google Scholar]
- Danckwardt S, Gantzert AS, Macher-Goeppinger S, Probst HC, Gentzel M, Wilm M, Grone HJ, Schirmacher P, Hentze MW, Kulozik AE. p38 MAPK controls prothrombin expression by regulated RNA 3′ end processing. Mol Cell. 2011;41(3):298–310. doi: 10.1016/j.molcel.2010.12.032. [DOI] [PubMed] [Google Scholar]
- Didichenko SA, Thelen M. Phosphatidylinositol 3-kinase c2alpha contains a nuclear localization sequence and associates with nuclear speckles. J Biol Chem. 2001;276(51):48135–48142. doi: 10.1074/jbc.M104610200. [DOI] [PubMed] [Google Scholar]
- Flotow H, Roach PJ. Role of acidic residues as substrate determinants for casein kinase I. J Biol Chem. 1991;266(6):3724–3727. [PubMed] [Google Scholar]
- Fujita T, Ryser S, Piuz I, Schlegel W. Up-regulation of P-TEFb by the MEK1-extracellular signal-regulated kinase signaling pathway contributes to stimulated transcription elongation of immediate early genes in neuroendocrine cells. Mol Cell Biol. 2008;28(5):1630–1643. doi: 10.1128/MCB.01767-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin GM, Nevins JR. Molecular analyses of two poly(A) site-processing factors that determine the recognition and efficiency of cleavage of the pre-mRNA. Mol Cell Biol. 1991;11(5):2432–2438. doi: 10.1128/mcb.11.5.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol. 2008;15(1):71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes NP, Bjerke G, Llorente B, Szostek SA, Emerson BM, Espinosa JM. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 2006;20(5):601–612. doi: 10.1101/gad.1398206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales ML, Mellman DL, Anderson RA. CKIalpha is associated with and phosphorylates star-PAP and is also required for expression of select star-PAP target messenger RNAs. J Biol Chem. 2008;283(18):12665–12673. doi: 10.1074/jbc.M800656200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross SD, Anderson RA. Casein kinase I: spatial organization and positioning of a multifunctional protein kinase family. Cell Signal. 1998;10(10):699–711. doi: 10.1016/s0898-6568(98)00042-4. [DOI] [PubMed] [Google Scholar]
- Gross SD, Loijens JC, Anderson RA. The casein kinase Ialpha isoform is both physically positioned and functionally competent to regulate multiple events of mRNA metabolism. J Cell Sci. 1999;112(Pt 16):2647–2656. doi: 10.1242/jcs.112.16.2647. [DOI] [PubMed] [Google Scholar]
- Hall-Pogar T, Zhang H, Tian B, Lutz CS. Alternative polyadenylation of cyclooxygenase-2. Nucleic Acids Res. 2005;33(8):2565–2579. doi: 10.1093/nar/gki544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck JN, Mellman DL, Ling K, Sun Y, Wagoner MP, Schill NJ, Anderson RA. A conspicuous connection: structure defines function for the phosphatidylinositol-phosphate kinase family. Crit Rev Biochem Mol Biol. 2007;42(1):15–39. doi: 10.1080/10409230601162752. [DOI] [PubMed] [Google Scholar]
- Hitti E, Iakovleva T, Brook M, Deppenmeier S, Gruber AD, Radzioch D, Clark AR, Blackshear PJ, Kotlyarov A, Gaestel M. Mitogen-activated protein kinase-activated protein kinase 2 regulates tumor necrosis factor mRNA stability and translation mainly by altering tristetraprolin expression, stability, and binding to adenine/uridine-rich element. Mol Cell Biol. 2006;26(6):2399–2407. doi: 10.1128/MCB.26.6.2399-2407.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang FL, Huang KP. Interaction of protein kinase C isozymes with phosphatidylinositol 4,5-bisphosphate. J Biol Chem. 1991;266(14):8727–8733. [PubMed] [Google Scholar]
- Humphrey T, Christofori G, Lucijanic V, Keller W. Cleavage and polyadenylation of messenger RNA precursors in vitro occurs within large and specific 3′ processing complexes. EMBO J. 1987;6(13):4159–4168. doi: 10.1002/j.1460-2075.1987.tb02762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries MJ, Limesand KH, Schneider JC, Nakayama KI, Anderson SM, Reyland ME. Suppression of apoptosis in the protein kinase Cdelta null mouse in vivo. J Biol Chem. 2006;281(14):9728–9737. doi: 10.1074/jbc.M507851200. [DOI] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci U S A. 2009;106(17):7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller W, Bienroth S, Lang KM, Christofori G. Cleavage and polyadenylation factor CPF specifically interacts with the pre-mRNA 3′ processing signal AAUAAA. EMBO J. 1991;10(13):4241–4249. doi: 10.1002/j.1460-2075.1991.tb05002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakopoulou CB, Nordvarg H, Virtanen A. A novel nuclear human poly(A) polymerase (PAP), PAP gamma. J Biol Chem. 2001;276(36):33504–33511. doi: 10.1074/jbc.M104599200. [DOI] [PubMed] [Google Scholar]
- Laishram RS, Anderson RA. The poly A polymerase Star-PAP controls 3′-end cleavage by promoting CPSF interaction and specificity toward the pre-mRNA. EMBO J. 2010;29(24):4132–4145. doi: 10.1038/emboj.2010.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laishram RS, Barlow CA, Anderson RA. CKI isoforms alpha and epsilon regulate Star-PAP target messages by controlling Star-PAP poly(A) polymerase activity and phosphoinositide stimulation. Nucleic Acids Res. 2011;39(18):7961–7973. doi: 10.1093/nar/gkr549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A, Abdelmohsen K, Pullmann R, Kawai T, Galban S, Yang X, Brewer G, Gorospe M. Posttranscriptional derepression of GADD45alpha by genotoxic stress. Mol Cell. 2006;22(1):117–128. doi: 10.1016/j.molcel.2006.03.016. [DOI] [PubMed] [Google Scholar]
