Abstract
Bacterial siderophores may enhance pathogenicity by scavenging iron but their expression has been proposed to exert a substantial metabolic cost. Here we describe a combined metabolomic-genetic approach to determine how mutations affecting the virulence-associated siderophore yersiniabactin affect the Escherichia coli primary metabolome. Contrary to expectations, we did not find yersiniabactin biosynthesis to correspond to consistent metabolomic shifts. Instead, we found that targeted deletion of ybtU or ybtA, dissimilar genes with similar roles in regulating yersiniabactin expression, were associated with a specific shift in arginine pathway metabolites during growth in minimal media. This interaction was associated with high arginine levels in the model uropathogen Escherichia coli UTI89 compared to its ybtU and ybtA mutants and the K12 strain MG1655, which lacks yersiniabactin-associated genes. Because arginine is not a direct yersiniabactin biosynthetic substrate, these findings show that virulence-associated secondary metabolite systems may shape bacterial primary metabolism independently of substrate consumption.
Keywords: metabolomics, siderophore, yersiniabactin, Escherichia coli, primary metabolism, ybtU, ybtA, arginine biosynthesis
INTRODUCTION
Siderophores are a chemically diverse class of secondary metabolites used by bacteria to chelate ferric iron. 1 Because they are secreted to diffuse freely in their surroundings, a bacterium that uses a ferric siderophore complex may not be the same individual that produced the siderophore. This circumstance has been proposed to result in the emergence of “cheater” bacteria in which siderophore biosynthesis is blocked by mutations while ferric siderophore uptake is preserved 2. Biosynthetic mutants have also been suggested to evolve where a particular siderophore type no longer confers a growth or survival advantage 2. In either circumstance, alleviation of a metabolic burden arising from siderophore biosynthesis is typically advanced to explain proliferation of these spontaneous mutants.2, 3 In cheater strains, the possibility that other siderophore-associated genes might retain important functions independently of siderophore synthesis or transport has not been directly investigated.
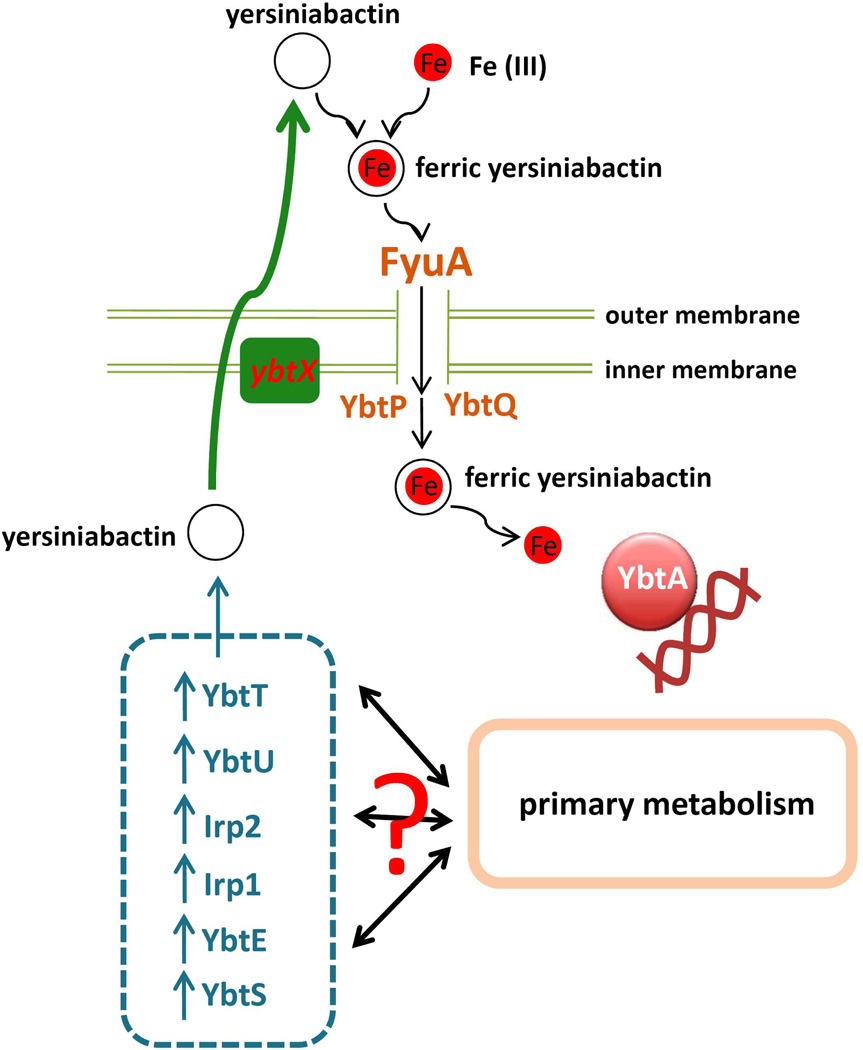
The Yersinia High Pathogenicity Island (HPI) directs synthesis, transport and regulation of the siderophore yersiniabactin in Enterobacteriaceae and biosynthetic mutants have been observed in uropathogenic (associated with urinary tract infection) E.coli isolates.4, 5 In vivo bacterial genetic studies in Yersinia species, biochemical studies of purified proteins, and in vitro recapitulation of the yersiniabactin biosynthetic pathway have provided a detailed explanation of yersiniabactin biosynthesis by nonribosomal peptide synthetase/polyketide synthase proteins and their accessory enzymes (eg. irp1, irp2, ybtS, ybtE, ybtU) (see Fig.1). 6–9 This molecular assembly line consumes chorismic acid, cysteine, malonyl-CoA, and a methyl donor to synthesize yersiniabactin, which is then secreted from the cell through an unknown route.10, 11 The thioesterase YbtT has been shown to be important for in vivo yersiniabactin production but was dispensible in an in vitro system. The outer membrane protein FyuA and the inner membrane proteins YbtP and YbtQ are subsequently associated with inward transport of extracellular ferric yersiniabactin. 12, 13 YbtX is a protein whose function has thus far defied identification, though its homology to AmpG has suggested a possible transport activity (see Fig.1). 14
Figure 1. Functions and interactions of Yersinia high pathogenicity island (HPI) genes.
Genes carried on the Yersinia HPI are involved in yersiniabactin synthesis, transport of extracellular ferric yersiniabactin complexes, and regulation of other HPI genes. In vivo interactions between these genes and the primary metabolome have been unclear.
