Abstract
While T cell memory is generally thought to require direct antigen exposure, we find an abundance of memory phenotype cells (20–90%, averaging over 50%) of CD4+ T cells specific for viral antigens in adults that have never been infected. These cells express the appropriate memory markers and genes, rapidly produce cytokines, and have clonally expanded. This contrasts with newborns where the same T cell receptor (TCR) specificities are almost entirely naïve, which may explain the vulnerability of young children to infections. One mechanism for this phenomenon is TCR cross-reactivity to environmental antigens and in support of this we find extensive cross-recognition by HIV-1 and influenza-reactive T lymphocytes to other microbial peptides and the expansion of one of these following influenza vaccination. Thus the presence of these memory phenotype T cells has significant implications for immunity to novel pathogens, child and adult health, and the influence of pathogen-rich versus hygienic environments.
Introduction
It is well known that memory T and B cells are critical for the most rapid and efficacious immune responses (Jameson and Masopust, 2009). This contrasts with naïve T cells, which can take a few days to over a week to mount a response (Flynn et al., 1998). Thus a principal goal of vaccine development is to trigger memory T cells, along with B cells and long-lived plasma cells specific for particular pathogens. While it has been thought that direct antigenic exposure is required for the formation of memory cells, recent work on CD8+ T cell precursors in mice has found that memory phenotype cells can also develop without specific exposure to their cognate antigen (Akue et al., 2012; Decman et al., 2012; Haluszczak et al., 2009; Rudd et al., 2011). These cells are thought to have developed their memory phenotype through homeostatic signals mediated via self peptide-major histocompatibility complex (MHC) interactions. Potentially, other mechanisms may also be involved, such as T cell activation through T cell receptor (TCR) cross-recognition of alternate ligand(s).
Recently, enrichment techniques combined with peptide-MHC (pMHC) tetramer staining have allowed the direct analysis of the T cell repertoire to an unprecedented degree, including cells that represent the preimmune repertoire (Moon et al., 2007). This has resulted in a wealth of information about the frequency of pre-immune T cells in mice and growing evidence that the T cell response is directly proportional to the antigen-specific naïve T cell pool (Kwok et al., 2012; Moon et al., 2007; Obar et al., 2008). However, far less is known about the human T cell repertoire at baseline, particularly pertaining to CD4+ T cells. Thus, we set out to comprehensively characterize the adult human CD4+ T cell repertoire using HLA-DR4 restricted epitopes and pMHC tetramer enrichment to examine the frequency and phenotype of precursor T cells recognizing self-antigens or microbial epitopes in exposed or unexposed individuals.
We find that for almost all the unexposed specificities surveyed, our pMHC tetramers detect frequencies in a fairly narrow range between 1 to 10 cells per million CD4+ T cells in 26 adult blood bank donors aged 28–80+. Surprisingly, T cells staining for tetramers derived from HIV-1, cytomegalovirus (CMV), and herpes simplex virus (HSV) epitopes often had a very high proportion of memory phenotype cells, up to 93%, (and on average over 50%) in individuals that had never been infected with these viruses. These cells not only had memory surface markers, they also expressed memory-associated genes, exhibited rapid cytokine production, and showed evidence of clonal expansion. Thus they have many of the expected characteristics of memory T cells and could offer survival advantage in the event of a cognate infection. In this context it seems particularly notable that at least some of these specificities are present in the umbilical blood cells of newborns but virtually all are of the naïve phenotype, suggesting that this might partially explain the vulnerability of young children to infectious diseases.
With respect to how these memory phenotype T cells are acquired, one possibility is homeostatic proliferation, where proliferating lymphocytes can acquire the characteristics of memory (Sprent and Surh, 2011). Another possibility is cross-reactivity with the many antigens in the environment, especially given the myriad organisms that humans and other species can be exposed to. In this context, it is well known that αβ T cell receptors have a strong propensity to be cross-reactive to different pMHC’s, likely due to their flexible binding sites (Newell et al., 2011; Reinherz et al., 1999; Reiser et al., 2003). Consistent with this possibility, these memory phenotype cells exhibited extensive cross-reactivity to homologous peptides derived from other microbial genomes. Furthermore, we used a seasonal influenza vaccine to directly show that immunization with 2009 H1N1 Influenza strain can stimulate T cells specific for a cross-reactive bacterial homolog. Thus cross-reactivity to environmental antigens could be a major mechanism in eliciting populations of memory T cells for infectious diseases that an individual has not yet encountered.
