Abstract
The Succinate Dehydrogenase (SDH) heterotetrameric complex catalyzes the oxidation of succinate to fumarate in the tricarboxylic acid (TCA) cycle and in the aerobic respiratory chains of eukaryotes and bacteria. Essential in this catalysis, is the covalently-linked cofactor flavin adenine dinucleotide (FAD) in subunit1 (Sdh1) of the SDH enzyme complex. The mechanism of FAD insertion and covalent attachment to Sdh1 is unknown. Our working concept of this flavinylation process has relied mostly on foundational works from the 1990s ago and by applying the principles learned from other enzymes containing a similarly linked FAD. The discovery of the flavinylation factor Sdh5, however, has provided new insight into the possible mechanism associated with Sdh1 flavinylation, bringing into question the autocatalytic mechanism associated with other flavoenzymes. This review focuses on encapsulating prior and recent advances towards understanding the mechanism associated with flavinylation of Sdh1 and how this flavinylation process affects the overall assembly of SDH.
Keywords: Succinate Dehydrogenase, Flavinylation, FAD, Cofactor, Sdh5, Sdh1
1.0 INTRODUCTION
Succinate dehydrogenase (SDH) is a hetero-tetrameric enzyme complex that catalyzes the oxidation of succinate to fumarate with the concomitant reduction of ubiquinone to ubiquinol. Formally, the enzyme is succinate:ubiquinone oxidoreductase, and the classic oxidation-reduction reaction it catalyzes is dependent on a flavin adenine dinucleotide (FAD) cofactor in subunit 1 (designated Sdh1 in yeast, SdhA in bacteria and SDHA in humans) (Figure 1). The second subunit of SDH (Sdh2 or SDHB) contains additional cofactors; three distinct iron-sulfur clusters whose function is to transfer the two electrons in one electron increments resulting from the dehydrogenation of succinate at the active-site FAD. Ultimately, the electrons reduce the quinone that is bound at the interface of Sdh2 and the membrane-spanning subunits (Sdh3 or SDHC and Sdh4 or SDHD), possibly with the involvement of a heme bound between the membrane subunits [1–4].
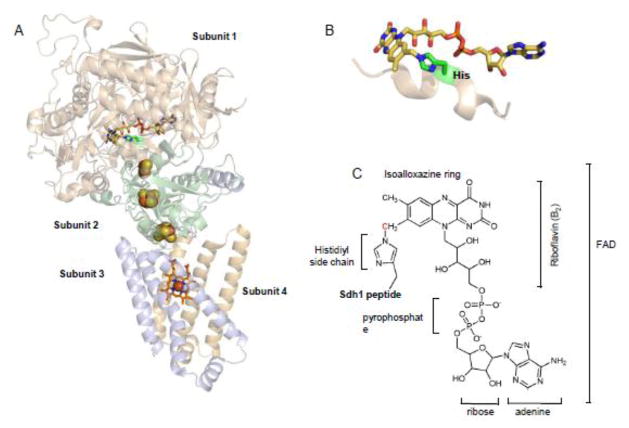
Fig 1.
The covalently-linked FAD cofactor in subunit 1 of the succinate dehydrogenase (SDH) complex. (A) The overall structure of SDH (avian; PDB 1YQ3 from reference [3] ) illustrating the distribution of cofactors. Subunit 1 (tan) contains FAD (stick: yellow; carbon: blue; nitrogen: red; oxygen). Subunit 2 (green) contains 3 Fe-S centers (spheres: yellow; sulfur: orange; iron). Subunits 3 and 4 (light orange and purple) contain heme b (stick: orange; carbon: blue; nitrogen: red; oxygen: red sphere; iron). (B) Close up of the FAD cofactor showing the covalently-linking His residue (green stick) along the Sdh1 peptide backbone. (C) Chemical structure of FAD molecule covalently-linked to Sdh1 that is composed of different chemical components as shown. The red carbon of the isoalloxazine ring indicates the sight of covalent attachment forming the N3-histidyl-8α FAD bond.
The cofactors of succinate dehydrogenase were first noted nearly 60 years ago, which included the discovery of an unusually tightly-bound form of FAD [5]. This observation is now considered to be the first identification of a covalently-bound flavin to a polypeptide structure [6]. The FAD is buried deeply in Sdh1 and the carbon-nitrogen covalent bond occurs via the side chain of a histidyl residue (Figure 1B). The catalytic chemistry of succinate oxidation yielding fumarate is absolutely dependent on this covalent bond [7,8]. Substitution of the His to a Ser residue in yeast Sdh1 yields an assembled, but catalytically defective SDH complex in the direction of succinate oxidation [8]. The reverse reaction of fumarate reduction can proceed, albeit at a very slow rate, illustrating the partiality of the covalent bond towards succinate oxidation.
In another histidyl-linked covalent flavoprotein, nicotine oxidase, replacement of the His to a Cys to block covalent flavinylation leads to a substantial amount of activity being retained as long as FAD is present for the reaction [9]. This suggests that at least in this enzyme, covalent attachment is not an absolute requirement for activity, which raises the question why the covalent attachment in Sdh1 is exceptional in its function relative to other covalent flavoproteins.
For most flavoproteins, the mode of flavin association is through non-covalent interactions with the peptide backbone that prevents diffusion of the cofactor [10,11]. The covalent linkage places SDH in the minority 5–10% of characterized flavoproteins. The overall mechanism of this covalent flavin attachment is unknown, but has been generally accepted that the attachment occurs autocatalytically (or self catalytically) as a result of in-vitro studies with several bacterial proteins. This notion, however, is now made more complex and intriguing with the discovery of Sdh5 in yeast (SdhE in bacteria and SDHAF2 in humans) [12] that is considered to be the first factor that is absolutely required for covalent flavinylation in vivo. Sdh5 is conserved from bacteria [13] to humans [12], and also in the Arabidopsis plant [14], and co-expressed with the subunits of SDH. It is clear Sdh5 and its bacterial and plant homologues are functionally important in flavinylation mechanism; however, the exact nature of this function is not yet understood.
The discovery of Sdh5 has furthered the interest in elucidating the flavinylation mechanism of Sdh1. In addition, the overall assembly of SDH has been brought into the forefront, as flavinylation and SDH assembly are intricately linked processes. SDH is an ideal model system to study such phenomena of cofactor maturation linked to overall complex assembly. The system is relatively simple comprising of just four subunits, which can be further compartmentalized to a soluble and membrane domains.
This review surveys the literature that have been focused on Sdh1 flavinylation and SDH assembly, covering what we have learned and discovered in the years proceeding the discovery of the first “tightly-bound” FAD by Singer, Kearney and Zastrow more than 60 years ago [5]. We begin with a general discussion of SDH, discuss where and how FAD is synthesized and transported to Sdh1, discuss the protein(s) and effectors required for flavinylation, and how SDH assembly proceeds in relation to Sdh1 flavinylation.
2.0 FAD AS A COFACTOR IN COMPLEX II
2.1 Overview of Succinate Dehydrogenase
The succinate dehydrogenase complex is also known as complex II in eukaryotes, which designates it as an integral component of the aerobic respiratory chain along with complex I (NADH dehydrogenase), complex III (cytochrome c reductase), and complex IV (cytochrome c oxidase). A distinguishing feature of complex II, from that of the other complexes, is that it also serves as part of the chain of eight enzymes forming the tricarboxylic acid (TCA) cycle. SDH catalyzes a classic oxidation-reduction, coupling the two-electron oxidation of succinate to fumarate with the reduction of ubiquinone to ubiquinol (succinate:ubiquinone oxidoreductase). The product fumarate is utilized in the next TCA cycle reaction catalyzed by fumarase; the co-product ubiquinol is oxidized by complex III in the electron transfer chain (ETC). The SDH enzyme is a hetero-tetrameric complex consisting of a hydrophilic catalytic domain and a membrane-spanning, ubiquinone-binding domain. The FAD-containing Sdh1 together with Sdh2 comprise the top portion of what has been aptly described as an overall “q-shaped” or “mushroom-shaped” complex [15,16]. The two membrane-spanning domains, Sdh3 and Sdh4, comprise the lower portion of the “q” as a heterodimer that act to tether Sdh1, via Sdh2, to the inner membrane. Approximately 40% of the surface area of Sdh2 is involved in interaction between Sdh1 and the membrane domain. Thus, the overall stability of the SDH complex is dependent to a large extent on this tethering subunit [17].
The Sdh2 subunit contains the Fe/S cofactors of SDH. The two electrons abstracted from the dehydrogenation of succinate at the active-site FAD are channeled sequentially through 3 distinct [2Fe–2S], [4Fe–4S], [3Fe–4S] iron-sulfur clusters in Sdh2. Ultimately, the electrons reduce ubiquinone, located in a cavity near the matrix-lipid interface comprised of Sdh2, Sdh3 and Sdh4 [16,17]. These cofactors form a long-range, near-linear electron conduit extending over 40 Å from the soluble catalytic domain of SDH to the membrane-spanning domain of the enzyme [16] for the exact purpose of driving electrons into the respiratory chain.