In this study we describe a combined metabolomic-bacterial genetic approach to identify interactions between the Yersinia HPI and the E.coli primary metabolome (see Fig.1). Our metabolomic approach focuses on both amino acid and central carbon metabolism, which are fundamental to living things and are conserved between different bacterial species. Bacterial pathogens may emphasize different primary metabolic pathways during environmental persistence, colonization, and infection. These differences may reflect a balance between nutrient availability, demands to synthesize the entire biomass of the cell and maintain barrier functions, and to the need to exploit different environmental niches during infection.15–19 We therefore sought insights into Yersinia HPI function by systematically monitoring amino acids and central carbon metabolites related to glycolysis, gluconeogenesis, and the TCA cycle. Contrary to expectation, hierarchical clustering analysis and principal components analysis of metabolomic data from Yersinia HPI mutants did not yield groupings based upon yersiniabactin production. Instead, stronger and more specific metabolomic shifts were correlated with ybtU and ybtA, Yersinia HPI genes associated with regulatory functions. These metabolomic shifts were consistent with enhanced arginine biosynthesis in E.coli carrying the Yersinia HPI and suggest that previously unappreciated metabolic functions may drive carriage of these virulence-associated genes.
MATERIALS AND METHODS
Chemicals and reagents
Acetonitrile (HPLC grade), formic acid (LC/MS grade) and water (LC/MS grade) were purchased from Fisher Scientific (Fisher Scientific, Pittsburg, PA, USA); the standard compounds of NAD+, NADH, NADP, NADPH, AMP, ADP, ATP, ribulose 5-phosphate (R 5-P), xylulose 5-phosphate (X 5-P), fructose 6-phosphate (F 6-P), glyceraldehyde 3-phosphate (G 3-P), phosphoenolpyruvate (PEP), lactate, pyruvate, citrate, (iso)citrate, alpha-ketoglutarate (Alpha-KG), succinate, malate, oxaloacetate (OAA), aspartate, glutaminate, glutamine, reduced glutathione (GSH), oxidized glutathione (GSSG), (iso)leucine, leucine, alanine, arginine, cysteine, methionine, proline, serine, threonine, tyrosine, phenylalanine, valine, histidine, N-acetylcysteine, hydroxybutyric acid were purchased from Sigma (Sigma-Aldrich Corp., Saint Louis, MO, USA). All other used reagents were all ACS grade reagents.
Bacterial strains and cultivation
Bacteria were routinely cultured in LB broth. Bacterial metabolites were analyzed following growth in an established minimal medium condition.4, 20 Briefly, 4 hour bacterial cultures grown in LB broth were diluted 1:100 into M63 medium supplemented with 0.2% glycerol and 10 mg/L nicotinic acid and incubated for 18 h at 37 °C in a rotary shaker.
Deletion strain construction
Deletion mutations were made using the previously described red recombinase method with pKD4 or pKD13 as a template and the primers as listed in Table S2.4, 21–22 PCR was performed with flanking primers to confirm the appropriate deletions. Antibiotic insertions were removed by transforming the mutant strains with pCP20 23 expressing the FLP recombinase.
Ultra-Fast Liquid Chromatography-Mass Spectrometry (LC-MS)
Ultra-fast liquid chromatography (UFLC) (Shimadzu, Kyoto, Japan) consisted of two LC-20AD XR pumps, a DGU-20A3 prominence vacuum degasser, an SIL-20AXR autosampler, a CTO-20A prominence column oven, and a CBM-20A communications bus module, coupled with a hybrid API 4000 QTrap (AB Sciex, Foster City, CA, USA) with an Turbo V ESI ionization source interface, and a computer platform equipped with a Solution Analyst software version 1.5.1 (AB Sciex, Foster City, CA, USA) was used for data acquisition and pre-processing.
Targeted Metabolomics analysis of hydrophilic metabolites was performed on a Acquity HSS T3 column (150 mm × 2.0 mm, 1.7 µm, waters) using a gradient: 0–27% B over 8 min, then B increase to 99% from 8 to 10 min (A: 0.1% formic acid in water; B: 0.1% formic acid in acetonitrile) at a flow rate of 0.3 mL/min. The samples were analyzed by UFLC/MS system in positive or negative ionization modes with an electrospray ionization voltage of 5500 V for positive mode and -4500 V for negative mode, nebulizer gas (air) and turbo gas (air) settings at 50 and 50 psi, and a turbosource gun temperature of 500°C. The curtain gas (nitrogen) was set at 25 psi, the collision cell pressure at low or high mode for different purposes. The MRM parameter for each metabolite is recorded in Table S1.
According to our previous protocol 4, LC-MS determination of yersiniabactin was performed on a Betasil C18 Column (50 mm × 2.1 mm, 5.0 µm, Thermo Scientific) with a gradient as follows: 2.0–65 % B over 10 min (A: 0.1% formic acid in water; B: 0.1% formic acid in acetonitrile) at a flow rate of 0.4 mL/min. MRM parameters are listed in Table S1.
Hydrophilic metabolite extraction
Firstly, The cell pellet was isolated from 50 ml of culture solution by spinning down to 115004 ×g at 4 °C for 10 min, then mixed with 1.2 mL of 80% ice-cold methanol by vortex for 30 seconds, and kept it on the dry ice for 30 minutes. Secondly, 20 dounces of homogenization was done to the sample and spun down to 23008×g at 4 °C for 10 min, the supernatant was collected to completely mix with 800 ul of acetonitrile kept on ice for 15 minutes. Thirdly, the supernatant was isolated to lyophilized. All above procedure should be performed within safety hood. Finally, the dried sample was re-dissolved with 1 ml of distill water, and 5 µl of solution was analyzed by LC-MS.
Yersiniabactin extraction
6 µL of 0.1 M ferric chloride and 50 µL of 13C-labeled internal standard 4 were added to 1 mL of cell supernatant to a final concentration of 0.1 mM. After 15 minute room temperature incubation, the precipitate was removed by centrifugation. The supernatant was applied to a 96-well SPE plate (United Chem Inc., PA, USA) with 50 mg C18, media per well that was preconditioned with 0.5 mL of methanol and 0.5 mL of water. Each well was washed with 0.5 mL of 20% methanol, and yersiniabactin was eluted with 0.5 mL of 80% methanol. 10 µl of elute was injected into LC-MS for analysis.