Results
Peptide-class II MHC tetramer staining and enrichment
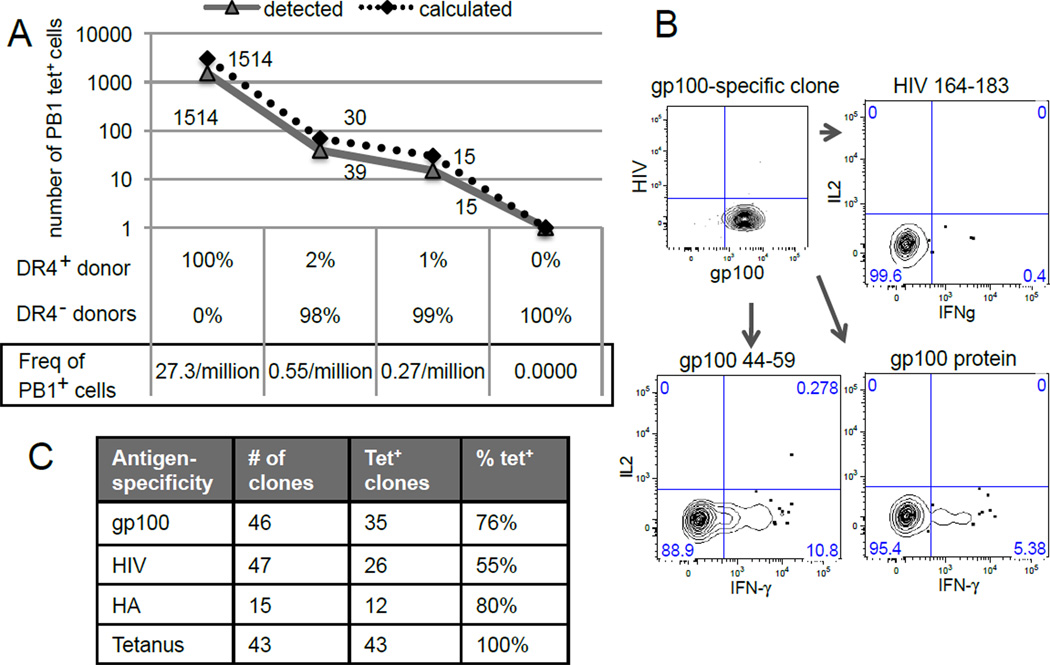
How well class II pMHC tetramers can identify rare unprimed T cells in humans has been unclear and therefore we first explored the technical limits of using this approach to analyze the naïve human T cell repertoire. HLA-DR4 (DRA, DRB1*0401) tetramers against a conserved epitope from the PB1 protein of the influenza virus were generated and used to measure the recovery of a known number of PB1-specific lymphocytes spiked into HLA-DR4− background cells (Day et al., 2003). Using this approach, we found more than 70% concordance between the numbers of cells captured experimentally versus what was expected (Fig. 1A). These tetramer labeled cells were clonally restricted to Vβ2 expression and could be detected in diluted samples containing as few as 3 per 10 million PB1-specific T cells (Fig. S1 and 1A).
Figure 1. Peptide-MHCII tetramer-based enrichment is highly sensitive and specific.
(A) HLA-DR4+ CD4+ lymphocytes were used as is or spiked into HLA-DR4− lymphocytes to achieve 50- and 100-fold dilutions in a total of 55 million CD4+ T cells. Undiluted HLA-DR4− lymphocytes were used as negative control. PB1 tetramer staining and enrichment were performed in parallel. Data are representative of one individual from 2 independent experiments. (B) A gp100-specific clone from single cell expansion that re-stained with gp100 tetramers but not with HIV-1 tetramers. IFN-γ is produced in response to stimulation by gp100 peptide, gp100 protein, but not to a control HIV-1 peptide. Data is representative of two gp100-specific clones. (C) Table showing T cell clones generated by expansion of single cells labeled with gp100, HIV-1, HA, or tetanus tetramers. The percentage of cells that retained staining with the same tetramer (% tet+) was calculated by dividing the number of tetramer positive clones by the total numbers of clones generated for each specificity (See also Fig. S1).