Complex II, through evolution and time, has managed to retain four – and only four – essential “core subunits” that are found in the bacterial counterpart succinate quinone reductase (SQR). This is in stark contrast to complexes III and IV, where their departures from the bacterial complexes have evolved to recruit several additional subunits. Thus, complex II has a very low number of subunits relative to its cofactors compared to mitochondrial complexes I, III, and IV, which provides a relatively simplified model system in studying the very complex, intricate and relatively unknown processes of cofactor insertion and respiratory complex maturation. For further details on SDH structure and function, we refer readers to the following reviews [18–21].
2.2 FAD and its link to Sdh1
The 70-kDa Sdh1 subunit structure consists of a Rossmann fold [17,22] with the FAD bound by its isoalloxazine ring to a histidyl residue (His90 in yeast). This covalent linkage is via a secondary amide bond between the N3 atom of the histidyl imidazole and the 8α-methyl group of the isoalloxazine ring (a N3-histidyl-8α-FAD linkage) (Figure 1C). The mechanism of covalent attachment of FAD is unknown. In general, the formation of covalent bonds requires free energy [23], and in the case of the covalent FAD linkage in Sdh1, energy-requiring activation of either the N3 atom of the imidazole side chain of histidine or the hydroxylation of the 8α-methyl group of the isoalloxazine ring is likely necessary [24,25]. It is unknown whether Sdh5 could facilitate such a reaction (see section 5.3).
Covalent flavinylation involving non-enzymatic mechanisms with no energy expenditure, however, has also been proposed [23,24]. Perhaps the most notable model involves an autocatalytic process facilitated by the apo-protein itself where the nucleophilic imidazole side chain attacks a quinone methide form of the of the isoalloxazine ring. The highly reactive electrophilic quinone methide intermediate could be generated from a proton abstraction from a base (e.g. Arg residues) nearby the isoalloxazine ring. Consistent with the involvement of this nucleophilic attack by a residue side chain, the only amino acids known to form the covalent linkage are nucleophiles (His, Tyr and Cys) [10,26].
Perhaps the best supporting evidence for an autocatalytic flavinylation mechanism comes from vanillyl-alcohol oxidase (VAO), a fungal enzyme that contains, like Sdh1, a N3-histidyl-8α-FAD linkage. An apo-form of VAO, produced in a riboflavin auxotrophic Escherichia coli strain, can be reconstituted with FAD in vitro resulting in full covalent incorporation and full recovery of oxidase activity [27]. FAD was found to bind to a preorganized binding site in a folded apo VAO with an overall structure very similar to holo-VAO [28]. The reaction involves a reduced FAD intermediate that can be captured by anaerobic mixing of apo VAO and oxidized. Molecular oxygen reoxidizes the FAD with the formation of the covalent bond and hydrogen peroxide.
This process is slow (in the time scale of minutes) to achieve full flavinylation, which could be an indication of less than optimal in-vitro reaction condition. This demonstration cannot rule out that other factors are involved in vivo, perhaps alternative electron acceptors or helper proteins. Interestingly, anaerobically grown E. coli cells expressing VAO produce fully covalently flavinylated form of the enzyme, suggesting that an alternative electron acceptor to molecular oxygen is involved [27].
2.3 Advantages of the covalent link in Sdh1
2.3.1 Increase in Redox Potential
A compiled survey of redox potentials of flavoproteins containing either a noncovalent or a covalent bond shows a clear trend indicating that the covalent nature of the bond increases the redox potential significantly [10], thus increasing the oxidative power of the enzyme. Identification of flavoproteins containing two covalent linkages (a N1-histidyl-C6-cystinyl-8α-FAD linkage) further support the overall trend with additional increases in potential with the extra covalent bond [29]. Similar observations can be seen using synthetic model compounds where the modified forms of riboflavin have higher midpoint potentials compared to the free forms [30].
In cases where the mutant form of the flavoprotein has the covalent bond impaired, the redox potential is dramatically reduced [10]. Replacing the linking His residue in VAO disrupts the covalent bond and decreases the FAD redox potential by approximately 120 mV [31]. The resulting enzyme, with a tightly but noncovalently bound FAD, shows poor activity with an order of magnitude decrease in the turnover number kcat. In Sdh1, the disruption of the covalent bond by substitution of the linking His residue to a Ser residue yields a catalytically nonfunctioning SDH enzyme in succinate oxidation [8]; which, like VAO, reveals that the covalent bond has a functional catalytic significance.
The anaerobic analogue of SDH in bacteria, fumarate reductase (FRD), has the same N3-histidyl-8α-FAD linkage in its FrdA subunit (Sdh1 equivalent). Fumarate reductase can catalyze succinate oxidation at about 30–40% of the rate of fumarate reduction. In this enzyme system, mutation of the linking His residue to a Ser has a similar effect as found in SDH: loss of succinate oxidation while maintaining to a large extent fumarate reduction [7]. Thus, the covalent attachment likely increases the FAD redox potential by about 60 mV to permit both succinate oxidation and fumarate reduction.
2.3.2 Stability of the SDH complex
Mutation of the covalently linking His90 to a Ser residue in Sdh1 of yeast prevents the formation of the FAD covalent linkage [8,32]. However, the authors concluded that Sdh1 still folds, forming a preorganized site that can still bind FAD noncovalently, similar to the previously mentioned FAD binding to a preorganized binding site in VAO [27,28] (section 2.2). This incorporation of FAD has been demonstrated experimentally using radiolabeled FAD that comigrates with the holo-SDH complex on a Blue Native gel [8,32]. A similar effect is observed when the FAD covalent linkage is disrupted by deletion of SDH5. Sdh1 assembles into a mature complex presumably with a noncovalently-bound FAD [32]. Thus, the covalent bond is not needed to induce proper folding of Sdh1 and subsequent assembly of the SDH complex.
The absence of the covalent bond in Sdh1 does, however, compromise the overall stability of the mature SDH complex. The steady-state levels of the mature SDH complex is lower when the covalent bond is disrupted either by mutation of the His90 residue or by deletion of Sdh5. This is probably not caused by an impaired import or processing of Sdh1 [8]; rather, the possibilities include an increase in protease sensitivity [33] or an inherently less stable holo-complex in vivo that is prone to disassembly [32].
In some flavoenzymes, removal of the covalent linkage results in incorrectly folded protein. In alditol oxidase for example, approximately half of the of the protein becomes insoluble when the covalently-linking His residue is mutated [34]. In contrast, mutant forms of VAO, with the covalently-linking His residue disrupted, can still fold properly similar to Sdh1. The role of the covalent bond in terms of protein folding therefore appears enzyme specific. Sdh1 falls into the category with VAO where a certain amount of folding robustness is present even in the absence of the covalent linkage.
3.0 THE BIOSYNTHESIS AND TRAFFICKING OF FAD to SDH
The formation of flavinylated Sdh1 is dependent on the availability of FAD in the same subcellular compartment as apo-Sdh1. Therefore, the synthesis and maintenance of proper FAD concentration in the mitochondrial matrix is critical for Sdh1 maturation. In yeast, this involves the activities of riboflavin kinase, FAD synthetase, and a putative mitochondrial FAD transporter, Flx1 (FLavin eXchange) protein. Of particular relevance to Sdh1 flavinylation is the latter protein Flx1, primarily due to the curious finding that the deletion of flx1 results in the loss of covalent flavinylation in Sdh1 [12,32]. Thus, this protein raises many questions regarding its role in Sdh1 flavinylation. A key in deciphering how this effect is mediated is knowing the cellular compartment where FAD is synthesized, and how (and in which direction) it is transported in eukaryotes. These topics are discussed in sections 3.1 and 3.2. Flx1 is discussed in detail in sections 3.3 and 3.4.
3.1 Biosynthesis of FAD
Flavins are derived from riboflavin (vitamin B2) by the addition of either a phosphate group or an ADP moiety to the vitamin’s ribityl side chain (Fig 1C). The conversion of riboflavin to FAD occurs through the sequential actions of two ATP-dependent enzymes. The genes encoding these enzymes were identified in S. cerevisiae as FMN1 (riboflavin kinase) [35] and FAD1 (FAD synthetase) [36]. The kinase phosphorylates the redox-active tricyclic isoalloxazine ring yielding flavin mononucleotide (FMN; riboflavin-5′-phosphate). Although several flavo4enzymes use FMN as a cofactor, the majority of the FMN is adenylated by FAD synthetase to yield FAD [11].
Recently, the crystal structure of yeast Fad1 in complex with FAD was determined [37] revealing a strong interaction between the phosphoribityl group of FAD and a pair of nearby arginine residues. Known as an “arginine grip,” the pair is highly conserved and appears to be a di-phosphate binding motif stabilizing either the substrates (ATP and FMN) and/or the product FAD, preventing the latter’s release. In fact, recombinant human FAD synthase is purified with a stoichiometric amount of FAD that is tightly, but non-covalently bound [38]. Furthermore, release of FAD from this recombinant protein required extensive urea denaturation. These observations suggest that the release of newly synthesized FAD could require conformational alterations that disrupt the arginine grip, perhaps mediated by protein-protein interaction with an FAD acceptor protein.