Chemometric Analysis
Chemometric analyses were performed using SIMCA-P+ version 12.0.1 (Umetrics) with unsupervised principal components analysis (PCA), and supervised partial least square discriminate analysis (PLS-DA). Variables were firstly normalized to factors of volume and CFU/sample, then were scaled to a Pareto distribution to ensure equal contributions from each variable to the models. To provide an overview of the data we used FDA Genomic Tool (ArrayTrack™) 24 to plot a heatmap. Log10 transformations of normalized, median-centered data were analyzed using Ward’s Minimum Variance method (Euclidian-Ward, auto-scaled) of hierarchical clustering.
Statistical analyses
Statistics and graphs were generated using GraphPad Prism 5.0 and Microsoft Excel.
RESULTS
Creation and characterization of Yersinia High Pathogenicity Island (HPI) mutants in a model uropathogenic E.coli strain
The model uropathogen UTI89 secreted secondary metabolome contains a combination of siderophores that is typical of uropathogens, notably including yersiniabactin. To evaluate the function of Yersinia HPI genes in this uropathogenic E.coli strain, we created defined gene deletion mutants lacking ybtS, ybtX, ybtQ, ybtP, ybtA, irp1, ybtU, ybtT, ybtE, fyuA (Fig.1). Growth of these strains at 37 C in LB broth and M63/glycerol/nicotinic acid minimal medium was not markedly impacted when assessed spectrophotometrically (OD600 nm) or with a viability assay based on number of colony-forming units (CFU)(Fig. S1).
Yersiniabactin production by defined HPI mutants
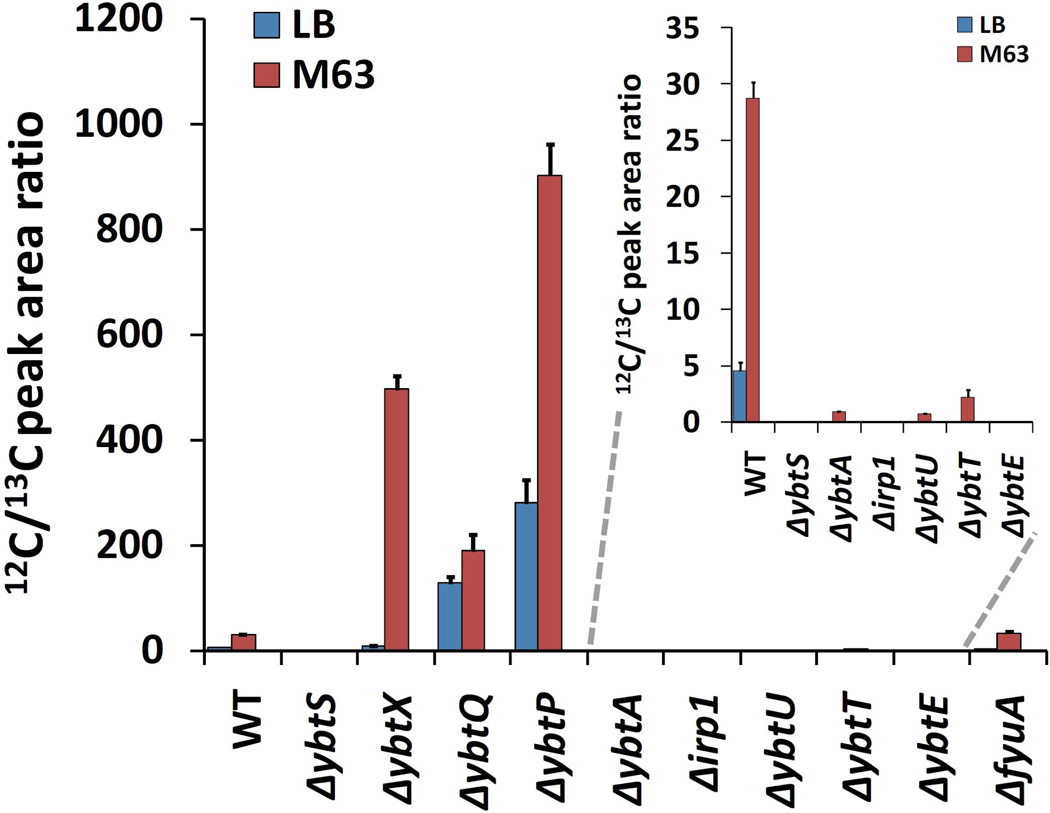
To determine how different HPI genes affect yersiniabactin production in UTI89, we quantified yersiniabactin production in 18 hour cultures of LB or a standardized minimal media using stable isotope dilution LC-MS/MS 5 (Fig.2). As previously observed, UTI89 produced yersiniabactin while UTI89ΔybtS did not. Similarly to UTI89ΔybtS, the ybtE, irp1 mutants were also yersiniabactin non-producers as expected from their direct roles in yersiniabactin biosynthesis. Detectable levels of yersiniabactin biosynthesis below that of wild type UTI89 were observed in ybtU, ybtT, and ybtA, consistent with inefficient biosynthesis, a regulatory defect, or partial complementation of the mutant by another protein 25. Yersiniabactin production was maintained or enhanced in ybtP, ybtQ, and fyuA, genes associated with ferric yersiniabactin uptake. UTI89ΔybtX also exhibited supraphysiologic yersiniabactin levels. While yersiniabactin production by ybtS, ybtA, irp1, ybtU, ybtT, and ybtE mutants mirrors those observed in Y.pestis mutants 25, supraphysiologic yersiniabactin production in ybtP, ybtQ, and ybtX mutants was unexpected and suggests altered regulation in these mutants.
Figure 2. Yersinia HPI mutants of a model urinary pathogenic E.coli strain exhibit increased or decreased yersiniabactin expression in a standard minimal media condition.
Strains were cultured in M63 minimal medium+glycerol for 18 hours at 37 C and yersiniabactin levels in the media were determined by stable isotope dilution quantification using 13C-labeled internal standard. Mutants strains lacking genes for transport (fyuA, ybtP, ybtQ) or a gene of unknown function (ybtX) exhibited preserved or elevated yersiniabactin levels. Results are expressed as LC-MS/MS peak area of unlabeled (12C) analyte to internal standard (13C).