To validate the specificity of this method with another tetramer, we also stained peripheral blood mononuclear cells (PBMCs) from an HLA-DR4+ blood donor with a peptide derived from the self-protein, gp100. Tetramer positive clones were generated, among which 76% (35/46) retained staining with gp100 tetramers. These clones responded to the gp100 peptide and recombinant gp100 protein by interferon-γ (IFN-γ) production, but not to a control HIV-1 peptide (Fig. 1B). Clones recognizing additional specificities from HIV-1, tetanus toxin, and influenza virus were also generated and 55–100% of those clones that grew retained staining with the same tetramer used initially (Fig. 1C). Taken together, these data indicate that tetramer binding is specific and can identify antigen-specific T cells with a sensitivity of one cell in several million.
The unprimed CD4+ T cell repertoire contains pre-existing memory phenotype T cells
To broadly survey antigen-specific CD4+ T cells, we selected self-peptides from gp100, fibrinogen (Fib), or preproinsulin (PPins) for their relevance in anti-tumor responses and autoimmunity (Nielen et al., 2005; Yuan et al., 2009; Zhang et al., 2008). Foreign antigens from gag p24 from HIV-1, pp65 from cytomegalovirus (CMV) and the VP16 protein of herpes simplex virus (HSV) were selected because highly sensitive serologic tests are available to clearly distinguish exposed versus unexposed individuals. The sensitivity of the HSV, HIV, and CMV serologic tests are 97%–100%, and with respect to HIV, the risk of a false negative is estimated to be less than 1 in 2.6 million blood donations nationwide (Lackritz et al., 1995). We also examined several epitopes from the influenza virus and tetanus toxin, to which most people have been exposed. Specific epitopes from each protein were chosen from the literature based on T cell stimulation and evidence that they are the product of natural processing and presentation (Alexander et al., 2010; Arif et al., 2004; Auger et al., 2005; Bronke et al., 2005; Diethelm-Okita et al., 1997; Lamb et al., 1982; Norris et al., 2004; Novak et al., 2001; Phan et al., 2003).
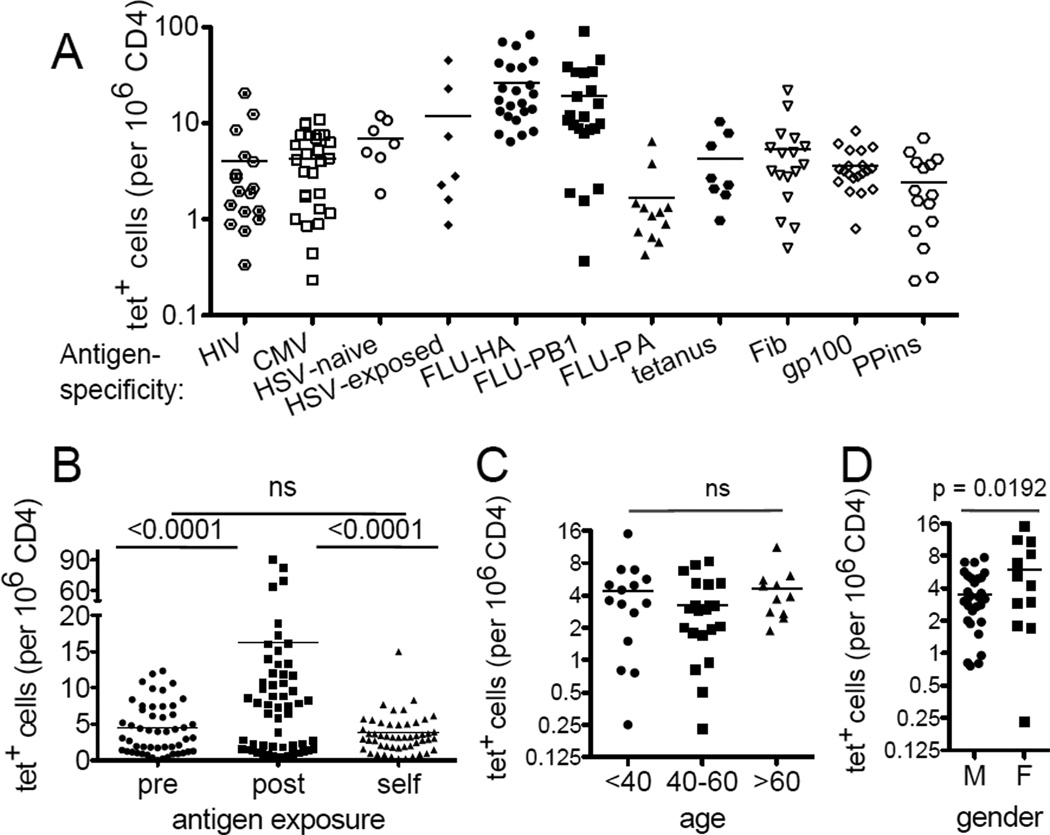
Tetramers were made using peptide exchange to examine antigen-specific CD4+ T cells from 26 HLA-DR4+ healthy blood donors (Fig. S2). We found that the frequency of precursor cells recognizing a self-antigen or an unexposed viral epitope generally ranged from 1 to 10 cells per million CD4+ T cells (Fig. 2A). Notably, we found that previous exposure did not necessarily result in a detectable expansion of cells