3.2 Where is FAD synthesized?
In bacteria, FAD is synthesized in the cytosol from a single, but a dual functioning enzyme possessing the activities of riboflavin kinase and FAD synthetase [39]. In yeast, the riboflavin kinase has been found in both the cytosol and in mitochondria [40]; however, the location of the FAD synthetase is not so clear due to conflicting results from two laboratories. Initial reports of FAD synthetase activity and location was from Alexander Tzagoloff's laboratory which found that the synthetase activity was present only in the cytosol [40]. This implies that FAD used in the mitochondrial matrix for flavinylation of Sdh1 must be transported in from the cytosol involving a carrier protein (proposed to be Flx1; see section 3.3).
Since the initial report of Fad1 activity in yeast from the Tzagoloff laboratory, the laboratory of Maria Barile has reported FAD synthetase activity to be present in the mitochondria of S. cerevisiae [41,42]. Additionally FAD synthetase has been found in the mitochondria of rat liver [43,44] and of the tobacco plant [45]. In humans, the ortholog of yeast FAD1 gene is FLAD1, which encode two transcript variants leading to two isoforms of the FAD synthetase. The two isoforms differ by an additional 97 amino acids at the amino terminus, containing a mitochondrial targeting sequence. This extra sequence is present only in isoform 1, and its mitochondrial localization has been demonstrated using antibodies specific for this isoform [46]. Isoform 2 of FLAD1 was demonstrated to localize to the cytosol. Thus, the possibility of a dual localized Fad1 in S. cerevisiae is not without precedent in other organisms. Results from both laboratories are discussed further below.
3.3 Flx1 – An Innie or an Outie?
The Flx1 (FLavin eXchange) protein belongs to the superfamily of mitochondrial carriers. Proteins in this family are located in the inner mitochondrial membrane and exchange substrates between the cytosol and the matrix [47]. The family is exclusive to eukaryotes, and the substrates that have been confirmed to be transported include nucleotides, amino acids, inorganic anions and intermediates of the TCA cycle such as succinate, oxaloacetate and malate [47]. Utilizing multiple sequence alignment analysis, Flx1 has been found to be homologous to the solute carrier proteins in mammals that transport ATP/ADP in the mitochondrial inner membrane [48]. Flx1 also bears homology to a carrier of vitamin B9 or folate [47]. A structural model of Flx1 based on homology to these carrier proteins suggest the presence of six transmembrane helices that together form a pore for substrate translocation [47,49,50] (Figure 2A).
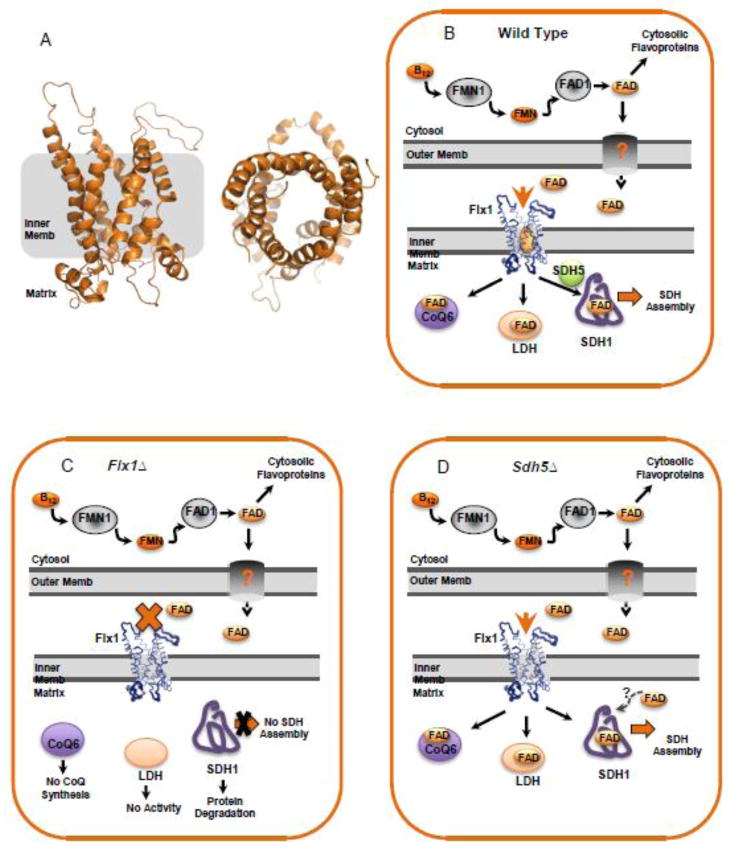
Fig 2.
The putative role of Flx1 in the trafficking of FAD in the mitochondria and its effect on Sdh1 flavinylation. (A) Model of Flx1 structure based on template PDB 2LCK, an integral membrane protein in the mitochondrial anion carrier protein family [47] showing a side view (left) and a top-down view (right). (B) A theoretical model of FAD trafficking based primarily on the proposed import role of Flx1 by Tzagoloff [40]. An alternative model has been put forth by the Barile group that proposes Flx1 as an exporter of FAD [42]. In the wild-type mitochondria of S. cerevisiae FAD is synthesized from riboflavin from the sequential actions of FMN1 and FAD1 in the cytosol. A certain portion of the FAD enters the mitochondria through an unknown outer membrane transporter and into the mitochondrial matrix through the carrier Flx1. The matrix FAD is utilized for the enzymes including CoQ6, LDH, and Sdh1. (C) In the absence of Flx1, FAD levels in the mitochondrial matrix are thought to decrease and the activities of CoQ6 and LDH are dramatically impaired. Moreover, SDH does not assemble presumably due to the lack of a Sdh1-FAD interaction leading to apo Sdh1 instability. flx1Δ cells do not render the IM completely impermeable; over expression of FAD1 partly complements the CoQ6 and LDH defect. (D) In the absence of Sdh5, SDH assembly still proceeds, presumably due to the fact that FAD can still bind to Sdh1.
The gene encoding Flx1 was first identified based on its ability to complement a respiratory defective mutant of S. cerevisiae characterized by a low ratio of mitochondrial FAD/FMN, restoring the two flavin levels to that found in WT cells [40]. Yeast cells with a FLX1 deletion are also characterized with a low mitochondrial FAD/FMN ratio and with decreased SDH and lipoamide dehydrogenase activities — two proteins that are FAD dependent (Figure 2C). Another matrix FAD-containing enzyme, Coq6 (a monooxygenase that is essential in coenzyme ubiquinone biosynthesis), is also negatively affected in a flx1 yeast mutant, resulting in a respiratory defect from the absence of ubiquinone (Q6) [51]. This Coq6 defect can be reversed by overexpression of Fad1 that restores Q6 levels. This strongly suggests that Fad1 can complement the matrix FAD deficiency in a flx1Δ mutant. Thus, the notion of Flx1 as an inner membrane importer of cytoplasmically produced FAD is consistent with the observed phenotypes associated with lower mitochondrial FAD levels in a flx1 mutant.
Further support for an import role came from two additional observations: (1) FAD synthetase activity was found only in the cytosolic fraction [36], and (2) prepared spherical submitochondrial particles (inside-out inner mitochondrial membrane with entrapped FAD) showed FAD efflux activity across the mitochondrial membranes in WT particles [40]. This FAD efflux rate was 2–3 times less in a flx1 mutant.
An alternative model for Flx1, put forth by the Barile laboratory, places the carrier protein as an exporter of mitochondrially-produced FAD to the cytosol. A necessary element to this idea is that FAD synthesis must occur in the mitochondria, either solely or in conjunction with cytosolic FAD synthesis. Indeed, the authors were able to show FAD synthesis activities in both the cytosol and in mitochondria of yeast [42], although the latter activity accounted a small fraction compared to that found in the cytosol. Furthermore, flx1Δ cells did not decrease mitochondrial FAD levels, but did inhibit export of FAD from the mitochondria.
The observed conflicting data on yeast flx1Δ cells may arise from inherent strains differences. The deletion in strain W303 results in limited matrix FAD levels implying an alternative route to matrix FAD. This route may arise from the presence of an alternative FAD carrier or limited levels of Fad1. The EBY157A flx1Δ cells may have an augmented level of an alternative FAD carrier or a greater fraction of Fad1 in an eclipsed localization within the matrix. Further studies are needed to resolve the role of Flx1 in matrix FAD metabolism.