Creation of a targeted metabolomic profiling approach based on primary metabolism
To identify interactions between Yersinia HPI genes and the primary metabolome, we developed a quantitative LC-MS/MS-based profiling approach targeting intracellular E.coli metabolites. The resulting liquid chromatography-multiple reaction-monitoring (MRM) analysis allows us to readily detect forty hydrophilic primary metabolites representing multiple central carbon metabolism pathways as well as amino acid metabolism (table S1, and Fig.S2), with excellent reproducibility and stability (Tables S3-5). Use of MRM in the triple quadrupole mode was employed to maximize sensitivity and reproducibility while taking advantage of this instrument’s broad linear response range.
Yersiniabactin production is not a strong determinant of primary metabolomic changes in HPI mutants
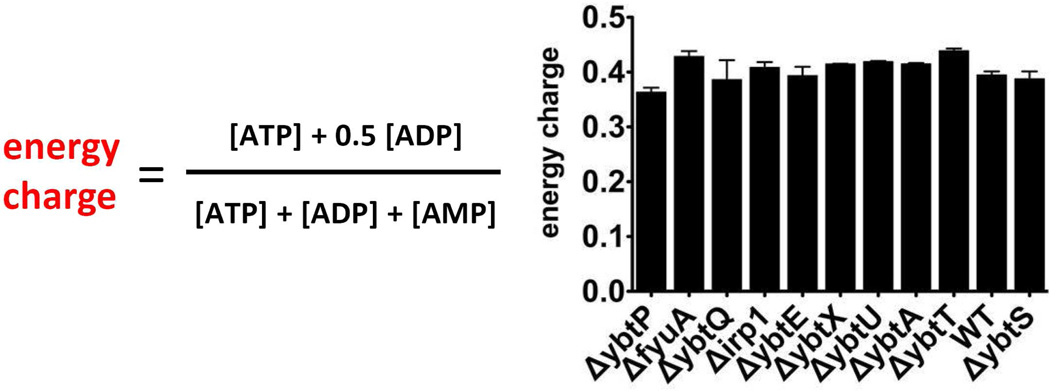
We first assessed relative cellular energy status by determining an energy charge parameter (Fig.3).26 Consistent with its role as a key homeostatic set point, energy charge was not markedly altered in any of the strains. This is consistent with the similar growth density for these cells and indicates that these cells were in a grossly similar physiological state at the time of metabolomic sampling.
Figure 3. Energy charge parameter for UTI89 and its Yersinia HPI mutants.
Strains exhibited similar energy charge values at the time of metabolomic profiling.
Primary metabolomic differences were evident between bacterial strains. To determine whether these differences are attributable to differences in yersiniabactin production, we categorized strains as yersiniabactin non-producers (ybtS, irp1 and ybtE), low-level producers (ybtU, ybtA, and ybtT), and high producers (wild type, fyuA, ybtX, ybtQ and ybtP). Although the supervised PLS-DA analysis reveals good separation of low-level producers from high-level producers, non-producers was not clearly separated from either of the other two groups (Fig.S3). Surprisingly, this analysis was more effective at differentiating strains with high and low production than it was at differentiating producers from non-producers. The very important variable plot derived from PLS-DA analysis validates twelve metabolites (cut off VIP values > 1) that account for groupwise separation. Of these, only phosphoenolpyruvate and phenylalanine exhibited a linear correlation with extracellular yersiniabactin levels. The reciprocal relationship between yersiniabactin production and phenylalanine levels may result by chance or could reflect diversion of aromatic biosynthetic precursors to salicylic acid production. Regardless, absence of a clear proportional relationship between yersiniabactin production and primary metabolites suggests unanticipated interactions between the Yersinia HPI and primary metabolism not explained by yersiniabactin production alone.
Unsupervised primary metabolomic clustering analyses of strains
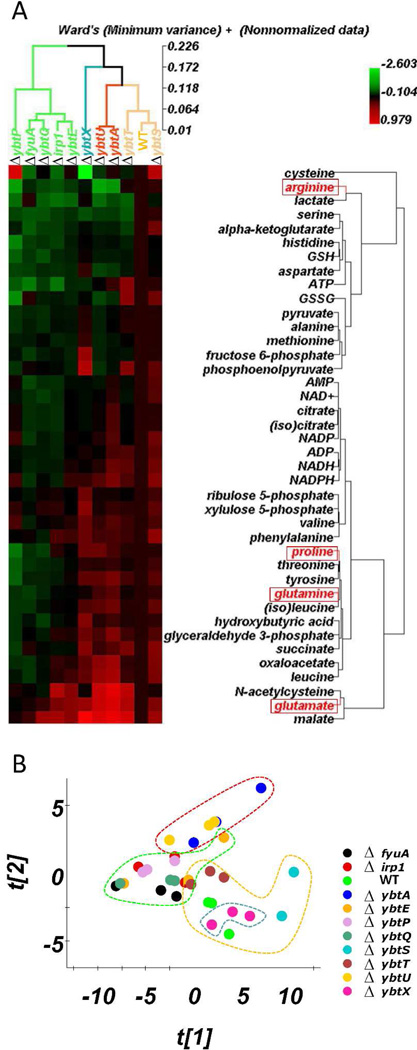
Rather than seek supervised metabolomic associations based on yersiniabactin production, we instead employed hierarchical clustering analysis (HCA) and principal components analysis (PCA) to identify metabolomic relatedness within the mutant strain panel (Figs 4A-B). A heat map was constructed from the metabolomic dataset in which HCA was used to organize the strain and metabolite axes (Fig.4A). As suggested by the supervised analysis of yersiniabactin producers and non-producers above, the HCA dendrogram did not cluster strains based on yersiniabactin production. Instead, biosynthetic genes were distributed between two main branches with no clear division between early and late biosynthetic genes. It is notably that wild type UTI89 was clustered closely with its yersiniabactin-null ybtS mutant. In contrast, genes associated with ferric yersiniabactin transport genes (fyuA, ybtP, ybtQ) clustered together into a single main branch, as expected a priori for genes with related transport functions.
Figure 4. Genome-metabolome correlations of defined Yersinia HPI mutants.
(A) A heat map was generated and arranged by hierarchical clustering analysis (HCA) in which mutants and metabolites are grouped based on metabolomic similarity. Dendrograms for both mutants and metabolites are depicted (B) Principal components analysis (PCA) of the same metabolomic profiles reveals distinct clusters of the ybtU and ybtA mutants from UTI89 and its ybtS and ybtT mutants as identified by HCA.