specific for that antigen, as shown by the similarity in the frequencies of HSV-specific T cells between seropositive individuals and those who are HSV negative. These data, as well as large inter-individual differences in T cells recognizing the two dominant flu epitopes, HA and PB1, highlight the heterogeneity of a T cell response in humans. Another notable feature in these data is the absence of a statistically significant difference between the frequencies of cells that recognize self antigens, PPins, Fib, or gp100 and those that bind to a naïve foreign antigen from HIV-1, CMV, or HSV in uninfected individuals (Fig. 2B). These self-antigen specific T cells were examined for potential age- or gender-dependent change, particularly because the risk of autoimmunity is generally higher in the elderly and in females. While we did not detect significantly more gp100-, Fib-, or PPins-specific T cells in older individuals (Fig. 2C), we did note a gender-specific difference, with 1.7-fold more gp100-, PPins-, and Fib-specific lymphocytes in females than males (Fig. 2D).
Figure 2. Frequency analysis of the human antigen-specific CD4+ lymphocytes.
All blood samples were obtained from individuals seronegative for HIV and CMV exposures. HSV exposure is as indicated (naïve vs exposed). (A) Frequencies of tetramer tagged cells per million CD4+ T cells. Each symbol represents an antigen-specific population from one individual and the bar indicates the mean of experiments performed independently using blood obtained at different times. (B) Comparison between T cells recognizing a novel foreign antigen (pre: HIV-1, CMV, HSV-naïve), exposed foreign antigen (post: Flu, tetanus, HSV-exposed), and self antigen (self: Fib, gp100, PPins). Figure summarizes data from all 26 individuals. (C) Precursor frequencies of self-specific lymphocytes that recognized gp100, Fib, or PPins in people ages <40 (n= 7), 40–60 (n = 13), >60 (n = 6). (D) Precursor frequency of self-specific lymphocytes that recognized gp100, Fib, or PPins in males (n = 17) and females (n = 9) (See also Fig. S2).
The absence of an obvious frequency difference between self and foreign-antigen specific populations suggest that negative selection is incomplete for the particular self-specificities examined. Alternatively, post-thymic antigen stimulation may have resulted in expansion of certain populations that masked a difference in their initial frequencies. To assess the latter, we analyzed the memory vs naïve phenotype of these tetramer positive cells using the classical memory marker, CD45RO (Appay et al., 2008).