3.4 Flx1 and its Effect on Flavinylation of Sdh1
A direct link between the proteins Flx1 and Sdh1 (such as protein-protein interaction) has not been demonstrated. Yet, it has become increasingly evident that Flx1 affects the flavinylation status of Sdh1 (or the overall stability of Sdh1) in a manner that has yet to be understood. In our on-going study with Flx1, we have shown that its deletion in yeast results in a decrease in mitochondrial FAD levels by ~50% [32], which is in line with the observation from Tzagoloff’s laboratory [40]. Additionally, we observe a near complete absence of the Sdh1 subunit, and consequently the SDH complex is undetectable as analyzed on a Blue-Native gel. We thus postulate that in a flx1Δ mutant, the resulting FAD limitation in the mitochondria (presumably in the matrix) leads to a flavinlyation defect of Sdh1 due to non-binding of FAD to Sdh1 (the mechanism for covalent bond formation remains functional in flx1Δ cells). Consequently, the unflavinylated apo-Sdh1 becomes unstable leading to an absence of the SDH complex (Figure 2C). We note, however, that the Rutter group showed that apo-Sdh1 was relatively stable in flx1Δ cells, but lacking almost completely its flavinylation [12]. Currently, we are unsure of the reason for this discrepancy in protein levels, but what is consistent between our group and Rutter’s is that these observations suggest a flavinylation impairment in Sdh1, resulting from FAD-limiting conditions in the mitochondrial matrix in flx1Δ cells.
Perhaps one of the most interesting aspects of the flx1 mutant is that the flavinylation defect in Sdh1 can be reversed partially by overexpression of Sdh5 [12]. This implies a FAD carrier/chaperone function of Sdh5, and that increasing this delivery vehicle can increase the “effective” concentration of FAD available for Sdh1. This concept means necessarily that some FAD is still available in the matrix of flx1Δ cells, perhaps through turnover and FAD release of FAD-containing enzymes in the matrix. Although, the flavinylation defect in Sdh1 can be partially reversed with Sdh5 overexpression, the respiratory growth defect is not restored indicating that Flx1 affects other cellular functions in addition to the SDH complex.
In an independent set of work by the Barile laboratory studying the relationship between Flx1 and Sdh1, covalent flavinylation of Sdh1 was found to be unaffected per se. The overall level of flavinylated Sdh1 (holo-Sdh1) however, was significantly lowered compared to WT cells [52]. Since the Barile laboratory proposes that Flx1 is an exporter of FAD [42], the loss of Sdh1 must result from something other than FAD limitation. This decrease is proposed to be a result of post-transcriptional control, with a mechanism involving the 5’-UTR of the mRNA, with possible involvement of flavin (FMN or FAD) as controlling the efficiency of translation [52]. Thus, the decreased level of Sdh1 in an flx1Δ mutant is proposed to result from decreased Sdh1 expression, rather than from FAD limitation in the mitochondria and subsequent degradation of Sdh1.
4.0 IN VIVO FLAVINYLATION OF SDH1 REQUIRES OTHER FACTORS
Role of Sdh5, Sdh2 and Sdh1 Carboxyl terminus
Unlike most flavoproteins such as VAO, monomeric sarcosine oxidase (MSOX) and p-cresol methylhydroxylase (PCMH), where covalent flavinylation has been demonstrated to be an autocatalytic process, flavinylation of Sdh1 appears to involve a more complex mechanism. This is perhaps due to the fact that Sdh1 is a mere single component of a larger protein complex, requiring careful coordination of protein folding, cofactor insertion, and finally the nucleation of all the subunits into an active SDH complex. For example, although only Sdh1 and Sdh5 are seemingly required to achieve covalent flavinylation, the absence of the Sdh2 subunit somehow affects dramatically the overall efficiency of the covalent modification (section 4.2). This indicates that covalent flavinylation and SDH assembly are intertwined at least in one step during the two processes.
The first evidence of a mediated flavinylation mechanism for Sdh1 came from the laboratory of Bernie Lemire in 1996. In this work, they noted that for flavinylation of Sdh1, “at least one matrix component appears to be required [53]” and flavinylation was found to be proportional to the concentration of the matrix fraction. This insight has been validated by the discovery of the Sdh5 protein in yeast; it is the first bona fide flavinylation protein factor to be identified.
Other mitochondrial flavoproteins may also require an additional protein(s) for covalent flavinylation. For example, dimethylglycine dehydrogenase from rat liver, also a mitochondrial matrix flavoprotein, is stimulated by a matrix protein factor for efficient covalent attachment of FAD [54]. However, in the absence of this yet unidentified protein factor, flavinylation still proceeds. What is remarkable about Sdh5 is that in its absence, covalent flavinylation is completely abolished. Thus, the mechanism involving Sdh5 is intriguing and begs the question of whether the flavinylation mechanism in Sdh1 is unique relative to other flavor proteins.
4.1 Sdh5 – The Small Protein with a Big Function
The protein Sdh5 from S. cerevisiae was discovered from a compendium of uncharacterized mitochondrial proteins with a very high degree of conservation in eukaryotes [12]. The small (~19 kDa) soluble protein is conserved from bacteria to humans and present also in plants [14], suggesting a high degree of functional significance. The bacterial homologue (designated SdhE) is smaller, but appears to have the same role as in yeast and in other eukaryotes. Sdh5 localizes to the mitochondrial matrix in eukaryotes and plants, and to the cytosol in bacteria. Although the exact function of Sdh5 has not been established, its deletion in yeast indicates that it is absolutely required for the covalent attachment of FAD to Sdh1 in vivo.
Yeast cells with a sdh5Δ genotype exhibit a respiratory defect stemming from a nonfunctioning complex II. Specifically, the covalent bond between the His90 residue of Sdh1 and FAD cannot form leaving a catalytically inactive protein [12]. Somewhat surprisingly, the steady-state levels of apo-Sdh1 persists, although decreased from WT levels [12]. The same effects are seen with a deletion in the bacterial homolog, sdhE, from Serratia [13]. Furthermore, in both yeast and bacterial systems, the SDH complex (as visualized on Blue-Native gel) can assemble in the absence of Sdh5/SdhE; albeit in the yeast system, the amount of the assembled complex is typically lower compared to the WT. In the bacterial system, the amount of the assembled complex on Blue Native gel was equal or even greater than the WT, leading the authors to conclude that the loss of SdhE does not affect the stability or the formation of SDH in Serratia.
4.1.1 Interaction of Sdh1 and Sdh5
The SDH complex in yeast, when analyzed on a Blue Native gel, migrates to a position corresponding to an apparent Mr of ~220 kDa [12]. Sdh5 is not a stable component of this complex; rather, the protein migrates to a position corresponding to an apparent Mr of a ~90 kDa – presumably a Sdh1-Sdh5 heterodimer [12]. This interaction was confirmed using tandem affinity purification of Sdh5 with polyhistidyl and hemagglutinin (HA) tags where Sdh1 was detected in the purification eluate [12]. Similar observations have been made with SdhE in Serratia using reciprocal purifications of SdhE and SdhA [13]. An interesting note with SdhE is that the population of SdhA in complex with SdhE was suggested to have a covalently bound FAD. The Sdh1-Sdh5 protein interaction is also important in maintaining each protein’s stability.
Deletion of SDH5 causes a significant decrease in the steady-state level of Sdh1. Inversely, deletion of SDH1 leads to a similar depletion of Sdh5 [12,32]. This interdependence on stability appears to result from a protein-protein interaction between Sdh1 and Sdh5. Substitution of the covalently linking His90 residue in Sdh1 to a Ser does not lead to the instability of Sdh5; rather, the steady-state level appears to be equal to, or sometimes greater than, the level found in WT cells [32]. Thus, a defect in covalent flavinylation does not lead to the instability of either Sdh1 or Sdh5.
The population of Sdh1 associated with Sdh5 is a fraction compared to Sdh1 found associated with the mature 220-kDa complex in WT cells. In the absence of the iron-sulfur protein Sdh2 subunit however, the steady-state level of Sdh5 accumulates [12,32]. This could indicate that the relative fraction of the Sdh1-Sdh5 complex also increases when SDH assembly is impaired.
An increase in Sdh5 steady-state level is also observed in the absence of Sdh3 [32], but a very important distinction between the effects of Sdh2 from Sdh3 is that the absence of the former leads to a reduction of flavinylated Sdh1 by an approximately 50% compared to WT level. Nevertheless, the Sdh1-Sdh5 complex appears to be discrete and does not include the associations of Sdh2 or the membrane subunits Sdh3 and Sdh4. This implies that the Sdh1-Sdh5 interaction, and the steps leading to the covalent flavinylation of Sdh1, is an early process prior to SDH complex assembly with the Sdh2 iron-sulfur protein and Sdh3-Sdh4 membrane domain.
4.1.2 The Solution Structure of Sdh5 from Yeast
The Sdh5 protein from Saccharomyces cerevisiae was recently cloned, expressed, and purified from E. coli and its solution structure determined using NMR [60]. As isolated, the recombinant protein, lacking 55 amino acids from the amino terminus (predicted disordered region), did not contain any noticeable amounts of FAD. Furthermore, addition of FAD to the purified protein did not induce chemical shift perturbations in its NMR spectrum, suggesting that the peptide backbone is not perturbed and that FAD does not bind in vitro, at least to the truncated protein. This of course does not rule out the possibility of FAD binding to Sdh5 in-vivo.
This demonstration of non-binding of FAD to Sdh5 is in contrast to SdhE from Serratia, where the addition of exogenous FAD to the purified protein resulted in covalent binding of the flavin. This binding was demonstrated by three techniques: a UV illuminated band on SDS-PAGE that corresponds to the mass of SdhA; the optical spectral characteristic of FAD-SdhA; and mass spectral analysis that identified a SdhE peptide with bound FAD [13]. The FAD-SdhE covalent interaction was likened to the heme chaperone CcmE that binds heme covalently for delivery and insertion into a c-type cytochrome [55,56].