The mutant with the lowest similarity to any other strain in the panel is UTI89ΔybtX. ybtX is predicted to encode a hydrophobic ~45 kD protein with membrane-spanning domains but its functional significance has been otherwise unclear as yersiniabactin production is maintained in the mutant and deletion mutants in other strains exhibit preserved ferric yersiniabactin utilization. 12 In this study, UTI89ΔybtX exhibits supraphysiologic yersiniabactin production along with an unexplained increase in phophoenolpyruvate and decrease in cysteine. Overall, metabolomic profiling continues to support a distinctive function for ybtX.
Interestingly, clustering analysis identified the AraC-type transcriptional regulator ybtA and the reductase ybtU as the most closely-related strains in this analysis. Although these genes encode proteins associated with different known biochemical activities, they exert similar effects upon the primary metabolome. A basis for this grouping is suggested by studies in Y.pestis that demonstrate similar regulatory effects of ybtU and ybtA such that both are necessary for expression of HMWP1, HMWP2, YbtE, and FyuA.6 Severely diminished yersiniabactin production in this study (Fig. 2) supports the same regulatory interaction in UTI89. Maintenance of this regulatory function in wild type Y.pestis and its ybtS and ybtT mutants provides a rationale for separate clustering of UTI89/UTI89ΔybtS/UTI89ΔybtT from UTI89ΔybtU/UTI89ΔybtA in both HCA and PCA analyses (Fig.4). Although the precise interactions underlying this putative regulatory pathway remain unclear, the transcription factor YbtA would be expected to play a central role, perhaps by interacting with promoter sites outside the HPI. In Y.pestis, a non-yersiniabactin signaling molecule(s) that modulates YbtA activity has been proposed and could explain the YbtU requirement. 27 Together, these results are consistent with a Yersinia HPI-directed, yersiniabactin-independent, regulatory interaction with the primary metabolome involving YbtA and YbtU [Fig.5–6].
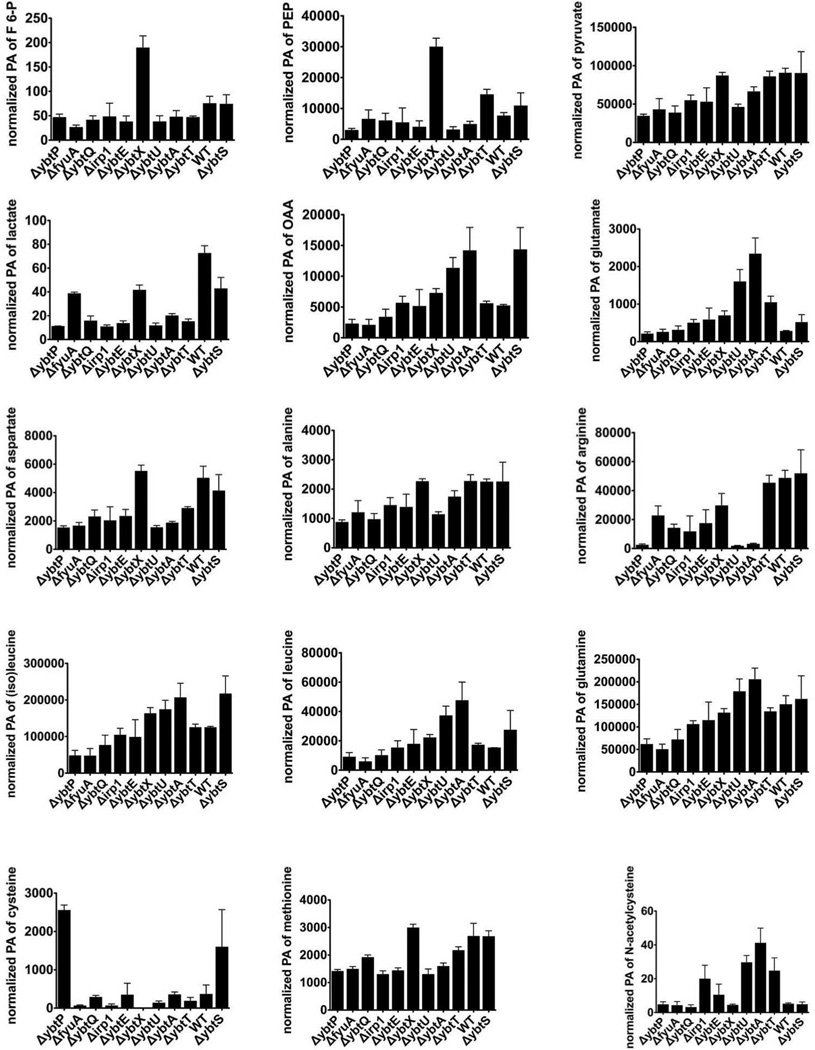
Figure 5. Individual metabolite levels for metabolites selected by VIP analysis (VIP values > 1) and cysteine.