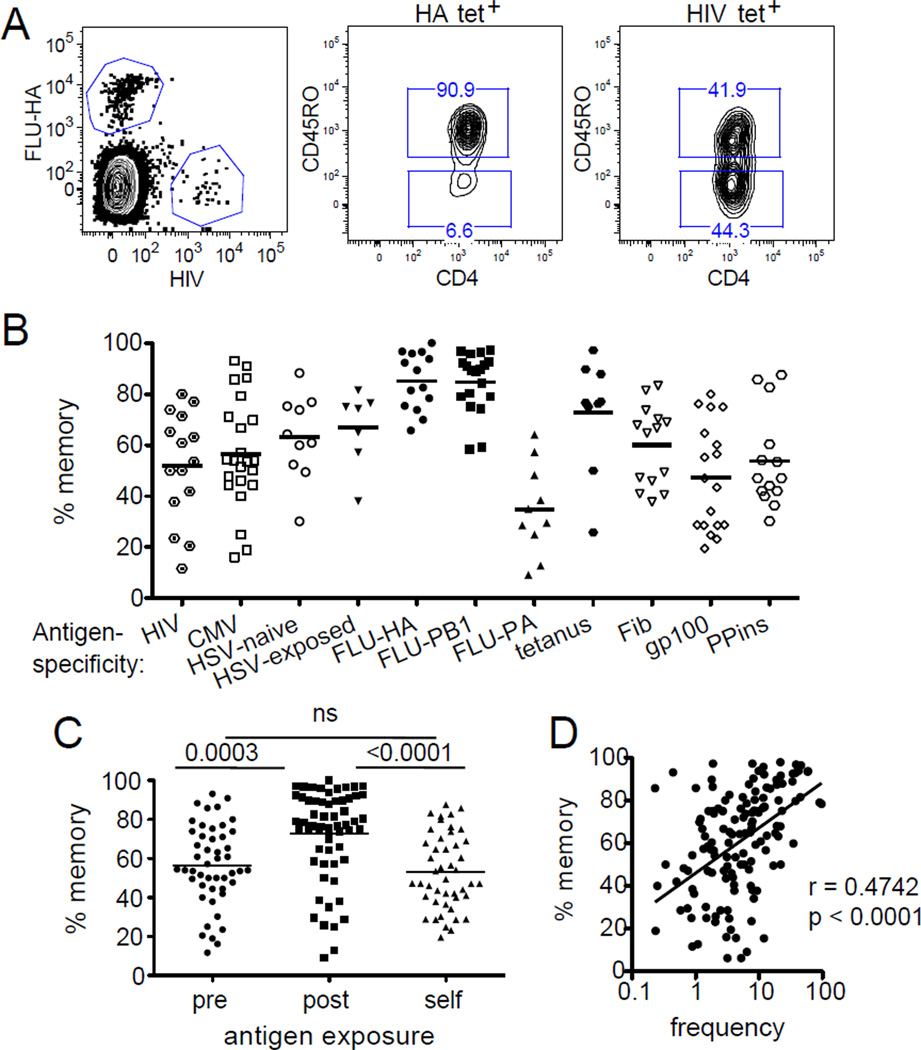
Unexpectedly, we found an abundance of HIV-1-, CMV-, and HSV-specific memory phenotype T cells in individuals who were seronegative for those viruses (Fig. 3A). The fraction of these memory phenotype T cells ranged from 12 – 93% between different individuals, with the average exceeding 50% (Fig. 3B–C). These memory phenotype T cells positively correlate with T cell frequency (Fig. 3D) and have similar frequencies of central memory phenotype cells by chemokine receptor CCR7 expression as influenza-specific T cells (Appay et al., 2008) (Fig. S3).
Figure 3. Characterization of the memory phenotype in the CD4+ T cell repertoire.
(A) Representative HA and HIV-1 tetramer labeling (left) and CD45RO antibody staining (right). Plots are representative of 15 individuals. (B) Percent of CD45RO+ memory cells within each tetramer tagged population. Each symbol represents an antigen-specific population from one individual and the bar indicates the mean of experiments performed independently using blood obtained at different times. (C) The percentage of memory phenotype among T cells recognizing a novel foreign antigen (pre: HIV-1, CMV, HSV-naïve), exposed foreign antigen (post: Flu, tetanus, HSV-exposed), and self antigen (self: Fib, gp100, PPins). Figure summarizes data from all 26 individuals. (D) Correlation plot combining all antigen-specific populations from all donors. Total cell frequency positively correlates with memory marker expression. Statistical analysis was performed using Spearman’s rank correlation. Regression was performed by least-square fit (See also Fig. S3).