Based on sequence homology, Sdh5 belongs to a large protein superfamily now classified as “flavinylation factors of SDH” or “Sdh5 superfamily.” The structures of three other proteins belonging to this family are also available as a result of structural genomics initiatives: YgfY from E. coli [57] along with two other proteins that are currently unnamed, NMA1147 from N. meningitides [58], and VC2471 from Vibrio cholerae. Compared to its bacterial counterparts, the eukaryotic Sdh5 has an extra stretch of residues comprised essentially of a strand at its amino terminus (after cleavage of the predicted mitochondrial targeting peptide [59]).
The overall structure of the core, a compact five α-helical bundle (Figure 3A), is highly conserved however. This suggests a strong functional conservation. In fact, the five α-helical structures of all four proteins are largely superimposable. Furthermore, many of the conserved amino acid residues of this superfamily, are located in a strikingly concentrated region on the surface of the α-helical core. This conserved surface patch is functionally important. Charge reversal of a highly conserved Arg68 residue to an Asp that lie in this conserved patch resulted in a complete absence of the UV-illuminated band corresponding to flavo-Sdh1 on a SDS-PAGE gel [60] (Figure 3B). In fact, yeast cells harboring this mutation in Sdh5 are respiratory defective and unable to grow in respiratory medium due to a nonfunctioning SDH. Most interestingly, this charge reversal results in an accumulation of the Sdh1-Sdh5 complex, effectively “trapping” the otherwise transient interaction of Sdh1 with Sdh5 that can be purified using an affinity tag on Sdh5 (unpublished result).
Fig 3.
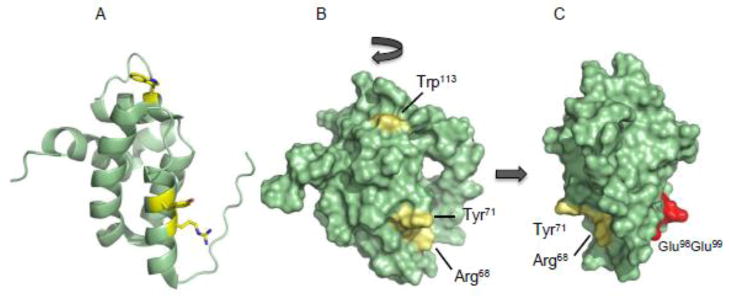
The NMR structure [60] of Sdh5 from yeast (PDB 2LM4). (A) Cartoon model showing the primarily helical characteristic of the protein and the residues (yellow sticks) when mutated leads to the loss of flavinylation in Sdh1. (B) Surface rendering showing the presence of the residues all on the same face of Sdh5. (C) Mutation of two glutamic acids that lie of on the other side of Sdh5 does not lead to the loss of covalent flavinylation in Sdh1.
Mutation of two other residues (a loosely conserved Tyr71 and a non-conserved Trp113) in the periphery of the highly conserved region also resulted in the loss of the covalent attachment of FAD (Figure 3B). This defect in flavinylation did not stem from unstable Sdh1 protein levels as The results of these Sdh5 substitutions illustrate the functional importance of this conserved region, which we speculate to be an interaction surface for Sdh1. Mutation of residues on the opposite side of this face had no effect on flavinylation of Sdh1 (Figure 3C).
4.2 The Role of the Carboxyl Terminus of Sdh1
The C-terminal domain of Sdh1 consists two loops interconnected by two β sheets; the entire domain sits atop the surface of Sdh1 [17]. The closest distance between this domain and the nearest edge of FAD is ~17Å. Truncation of 70 residues in this C-terminal domain prevents the covalent attachment of FAD [61]. Addition of purification tags such as polyhistidyl or hemagglutinin tags at the C terminus of Sdh1 renders cells inviable on respiratory medium (unpublished results), further highlighting the importance of this domain. One contributing factor in the importance of the C terminus is a set of key essential residues located in this region [32]. Mutation of a Cys630Arg638 pair to alanine residues located at the very tail end of the C terminus prevents flavinylation. Additionally, mutation of Arg582 to Ala located near, but not in the C-terminal tail, also prevents flavinylation. The flavinylation defects in these two sets of mutations prevent also the assembly of the tetrameric SDH complex. A second-site suppressor mutation of the R582A mutant can restore covalent flavinylation and assembly of SDH. The resulting suppressor mutation, a M599R substitution in Sdh1, has the effect of restoring the lost positive charge in the C-terminal region.
The finding that the SDH assembly is impaired in the C-terminal mutants is revealing in our view. Sdh1 lacking a covalently-bound flavin should not lead to an assembly defect. This has been clearly shown in the Sdh1 H90S mutant [8] and in a yeast strain lacking SDH5 [32] and in the bacteria Serratia lacking SdhE [13]. Based on these observations, we postulate that FAD binding to Sdh1, but not covalent attachment, is required to stabilize the Sdh1conformation enabling association with Sdh2 and the membrane-spanning subunits. If true, then the C-terminal mutants prevent the binding of FAD to Sdh1.
SDH assembly is also impaired in flx1Δ cells [32], which is characterized by attenuated levels of mitochondrial matrix FAD [32,40]. This observation supports the notion that FAD association with Sdh1 is a prerequisite for SDH assembly. Thus, the C-terminal positively charged Arg residues could be important for the recruitment and/or guidance of the dianionic FAD to the binding site.
4.3 The Role of Sdh2 in the Flavinylation of Sdh1
The covalent flavinylation of Sdh1, requiring its interaction with Sdh5, appears to be a process that is discrete from its interaction with other SDH subunits. This notion is based on the observation that covalent flavinylation can be achieved by co-expressing yeast Sdh1 and Sdh5 in a heterologous E. coli host [12]. Yet, deletion of SDH2 in yeast leads to a reduction of covalent flavinylation by approximately half compared to WT cells, although total Sdh1 protein levels remain relatively unchanged [32,61]. This reduction is specific to Sdh2, as deletions of SDH3 or SDH4 genes have relatively a minor effect. If indeed the covalent attachment is discreet to Sdh1 and Sdh5, then the decreased flavinylation in sdh2Δ cells is a curious effect.
The exact role of Sdh2 in promoting the efficiency of flavinylation in vivo is unclear. It has been suggested that the subunit aids in the folding of apo-Sdh1[61], adopting a requisite structure for FAD interaction and subsequent covalent bond formation [32]. However, flavinylation can still occur without Sdh2. Thus, if a requisite, prefolded structure is required for flavinylation, then Sdh1 must be able to fold without the aid of Sdh2. An interesting question that arises from noting the effect of Sdh2 is whether Sdh1 forms a complex with Sdh5 before or after its interaction with Sdh2. The notion that Sdh2 aids in the folding of apo-Sdh1 prior to flavinylation suggests that the interaction with Sdh5 would follow the Sdh1-Sdh2 interaction. Therefore, a coIP of Sdh5 should yield both Sdh1 and Sdh2, but thus far only a Sdh1-Sdh5 interaction has been reported [12,32]. Deciphering the exact role of Sdh2 in covalent flavinylation should lead to an understanding the temporal relationship between FAD insertion and subunit interaction and final SDH assembly.
5.0 SUMMARY OF EVENTS IN SDH FLAVINYLATION AND ASSEMBLY
The review thus far has presented prior and recent advances in flavinylation of Sdh1, assembly of SDH complex, and in characterization of Sdh5. We summarize these findings below in a conceptual model of events in flavinylation and assembly of SDH.
5.1 Import and Processing
The maturation of Sdh1, similar to other mitochondrial matrix proteins, starts with import from the cytosol by the TOM and TIM23 complexes, directed by amino terminal targeting presequence. Removal of this presequence – predicted to be by the proteases MPP and Oct1 [62] – is a necessary step before further maturation can occur. Therefore, FAD attachment requires the proteolytically processed Sdh1 protein [61,63]. Interestingly, an engineered amino terminal truncate of Sdh1, lacking its presequence, is flavinylation incompetent when assayed in vitro for FAD attachment [53]. Therefore, the authors of the study concluded that the process of presequence cleavage is itself, required for flavinylation.
5.2 Fold and Formation of a Structure
After import and presequence processing, flavin attachment occurs in the mitochondrial matrix requiring several components found in this compartment: ATP, Mg2+, TCA cycle intermediates succinate and/or fumarate, and Sdh5. Upon entry into the matrix, the apo-Sdh1 folds, perhaps into an intermediate structure, but likely bearing similarity to a mature fold. Alternatively, the apo Sdh1 may adopt a fully folded, mature state.
Two findings support the idea of a prefolded state prior to binding FAD. First, TCA cycle intermediate succinate is required for flavinylation suggesting that the active site, competent to bind succinate, has already formed [61,64]. Second, C-terminal truncates and C-terminal mutants preclude covalent flavinylation. If only the unfolded state was required, then only the residues around the N terminus should affect covalent flavinylation [32,61].