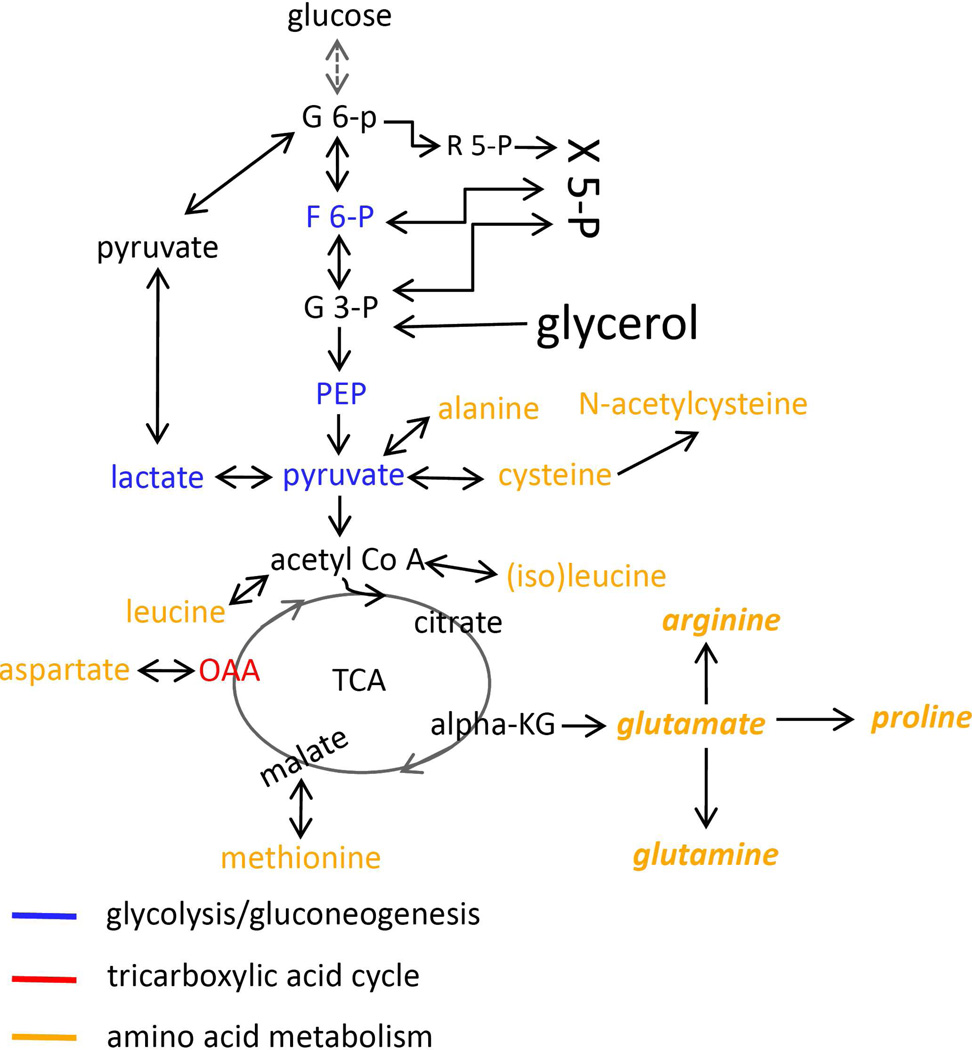
Figure 6. Primary metabolic pathways affected by Yersinia HPI deletion mutations.
Levels of metabolites listed in yellow, blue or red were affected by Yersinia HPI mutants. Amino acid metabolism, glycolysis/ gluconeogenesis cycle and TCA cycle are mostly affected by those HPI mutants.
The transcription factor gene ybtA and the reductase gene ybtU specifically affect arginine levels
To understand how this ybtU/ybtA-associated regulatory pathway affects central metabolism, we compared heat map and univariate analysis data (Fig.4A) between the UTI89ΔybtU/UTI89ΔybtA and UTI89ΔybtT/wild type/UTI89ΔybtS groups. Clear groupwise differences were evident in the levels of arginine, glutamate, malate, and N-acetyl-cysteine. These metabolites account for some of the most striking differences in this the strain panel. Among these, arginine was markedly deficient in the ybtU/ybtA mutants, with a marked decline also evident in the ybtP mutant (Fig.4A, 7C).
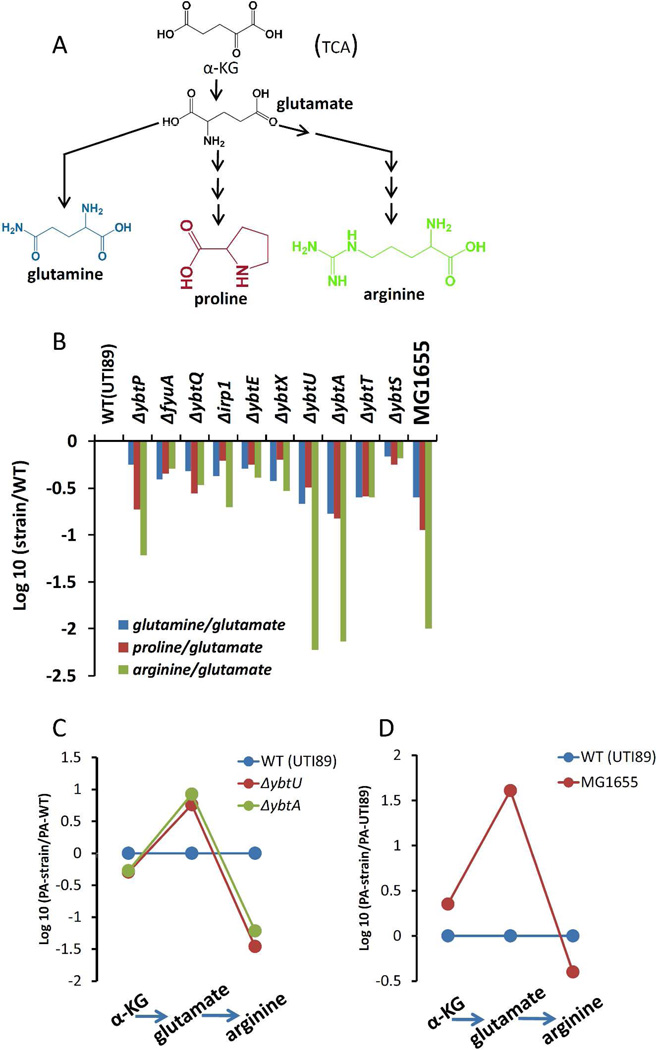
Figure 7. ybtU and ybtA mutants exhibit metabolite levels consistent with selectively diminished arginine biosynthesis.
(A) The glutamate family of amino acids glutamine, proline, and arginine are synthesized from glutamate. (B) Among glutamate family amino acids, the arginine/glutamate ratio (product/precursor ratio expressed on a log scale) is reduced by over 100-fold in ybtA and ybtU mutants relative to other HPI mutants and to a similar degree in K12 E.coli strain MG1655, which lacks Yersinia HPI genes. Glutamine and proline are much less markedly affected. (C) Relative (compared to wild type UTI89) metabolite levels along the arginine biosynthesis pathways of UTI89ΔybtU and UT89ΔybtA. Accumulation of glutamate (substrate) levels together with diminished arginine (product) levels are consistent with a relative metabolic block in arginine biosynthesis. Relatively stable levels of alpha-ketoglutarate (α-KG), from which glutamate is synthesized, likely reflect its carefully regulated steady state levels as a TCA cycle intermediate. (D) Arginine biosynthesis pathway metabolites in MG1655 exhibit shifts in glutamate and arginine levels comparable to those observed in UTI89ΔybtU and UTI89ΔybtA.