- Lee MH, Bell RM. Mechanism of protein kinase C activation by phosphatidylinositol 4,5-bisphosphate. Biochemistry. 1991;30(4):1041–1049. doi: 10.1021/bi00218a023. [DOI] [PubMed] [Google Scholar]
- Lee MH, Schedl T. The C elegans Research Community, editor. RNA-binding proteins. WormBook. 2006 doi: 10.1895/wormbook.1.79.1. doi/10.1895/wormbook.1.79.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Li W, Laishram RS, Ji Z, Barlow CA, Tian B, Anderson RA. Star-PAP control of BIK expression and apoptosis is regulated by nuclear PIPKIalpha and PKCdelta signaling. Mol Cell. 2012;45(1):25–37. doi: 10.1016/j.molcel.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol. 2007;8(6):479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci. 2008;65(7-8):1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature. 2006;444(7121):953–956. doi: 10.1038/nature05363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Doublie S, Keller W. Determinants of substrate specificity in RNA-dependent nucleotidyl transferases. Biochim Biophys Acta. 2008;1779(4):206–216. doi: 10.1016/j.bbagrm.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Keller W. Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and catalytic domain, homologous to the family X polymerases, and to other nucleotidyltransferases. EMBO J. 1996;15(10):2593–2603. [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF. More than one way to make a tail. EMBO J. 2010;29(24):4066–4067. doi: 10.1038/emboj.2010.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman DL, Gonzales ML, Song C, Barlow CA, Wang P, Kendziorski C, Anderson RA. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature. 2008;451(7181):1013–1017. doi: 10.1038/nature06666. [DOI] [PubMed] [Google Scholar]
- Moore JP, Weber M, Searles CD. Laminar shear stress modulates phosphorylation and localization of RNA polymerase II on the endothelial nitric oxide synthase gene. Arterioscler Thromb Vasc Biol. 2010;30(3):561–567. doi: 10.1161/ATVBAHA.109.199554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy KG, Manley JL. The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes Dev. 1995;9(21):2672–2683. doi: 10.1101/gad.9.21.2672. [DOI] [PubMed] [Google Scholar]
- Nagaike T, Suzuki T, Katoh T, Ueda T. Human mitochondrial mRNAs are stabilized with polyadenylation regulated by mitochondria-specific poly(A) polymerase and polynucleotide phosphorylase. J Biol Chem. 2005;280(20):19721–19727. doi: 10.1074/jbc.M500804200. [DOI] [PubMed] [Google Scholar]
- Osborne SL, Thomas CL, Gschmeissner S, Schiavo G. Nuclear PtdIns(4,5)P2 assembles in a mitotically regulated particle involved in pre-mRNA splicing. J Cell Sci. 2001;114(Pt 13):2501–2511. doi: 10.1242/jcs.114.13.2501. [DOI] [PubMed] [Google Scholar]
- Payrastre B, Nievers M, Boonstra J, Breton M, Verkleij AJ, Van Bergen en Henegouwen PM. A differential location of phosphoinositide kinases, diacylglycerol kinase, and phospholipase C in the nuclear matrix. J Biol Chem. 1992;267(8):5078–5084. [PubMed] [Google Scholar]
- Proft M, Mas G, de Nadal E, Vendrell A, Noriega N, Struhl K, Posas F. The stress-activated Hog1 kinase is a selective transcriptional elongation factor for genes responding to osmotic stress. Mol Cell. 2006;23(2):241–250. doi: 10.1016/j.molcel.2006.05.031. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr Opin Cell Biol. 2004;16(3):272–278. doi: 10.1016/j.ceb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386(6625):569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- Raabe T, Bollum FJ, Manley JL. Primary structure and expression of bovine poly(A) polymerase. Nature. 1991;353(6341):229–234. doi: 10.1038/353229a0. [DOI] [PubMed] [Google Scholar]
- Schmidt MJ, Norbury CJ. Polyadenylation and beyond: emerging roles for noncanonical poly(A) polymerases. Wiley Interdiscip Rev RNA. 2010;1(1):142–151. doi: 10.1002/wrna.16. [DOI] [PubMed] [Google Scholar]
- Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR, 3rd, Frank J, Manley JL. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell. 2009;33(3):365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg SF. Distinctive activation mechanisms and functions for protein kinase Cdelta. Biochem J. 2004;384(Pt 3):449–459. doi: 10.1042/BJ20040704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson AL, Norbury CJ. The Cid1 family of non-canonical poly(A) polymerases. Yeast. 2006;23(13):991–1000. doi: 10.1002/yea.1408. [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Ryner LC, Manley JL. Four factors are required for 3′-end cleavage of pre-mRNAs. Genes Dev. 1989;3(11):1711–1724. doi: 10.1101/gad.3.11.1711. [DOI] [PubMed] [Google Scholar]
- Trippe R, Guschina E, Hossbach M, Urlaub H, Luhrmann R, Benecke BJ. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA. 2006;12(8):1494–1504. doi: 10.1261/rna.87706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E. Purification and characterization of a mammalian polyadenylate polymerase involved in the 3′ end processing of messenger RNA precursors. J Biol Chem. 1991;266(5):3131–3139. [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63(2):405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhelkovsky AM, Kessler MM, Moore CL. Structure-function relationships in the Saccharomyces cerevisiae poly(A) polymerase. Identification of a novel RNA binding site and a domain that interacts with specificity factor(s) J Biol Chem. 1995;270(44):26715–26720. doi: 10.1074/jbc.270.44.26715. [DOI] [PubMed] [Google Scholar]