5.3 Association with Sdh5 and Covalent Bond Formation
In the concept of a prefolded state described above, Sdh5 likely interacts with a folded Sdh1. At this stage, Sdh5 could sequester mitochondrial matrix FAD for a coordinated delivery to apo-Sdh1. The demonstration in Serratia that purified SdhE could covalently bind exogenous FAD certainly raises this possibility. Furthermore, the fact that in flx1Δ cells, Sdh5 overexpression can partially restore flavinylation of Sdh1 leaves open the possibility that Sdh5 is indeed a FAD chaperone.
Discrepant results exist on whether Sdh5/SdhE stably associate with FAD. Whereas a fraction of SdhE was reported to contain a covalent FAD adduct, purified recombinant yeast Sdh5 did not contain any noticeable amounts of FAD as isolated and exogenous additions did not perturb the Sdh5 backbone structure as monitored by NMR (see section 4.1.2). Cells lacking Sdh5 or SdhE still assemble SDH without any covalent flavinylation of Sdh1/SdhA. If FAD binding (noncovalent) to Sdh1 is indeed required for assembly to proceed, then delivery of FAD to Sdh1 must still take place in the absence of Sdh5/SdhE. Consistent with this view, FAD limitation in the mitochondrial matrix by deletion of flx1 prevents assembly of SDH, seemingly since there is no interaction of FAD and Sdh1 [32].
Another possible function of Sdh5 is that it may assist in the activation of the FAD isoalloxazine ring to the highly reactive quinone methide form suitable for a nucleophilic attack by the covalently-linking His90 residue [23,24]. This activation reaction can be catalyzed by bases (arginines) nearby the isoalloxazine ring perhaps present in Sdh5. Activation of the N3 atom of the histidyl imidazole side chain can also be considered [24].
5.4 Association with Sdh2
The Sdh1-Sdh2 association could be considered the second most critical interaction next to Sdh1’s association with Sdh5. This is because in the absence of the Sdh1-Sdh2 interaction, covalent flavinylation appears impaired [61,32]. We tentatively place the Sdh1-Sdh2 complex after the Sdh1-Sdh5 association and post covalent flavinylation. A key element in this assignment is that a Sdh1-Sdh5-Sdh2 tricomplex has yet to be observed. This complex should exist if a Sdh1-Sdh2 complex precedes a Sdh5 association. It is possible that Sdh2 further stabilizes holo-Sdh1 preventing either degradation or aggregation, especially in light of the fact that a significant amount of Sdh2’s surface area is involved in interaction with Sdh1 [17].
5.5 Assembly of SDH Post-Flavinylation
Assembly factors or chaperoning are required for the maturation of the SDH complex post flavinylation of Sdh1. For example, Hsp60 (a catalytic chaperone that assist in folding of new monomeric proteins and oligomeric complexes) has been found associated with Sdh1 [53]. However, immunodepletion of this chaperone from the matrix did not affect flavinylation. Tcm62, with some sequence similarity to yeast Hsp60, and to E. coli GroEL [65,66] is also proposed to serve a chaperonin function in the assembly of SDH [65]. Tcm62 forms a complex containing at least Sdh1, Sdh2, and Sdh3 subunits [65]. Overexpression of Tcm62 results in an accumulation of Sdh2 subunit that can be found in an aggregated form possibly indicating an effect directly with the iron sulfur subunit.
The recruitment of Sdh3 and Sdh4 likely constitutes the terminal step in the assembly of SDH, as deletions of genes encoding these subunits do not affect covalent flavinylation. The recruitment of the soluble catalytic dimer to the membranes via Sdh3 and Sdh4 are unknown.
5.5 Concluding Remarks
The covalently-linked FAD in SDH is essential for the catalytic function of the SDH complex. Formation of the covalent bond between FAD and the histidyl residue of Sdh1 is dependent on the assembly factor Sdh5, although its mechanistic role of has yet to be defined. The dependency of covalent insertion of FAD on Sdh5 raises the question whether other assembly factors are needed for FAD insertion or covalent addition in other flavoproteins.
The elucidation of the solution structure of Sdh5 and the identification of the C-terminal segment of Sdh1 as a key determinant in FAD binding raises intriguing new questions about the formation of FAD center in SDH. FAD binding to Sdh1 is also key to the assembly process of the tetrameric enzyme. The conservation of the process between prokaryotes and eukaryotes creates new opportunities and systems to elucidate the mechanistic details. Intriguing questions left to resolve include: “Does Sdh5 deliver FAD to Sdh1?”, “What role does FAD binding have on the conformation of Sdh1 during biogenesis?”, “How does the Sdh1 C-terminal Arg motif contribute to flavinylation?”, “What is the role of Sdh2 in the flavinylation of Sdh1?”, and “What is the role of the Flx1 carrier protein in Sdh1 flavinylation?”. Foundational results are beginning to emerge, but we are far from definitive answers to these questions. The field is poised in being able to meet these challenging questions. The topic is of health relevance as impairment of flavinylation either by mutations in SDHAF2 (Sdh5) or SDHA (Sdh1) predisposes humans to a range of tumors ranging from paragangliomas, pheochromocytomas, gastrointestinal stromal tumors and neuroblastomas.
Fig 4.
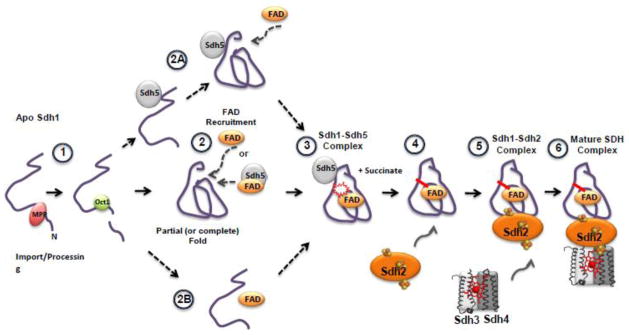
A general conceptual model of Sdh1 flavinylation and subsequent assembly into the SDH complex. (1) After import of Sdh1 into the matrix, processing of the presequence (predicted to be by proteases MPP and Oct1 [62]) occurs. Flavinylation has been shown to occur only to the processed peptide [61,63]. (2) Apo-Sdh1 presumably folds into a structure resembling that of holo-Sdh1 [32,61,64]. After the organized structure has formed, the FAD binding pocket could accept the cofactor and succinate, stabilizing Sdh1 from degradation. FAD recruitment may occur via Sdh5-mediated delivery or as “free” FAD. Alternative pathways after processing include (2A) Sdh5 mediated folding of Sdh1, (2B) FAD binding to a nascent structure of Sdh1 followed by folding into a holo-structure. (3) Formation of the Sdh1-Sdh5 complex and subsequent covalent bond formation. This complex can be purified or visualized using native gel electrophoresis. TCA cycle intermediates like succinate greatly enhances the covalent bond formation [53]. (4) Association with Sdh2. Currently, a trimeric complex consisting of Sdh1-Sdh2-Sdh5 has not been identified. (5) Association with the Sdh3 and Sdh4 membrane subunits and the (6) formation of the mature SDH complex anchored in the mitochondrial inner membrane.
Highlights.
The mechanism of FAD covalent attachment to Sdh1 has been a long-standing question.
The discovery of Sdh5 suggests that Sdh1 flavinylation is a complex, non-autocatalytic process.
FAD and succinate binding to Sdh1 and its binding to Sdh5 and Sdh2 are important in flavinylation.
A conceptual model is presented based on prior/current findings in flavinylation/SDH assembly.