To determine whether the striking ybtU/ybtA-associated decrease in arginine derives from decreased arginine biosynthesis or increased arginine consumption, we followed metabolites in the arginine biosynthetic pathway and monitored related amino acids pathways as control groups (Fig.7A). Together with glutamine and proline, arginine is part of a biosynthetic family of amino acids synthesized from glutamate. To determine if limited substrate availability explains the decrease in arginine levels, we examined glutamate levels. Instead, glutamate levels were increased in ybtU/ybtA mutants (Fig.7C). Relative to other glutamine and proline, arginine levels were the most markedly suppressed in the mutants, consistent with a selective decrease in arginine biosynthesis (Fig.7B). In the product/precursor relationship, the arginine/glutamate peak area ratio in wild type UTI89 is over 150-fold greater than in the ybtU and ybtA mutants (176.1 vs. 1.1 and 1.3). Decreases in arginine levels together with glutamate precursor accumulation is consistent with a relative arginine biosynthetic block in ybtU/ybtA mutants. Among the explanations for this block is transcriptional downregulation of arginine biosynthetic genes or a relative deficiency of other arginine biosynthetic substrates.
To determine the arginine biosynthetic metabolite profile in a strain lacking not only ybtU/ybtA but also the entire Yersinia HPI, we compared the metabolomic profile of wild type UTI89 to MG1655, a K12 E.coli strain. Relative to wild type UTI89, MG1655 exhibited a depressed arginine/glutamate ratio similar to that observed in the UTI89 ybtU/ybtA mutants (Figs 7B and D). While other genomic differences between MG1655 and UTI89 may contribute, diminished arginine levels and MG1655-like arginine biosynthetic profile in ybtU and ybtA mutants suggests that these genes play important roles in altering primary E.coli metabolism beyond what can be directly explained by yersiniabactin biosynthesis alone.
DISCUSSION
Our results show that yersiniabactin-associated genes can alter the primary metabolome independently of yersiniabactin production. While consumption of metabolic substrates for yersiniabactin must constitute a metabolic burden, its effect upon the primary metabolome were subtle in this study. Instead, clustering analysis identified consistent changes in the arginine biosynthesis pathway associated with ybtU and ybtA. These findings provide evidence that virulence-associated siderophore genes can alter primary metabolism in unexpected ways. Rather than representing a total loss of function, bacterial “cheater“ strains may retain virulence-associated primary metabolic functions not directly associated with siderophore production.
Indications that arginine may be a particularly consequential amino acid in urinary pathogens is suggested in previous genetic studies. A transposon mutagenesis study previously identified the arginine biosynthetic gene argC as being upregulated during urinary growth and important for virulence in a mouse model. 28 Another arginine biosynthesis gene, argL, emerged from a genome sequencing study as exhibiting evidence of positive selection among uropathogenic E.coli isolates including UTI89.29 Consistent with these findings is the increased expression of the arginine pathway enzyme ArgG observed by cytoplasmic proteome profiling of a uropathogen grown ex vivo in human urine.30 Why, among amino acid pathways, arginine exhibits pathogenic associations remains unclear.
Metabolomic grouping of ybtU and ybtA mutants is not evident from functional annotation of these genes, although this grouping parallels their similar effects on Yersinia HPI protein expression. It has been proposed that metabolites produced through YbtU-mediated reduction act as allosteric activators of the AraC-type transcription factor YbtA. This general relationship could explain the similar effect on arginine biosynthesis observed in both mutants. That arginine levels are not similarly impacted in the ybtS mutant suggests that this putative signaling molecule(s) does not require salicylate for its biosynthesis. Future studies will explore more directly the interactions between arginine biosynthesis and ybtU/ybtA.
Together, this study demonstrates the usefulness of combining metabolomics with bacterial genetic approaches to better understand gene function. The explosion of genetic information from metagenomic studies is identifying large numbers of gene candidates with unknown, or partially known, functions. Metabolomic analyses provide a means to compare genetic interactions or functional similarities by monitoring conserved metabolites present in most organisms. In this case, hierarchical clustering analysis of the results suggested functional groupings that were not immediately apparent but could be subsequently confirmed by PCA. A similar approach might be taken to characterize larger numbers of genes and to explore their function in E.coli or other host strains.
Supplementary Material
ACKNOWLEDGEMENTS
Funding was provided by National Institutes of Health grants NIH grants HD001459-09, UL1 RR024992, HL101263-01, DK064540-09, and DK082315. JPH was supported by a Career Award for Medical Scientists from the Burroughs Wellcome Fund. We also thank Chia Hung, Jennifer Walker, Tom Hannan, and Jan Crowley for technical assistance and Scott Hultgren for helpful discussions.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org
REFERENCES
- 1.Neilands JB. Siderophores: structure and function of microbial iron transport compounds. J Biol Chem. 1995;270(45):26723–26726. doi: 10.1074/jbc.270.45.26723. [DOI] [PubMed] [Google Scholar]
- 2.Griffin AS, West SA, Buckling A. Cooperation and competition in pathogenic bacteria. Nature. 2004;430(7003):1024–1027. doi: 10.1038/nature02744. [DOI] [PubMed] [Google Scholar]
- 3.Gehring AM, DeMoll E, Fetherston JD, Mori I, Mayhew GF, Blattner FR, Walsh CT, Perry RD. Iron acquisition in plague: modular logic in enzymatic biogenesis of yersiniabactin by Yersinia pestis. Chem Biol. 1998;5(10):573–586. doi: 10.1016/s1074-5521(98)90115-6. [DOI] [PubMed] [Google Scholar]
- 4.Henderson JP, Crowley JR, Pinkner JS, Walker JN, Tsukayama P, Stamm WE, Hooton TM, Hultgren SJ. Quantitative metabolomics reveals an epigenetic blueprint for iron acquisition in uropathogenic Escherichia coli. PLoS Pathog. 2009;5(2):e1000305. doi: 10.1371/journal.ppat.1000305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carniel E. The Yersinia high-pathogenicity island. Int Microbiol. 1999;2(3):161–167. [PubMed] [Google Scholar]
- 6.Geoffroy VA, Fetherston JD, Perry RD. Yersinia pestis YbtU and YbtT are involved in synthesis of the siderophore yersiniabactin but have different effects on regulation. Infect Immun. 2000;68(8):4452–4461. doi: 10.1128/iai.68.8.4452-4461.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gehring AM, Mori II, Perry RD, Walsh CT. The nonribosomal peptide synthetase HMWP2 forms a thiazoline ring during biogenesis of yersiniabactin, an iron-chelating virulence factor of yersinia pestis. Biochemistry. 1998;37(48):17104. doi: 10.1021/bi9850524. [DOI] [PubMed] [Google Scholar]
- 8.Guilvout I, Mercereau-Puijalon O, Bonnefoy S, Pugsley AP, Carniel E. Highmolecular- weight protein 2 of Yersinia enterocolitica is homologous to AngR of Vibrio anguillarum and belongs to a family of proteins involved in nonribosomal peptide synthesis. J Bacteriol. 1993;175(17):5488–5504. doi: 10.1128/jb.175.17.5488-5504.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bearden SW, Fetherston JD, Perry RD. Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect Immun. 1997;65(5):1659–1668. doi: 10.1128/iai.65.5.1659-1668.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crosa JH, Walsh CT. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol Mol Biol Rev. 2002;66(2):223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller DA, Luo L, Hillson N, Keating TA, Walsh CT. Yersiniabactin synthetase: a four-protein assembly line producing the nonribosomal peptide/polyketide hybrid siderophore of Yersinia pestis. Chem Biol. 2002;9(3):333–344. doi: 10.1016/s1074-5521(02)00115-1. [DOI] [PubMed] [Google Scholar]
- 12.Fetherston JD, Bertolino VJ, Perry RD. YbtP and YbtQ: two ABC transporters required for iron uptake in Yersinia pestis. Mol Microbiol. 1999;32(2):289–299. doi: 10.1046/j.1365-2958.1999.01348.x. [DOI] [PubMed] [Google Scholar]
- 13.Brem D, Pelludat C, Rakin A, Jacobi CA, Heesemann J. Functional analysis of yersiniabactin transport genes of Yersinia enterocolitica. Microbiology. 2001;147(Pt 5):1115–1127. doi: 10.1099/00221287-147-5-1115. [DOI] [PubMed] [Google Scholar]
- 14.Lindquist S, Weston-Hafer K, Schmidt H, Pul C, Korfmann G, Erickson J, Sanders C, Martin HH, Normark S. AmpG, a signal transducer in chromosomal betalactamase induction. Mol Microbiol. 1993;9(4):703–715. doi: 10.1111/j.1365-2958.1993.tb01731.x. [DOI] [PubMed] [Google Scholar]
- 15.Zhang N, Gur A, Gibon Y, Sulpice R, Flint-Garcia S, McMullen MD, Stitt M, Buckler ES. Genetic analysis of central carbon metabolism unveils an amino acid substitution that alters maize NAD-dependent isocitrate dehydrogenase activity. PLoS One. 2010;5(4):e9991. doi: 10.1371/journal.pone.0009991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noor E, Eden E, Milo R, Alon U. Central carbon metabolism as a minimal biochemical walk between precursors for biomass and energy. Mol Cell. 2010;39(5):809–820. doi: 10.1016/j.molcel.2010.08.031. [DOI] [PubMed] [Google Scholar]
- 17.de Carvalho LP, Fischer SM, Marrero J, Nathan C, Ehrt S, Rhee KY. Metabolomics of Mycobacterium tuberculosis reveals compartmentalized co-catabolism of carbon substrates. Chem Biol. 2010;17(10):1122–1131. doi: 10.1016/j.chembiol.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 18.Marrero J, Rhee KY, Schnappinger D, Pethe K, Ehrt S. Gluconeogenic carbon flow of tricarboxylic acid cycle intermediates is critical for Mycobacterium tuberculosis to establish and maintain infection. Proc Natl Acad Sci U S A. 2010;107(21):9819–9824. doi: 10.1073/pnas.1000715107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amador-Noguez D, Feng XJ, Fan J, Roquet N, Rabitz H, Rabinowitz JD. Systems-level metabolic flux profiling elucidates a complete, bifurcated tricarboxylic acid cycle in Clostridium acetobutylicum. J Bacteriol. 2010;192(17):4452–4461. doi: 10.1128/JB.00490-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valdebenito M, Bister B, Reissbrodt R, Hantke K, Winkelmann G. The detection of salmochelin and yersiniabactin in uropathogenic Escherichia coli strains by a novel hydrolysis-fluorescence-detection (HFD) method. Int J Med Microbiol. 2005;295(2):99–107. doi: 10.1016/j.ijmm.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 21.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy KC, Campellone KG. Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol Biol. 2003;4:11. doi: 10.1186/1471-2199-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158(1):9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 24.Tong W, Cao X, Harris S, Sun H, Fang H, Fuscoe J, Harris A, Hong H, Xie Q, Perkins R, Shi L, Casciano D, ArrayTrack--supporting toxicogenomic research at the US. Food and Drug Administration National Center for Toxicological Research. Environ Health Perspect. 2003;111(15):1819–1826. doi: 10.1289/ehp.6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perry RD, Fetherston JD. Yersiniabactin iron uptake: mechanisms and role in Yersinia pestis pathogenesis. Microbes Infect. 2011;13(10):808–817. doi: 10.1016/j.micinf.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersen KB, von Meyenburg K. Charges of nicotinamide adenine nucleotides and adenylate energy charge as regulatory parameters of the metabolism in Escherichia coli. J Biol Chem. 1977;252(12):4151–4156. [PubMed] [Google Scholar]
- 27.Miller MC, Fetherston JD, Pickett CL, Bobrov AG, Weaver RH, DeMoll E, Perry RD. Reduced synthesis of the Ybt siderophore or production of aberrant Ybt-like molecules activates transcription of yersiniabactin genes in Yersinia pestis. Microbiology. 2010;156(Pt 7):2226–2238. doi: 10.1099/mic.0.037945-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russo TA, Carlino UB, Mong A, Jodush ST. Identification of genes in an extraintestinal isolate of Escherichia coli with increased expression after exposure to human urine. Infect Immun. 1999;67(10):5306–5314. doi: 10.1128/iai.67.10.5306-5314.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen SL, Hung CS, Xu J, Reigstad CS, Magrini V, Sabo A, Blasiar D, Bieri T, Meyer RR, Ozersky P, Armstrong JR, Fulton RS, Latreille JP, Spieth J, Hooton TM, Mardis ER, Hultgren SJ, Gordon JI. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc Natl Acad Sci U S A. 2006;103(15):5977–5982. doi: 10.1073/pnas.0600938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alteri CJ, Smith SN, Mobley HL. Fitness of Escherichia coli during urinary tract infection requires gluconeogenesis and the TCA cycle. PLoS Pathog. 2009;5(5):e1000448. doi: 10.1371/journal.ppat.1000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.