Acknowledgments
We thank Drs. Mi-Young Jeong and Richard Dela Cruz for critical comments during the preparation of the review. We acknowledge support from the National Institutes of Health ES03817 to D.R.W. H.J.K. was supported by training grant T32 DK007115.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Oyedotun KS, Sit CS, Lemire BD. The Saccharomyces cerevisiae succinate dehydrogenase does not require heme for ubiquinone reduction. Biochim Biophys Acta. 2007;1767:1436–1445. doi: 10.1016/j.bbabio.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 2.Tran QM, Rothery RA, Maklashina E, Cecchini G, Weiner JH. Escherichia coli succinate dehydrogenase variant lacking the heme b. Proc Nat Acad Sci. 2007;104:18007–18012. doi: 10.1073/pnas.0707732104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maklashina E, Rajagukguk S, McIntire WS, Cecchini G. Mutation of the heme axial ligand of Escherichia coli succinate-quinone reductase: implications for heme ligation in mitochondrial complex II from yeast. Biochim Biophys Acta. 2010;1797:747–754. doi: 10.1016/j.bbabio.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim HJ, Khalimonchuk O, Smith PM, Winge DR. Structure, function, and assembly of heme centers in mitochondrial respiratory complexes. Biochim Biophys Acta. 2012;1823:1604–1616. doi: 10.1016/j.bbamcr.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singer TP, Kearney EB, Zastrow N. Isolation and properties of succinic dehydrogenase. Biochim Biophys Acta. 1955;17:154–155. doi: 10.1016/0006-3002(55)90340-1. [DOI] [PubMed] [Google Scholar]
- 6.Kearney EB. Studies on succinic dehydrogenase XII Flavin component of the mammalian enzyme. J Biol Chem. 1960;235:865–877. [PubMed] [Google Scholar]
- 7.Blaut M, Whittaker K, Valdovinos A, Ackrell BA, Gunsalus RP, Cecchini G. Fumarate reductase mutants of Escherichia coli that lack covalently bound flavin. J Biol Chem. 1989;264:13599–13604. [PubMed] [Google Scholar]
- 8.Robinson KM, Rothery RA, Weiner JH, Lemire BD. The covalent attachment of FAD to the flavoprotein of Saccharomyces cerevisiae succinate dehydrogenase is not necessary for import and assembly into mitochondria. Eur J Biochem. 1994;222:983–990. doi: 10.1111/j.1432-1033.1994.tb18949.x. [DOI] [PubMed] [Google Scholar]
- 9.Mauch L, Bichler V, Brandsch R. Site-directed mutagenesis of the FAD-binding histidine of 6-hydroxy-D-nicotine oxidase Consequences on flavinylation and enzyme activity. FEBS Lett. 1989;257:86–88. doi: 10.1016/0014-5793(89)81792-2. [DOI] [PubMed] [Google Scholar]
- 10.Heuts DPHM, Scrutton NS, McIntire WS, Fraaije MW. What's in a covalent bond? On the role and formation of covalently bound flavin cofactors. FEBS J. 2009;276:3405–3427. doi: 10.1111/j.1742-4658.2009.07053.x. [DOI] [PubMed] [Google Scholar]
- 11.Macheroux P, Kappes B, Ealick SE. Flavogenomics--a genomic and structural view of flavin-dependent proteins. FEBS J. 2011;278:2625–2634. doi: 10.1111/j.1742-4658.2011.08202.x. [DOI] [PubMed] [Google Scholar]
- 12.Hao HX, Khalimonchuk O, Schraders M, Dephoure N, Bayley JP, Kunst H, Devilee P, Cremers CWRJ, Schiffman JD, Bentz BG, Gygi SP, Winge DR, Kremer H, Rutter J. SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science. 2009;325:1139–1142. doi: 10.1126/science.1175689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McNeil MB, Clulow JS, Wilf NM, Salmond GPC, Fineran PC. SdhE Is a Conserved Protein Required for Flavinylation of Succinate Dehydrogenase in Bacteria. J Biol Chem. 2012;287:18418–18428. doi: 10.1074/jbc.M111.293803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang S, Taylor NL, Ströher E, Fenske R, Harvey Millar A. Succinate dehydrogenase assembly factor 2 is needed for assembly and activity of mitochondrial complex II and for normal root elongation in Arabidopsis. Plant J. 2012 doi: 10.1111/tpj.12041. in press. [DOI] [PubMed] [Google Scholar]
- 15.Iverson TM, Luna-Chavez C, Cecchini G, Rees DC. Structure of the Escherichia coli fumarate reductase respiratory complex. Science. 1999;284:1961–1966. doi: 10.1126/science.284.5422.1961. [DOI] [PubMed] [Google Scholar]
- 16.Yankovskaya V, Horsefield R, Törnroth S, Luna-Chavez C, Miyoshi H, Léger C, Byrne B, Cecchini G, Iwata S. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science. 2003;299:700–704. doi: 10.1126/science.1079605. [DOI] [PubMed] [Google Scholar]
- 17.Sun F, Huo X, Zhai Y, Wang A, Xu J, Su D, Bartlam M, Rao Z. Crystal structure of mitochondrial respiratory membrane protein complex II. Cell. 2005;121:1043–1057. doi: 10.1016/j.cell.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 18.Iverson TM, Luna-Chavez C, Schröder I, Cecchini G, Rees DC. Analyzing your complexes: structure of the quinol-fumarate reductase respiratory complex. Curr Opin Struct Biol. 2000;10:448–455. doi: 10.1016/s0959-440x(00)00113-5. [DOI] [PubMed] [Google Scholar]
- 19.Lemire BD, Oyedotun KS. The Saccharomyces cerevisiae mitochondrial succinate:ubiquinone oxidoreductase. Biochim Biophys Acta. 2002;1553:102–116. doi: 10.1016/s0005-2728(01)00229-8. [DOI] [PubMed] [Google Scholar]
- 20.Rutter J, Winge DR, Schiffman JD. Succinate dehydrogenase - Assembly, regulation and role in human disease. Mitochondrion. 2010;10:393–401. doi: 10.1016/j.mito.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iverson TM, Maklashina E, Cecchini G. Structural Basis for Malfunction in Complex II. 2012;287:35430–8. doi: 10.1074/jbc.R112.408419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dym O, Eisenberg D. Sequence-structure analysis of FAD-containing proteins. Protein Sci. 2001;10:1712–1728. doi: 10.1110/ps.12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Decker KF. Biosynthesis and function of enzymes with covalently bound flavin. Annu Rev Nutr. 1993;13:17–41. doi: 10.1146/annurev.nu.13.070193.000313. [DOI] [PubMed] [Google Scholar]
- 24.Walsh C. Flavin coenzymes: at the crossroads of biological redox chemistry. Acc Chem Res. 1980;13:148–155. [Google Scholar]
- 25.Jin J, Mazon H, van den Heuvel RHH, Heck AJ, Janssen DB, Fraaije MW. Covalent flavinylation of vanillyl-alcohol oxidase is an autocatalytic process. FEBS J. 2008;275:5191–5200. doi: 10.1111/j.1742-4658.2008.06649.x. [DOI] [PubMed] [Google Scholar]
- 26.Singer TP, McIntire WS. Covalent attachment of flavin to flavoproteins: occurrence, assay, and synthesis. Meth Enzymol. 1984;106:369–378. doi: 10.1016/0076-6879(84)06039-0. [DOI] [PubMed] [Google Scholar]
- 27.Jin J, Mazon H, van den Heuvel RHH, Heck AJ, Janssen DB, Fraaije MW. Covalent flavinylation of vanillyl-alcohol oxidase is an autocatalytic process. FEBS J. 2008;275:5191–5200. doi: 10.1111/j.1742-4658.2008.06649.x. [DOI] [PubMed] [Google Scholar]
- 28.Fraaije MW, van Den Heuvel RH, van Berkel WJ, Mattevi A. Structural analysis of flavinylation in vanillyl-alcohol oxidase. J Biol Chem. 2000;275:38654–38658. doi: 10.1074/jbc.M004753200. [DOI] [PubMed] [Google Scholar]
- 29.Huang CH, Lai WL, Lee MH, Chen CJ, Vasella A, Tsai YC, Liaw SH. Crystal structure of glucooligosaccharide oxidase from Acremonium strictum: a novel flavinylation of 6-S-cysteinyl, 8-alpha-N1-histidyl FAD. J Biol Chem. 2005;280:38831–38838. doi: 10.1074/jbc.M506078200. [DOI] [PubMed] [Google Scholar]
- 30.Williamson G, Edmondson DE. Effect of pH on oxidation-reduction potentials of 8-alpha-N-imidazole-substituted flavins. Biochemistry. 1985;24:7790–7797. doi: 10.1021/bi00347a043. [DOI] [PubMed] [Google Scholar]
- 31.Fraaije MW. Covalent Flavinylation Is Essential for Efficient Redox Catalysis in Vanillyl-alcohol Oxidase. J Biol Chem. 1999;274:35514–35520. doi: 10.1074/jbc.274.50.35514. [DOI] [PubMed] [Google Scholar]
- 32.Kim HJ, Jeong M-Y, Na U, Winge DR. Flavinylation and assembly of succinate dehydrogenase are dependent on the C-terminal tail of the flavoprotein subunit. J Biol Chem. 2012 doi: 10.1074/jbc.M112.405704. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brandsch R, Bichler V, Krauss B. Binding of FAD to 6-hydroxy-D-nicotine oxidase apoenzyme prevents degradation of the holoenzyme. Biochem J. 1989;258:187–192. doi: 10.1042/bj2580187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heuts DPHM, van Hellemond EW, Janssen DB, Fraaije MW. Discovery, characterization, and kinetic analysis of an alditol oxidase from Streptomyces coelicolor. J Biol Chem. 2007;282:20283–20291. doi: 10.1074/jbc.M610849200. [DOI] [PubMed] [Google Scholar]
- 35.Santos MA, Jimenez A, Revuelta JL. Molecular characterization of FMN1, the structural gene for the monofunctional flavokinase of Saccharomyces cerevisiae. J Biol Chem. 2000;275:28618–28624. doi: 10.1074/jbc.M004621200. [DOI] [PubMed] [Google Scholar]
- 36.Wu M, Repetto B, Glerum DM, Tzagoloff A. Cloning and characterization of FAD1, the structural gene for flavin adenine dinucleotide synthetase of Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:264–271. doi: 10.1128/mcb.15.1.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leulliot N, Blondeau K, Keller J, Ulryck N, Quevillon-Cheruel S, van Tilbeurgh H. Crystal structure of yeast FAD synthetase (Fad1) in complex with FAD. J Mol Biol. 2010;398:641–646. doi: 10.1016/j.jmb.2010.03.040. [DOI] [PubMed] [Google Scholar]
- 38.Torchetti EM, Bonomi F, Galluccio M, Gianazza E, Giancaspero TA, Iametti S, Indiveri C, Barile M. Human FAD synthase (isoform 2): a component of the machinery that delivers FAD to apo-flavoproteins. FEBS J. 2011;278:4434–4449. doi: 10.1111/j.1742-4658.2011.08368.x. [DOI] [PubMed] [Google Scholar]
- 39.Kearney EB, Goldenberg J, Lipsick J, Perl M. Flavokinase and FAD synthetase from Bacillus subtilis specific for reduced flavins. J Biol Chem. 1979;254:9551–9557. [PubMed] [Google Scholar]
- 40.Tzagoloff A, Jang J, Glerum DM, Wu M. FLX1 codes for a carrier protein involved in maintaining a proper balance of flavin nucleotides in yeast mitochondria. J Biol Chem. 1996;271:7392–7397. doi: 10.1074/jbc.271.13.7392. [DOI] [PubMed] [Google Scholar]
- 41.Pallotta ML, Brizio C, Fratianni A, De Virgilio C, Barile M, Passarella S. Saccharomyces cerevisiae mitochondria can synthesise FMN and FAD from externally added riboflavin and export them to the extramitochondrial phase. FEBS Lett. 1998;428:245–249. doi: 10.1016/s0014-5793(98)00544-4. [DOI] [PubMed] [Google Scholar]
- 42.Bafunno V, Giancaspero TA, Brizio C, Bufano D, Passarella S, Boles E, Barile M. Riboflavin uptake and FAD synthesis in Saccharomyces cerevisiae mitochondria: involvement of the Flx1p carrier in FAD export. J of Biol Chem. 2004;279:95–102. doi: 10.1074/jbc.M308230200. [DOI] [PubMed] [Google Scholar]
- 43.Barile M, Passarella S, Bertoldi A, Quagliariello E. Flavin adenine dinucleotide synthesis in isolated rat liver mitochondria caused by imported flavin mononucleotide. Arch Biochem Biophys. 1993;305:442–447. doi: 10.1006/abbi.1993.1444. [DOI] [PubMed] [Google Scholar]
- 44.Barile M, Brizio C, Valenti D, De Virgilio C, Passarella S. The riboflavin/FAD cycle in rat liver mitochondria. Eur J Biochem. 2000;267:4888–4900. doi: 10.1046/j.1432-1327.2000.01552.x. [DOI] [PubMed] [Google Scholar]
- 45.Giancaspero TA, Locato V, de Pinto MC, De Gara L, Barile M. The occurrence of riboflavin kinase and FAD synthetase ensures FAD synthesis in tobacco mitochondria and maintenance of cellular redox status. FEBS J. 2009;276:219–231. doi: 10.1111/j.1742-4658.2008.06775.x. [DOI] [PubMed] [Google Scholar]
- 46.Torchetti E, Brizio C, Colella M, Galluccio M. Mitochondrial localization of human FAD synthetase isoform 1. Mitochondrion. 2010;10:263–274. doi: 10.1016/j.mito.2009.12.149. [DOI] [PubMed] [Google Scholar]
- 47.Robinson AJ, Kunji ERS. Mitochondrial carriers in the cytoplasmic state have a common substrate binding site. Proc Nat Acad Sci. 2006;103:2617–2622. doi: 10.1073/pnas.0509994103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–9. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pebay-Peyroula E, Dahout-Gonzalez C, Kahn R, Trézéguet V, Lauquin GJM, Brandolin G. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature. 2003;426:39–44. doi: 10.1038/nature02056. [DOI] [PubMed] [Google Scholar]
- 50.Robinson AJ, Overy C, Kunji ERS. The mechanism of transport by mitochondrial carriers based on analysis of symmetry. Proc Nat Acad Sci. 2008;105:17766–17771. doi: 10.1073/pnas.0809580105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ozeir M, Mühlenhoff U, Webert H, Lill R, Fontecave M, Pierrel F. Coenzyme Q biosynthesis: Coq6 is required for the C5-hydroxylation reaction and substrate analogs rescue Coq6 deficiency. Chem Biol. 2011;18:1134–1142. doi: 10.1016/j.chembiol.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 52.Giancaspero TA, Wait R, Boles E, Barile M. Succinate dehydrogenase flavoprotein subunit expression in Saccharomyces cerevisiae--involvement of the mitochondrial FAD transporter, Flx1p. FEBS J. 2008;275:1103–1117. doi: 10.1111/j.1742-4658.2008.06270.x. [DOI] [PubMed] [Google Scholar]
- 53.Robinson KM, Lemire BD. A requirement for matrix processing peptidase but not for mitochondrial chaperonin in the covalent attachment of FAD to the yeast succinate dehydrogenase flavoprotein. J Biol Chem. 1996;271:4061–4067. doi: 10.1074/jbc.271.8.4061. [DOI] [PubMed] [Google Scholar]
- 54.Brizio C, Otto A, Brandsch R, Passarella S, Barile M. A protein factor of rat liver mitochondrial matrix involved in flavinylation of dimethylglycine dehydrogenase. Eur J Biochem. 2000;267:4346–4354. doi: 10.1046/j.1432-1327.2000.01464.x. [DOI] [PubMed] [Google Scholar]
- 55.Schulz H, Hennecke H. Prototype of a heme chaperone essential for cytochrome c maturation. Science. 1998;281:1197–200. doi: 10.1126/science.281.5380.1197. [DOI] [PubMed] [Google Scholar]
- 56.Richard-Fogal C, Kranz RG. The CcmC:heme:CcmE complex in heme trafficking and cytochrome c biosynthesis. J Mol Biol. 2010;401:350–362. doi: 10.1016/j.jmb.2010.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lim K, Doseeva V, Demirkan ES, Pullalarevu S, Krajewski W, Galkin A, Howard A, Herzberg O. Crystal structure of the YgfY from Escherichia coli, a protein that may be involved in transcriptional regulation. Proteins. 2005;58:759–763. doi: 10.1002/prot.20337. [DOI] [PubMed] [Google Scholar]
- 58.Liu G, Sukumaran DK, Xu D, Chiang Y, Acton T, Goldsmith-Fischman S, Honig B, Montelione GT, Szyperski T. NMR structure of the hypothetical protein NMA1147 from Neisseria meningitidis reveals a distinct 5-helix bundle. Proteins. 2004;55:756–758. doi: 10.1002/prot.20009. [DOI] [PubMed] [Google Scholar]
- 59.Vögtle FN, Wortelkamp S, Zahedi RP, Becker D, Leidhold C, Gevaert K, Kellermann J, Voos W, Sickmann A, Pfanner N, Meisinger C. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell. 2009;139:428–439. doi: 10.1016/j.cell.2009.07.045. [DOI] [PubMed] [Google Scholar]
- 60.Eletski A, Jeong M-Y, Kim HJ, Xiao R, Lee H-W, Pagliarini DJ, Prestegard JH, Winge DR, Montelione GT, Szyperski TA. Solution NMR Structure of Yeast Succinate Dehydrogenase Flavinylation Factor Sdh5 Reveals a Putative Sdh1 Binding Site. Biochemistry. 2012 doi: 10.1021/bi301171u. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robinson KM, Lemire BD. Covalent attachment of FAD to the yeast succinate dehydrogenase flavoprotein requires import into mitochondria, presequence removal, and folding. J Biol Chem. 1996;271:4055–4060. doi: 10.1074/jbc.271.8.4055. [DOI] [PubMed] [Google Scholar]
- 62.Schmidt O, Pfanner N, Meisinger C. Mitochondrial protein import: from proteomics to functional mechanisms. Nat Rev Mol Cell Biol. 2010;11:655–667. doi: 10.1038/nrm2959. [DOI] [PubMed] [Google Scholar]
- 63.Robinson KM, Lemire BD. Isolation and nucleotide sequence of the Saccharomyces cerevisiae gene for the succinate dehydrogenase flavoprotein subunit. J Biol Chem. 1992;267:10101–10107. [PubMed] [Google Scholar]
- 64.Brandsch R, Bichler V. Covalent cofactor binding to flavoenzymes requires specific effectors. Eur J Biochem. 1989;182:125–128. doi: 10.1111/j.1432-1033.1989.tb14808.x. [DOI] [PubMed] [Google Scholar]
- 65.Dibrov E, Fu S, Lemire BD. The Saccharomyces cerevisiae TCM62 gene encodes a chaperone necessary for the assembly of the mitochondrial succinate dehydrogenase (complex II) J Biol Chem. 1998;273:32042–32048. doi: 10.1074/jbc.273.48.32042. [DOI] [PubMed] [Google Scholar]
- 66.Klanner C, Neupert W, Langer T. The chaperonin-related protein Tcm62 ensures mitochondrial gene expresssion under heat stress. FEBS Lett. 2000;470:365–369. doi: 10.1016/s0014-5793(00)01322-3. [DOI] [PubMed] [Google Scholar]