HIV-1-specific CD4+ T cells from seronegative blood donors have the typical properties of memory T cells
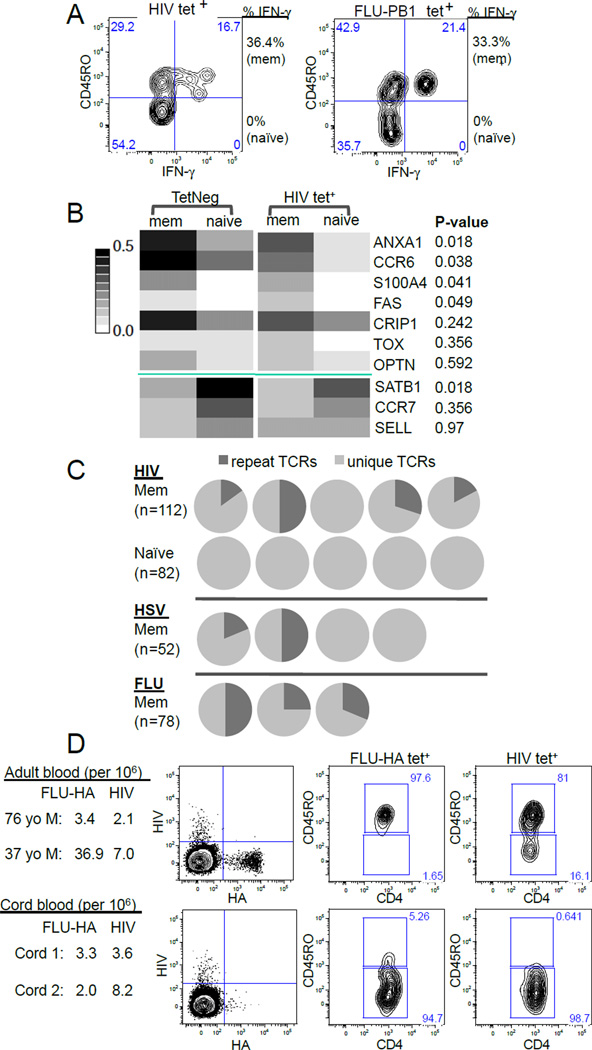
We also asked whether these specific memory phenotype T cells from unexposed individuals had known properties of functional memory cells. First we examined their cytokine responses, as memory cells are known to produce cytokines more rapidly and in larger amounts than naïve T cells (Mascher et al., 1999). T cells labeled with the HIV-1 tetramers were identified from healthy, seronegative, blood donors. Stimulation of these cells for 3 hours by phorbol myristate acetate (PMA) and ionomycin showed rapid IFN-γ production by the CD45RO+ expressing cells, but much less by the CD45RO− T cells. This was comparable to the influenza-specific T cells tested in parallel (Fig. 4A).
Figure 4. Memory phenotype T cells in adults have the typical memory features and are absent in cord blood.
(A) Contour plots showing IFN-γ response of HIV-1- and PB1-specific T cells to stimulation by PMA and ionomycin. Experiments were repeated twice using blood from 2 different individuals. (B) Gene expression of tetramer negative and HIV-1 tetramer labeled cells (TetNeg mem n = 48, TetNeg naïve n = 46, HIV-1 mem n = 53, HIV-1 naïve n = 52). Heatmap summarizes the fraction of cells expressing a particular gene out of the total number of cells assayed. The genes have been grouped by whether they associate with memory (top) or naïve T cells (bottom) and then ordered by ascending p value that compares the differences between HIV memory and HIV naïve T cells. (C) TCRβ sequencing of tetramer labeled CD45RO+ and CD45RO− cells. Each pie chart represents TCR sequences from one individual. Dark gray: the fraction of cells expressing a TCR identical to that of another cell. Light gray: cells expressing unique TCRs. HIV-1 and HSV-specific T cells were obtained from individuals negative for these infections. (D) HA- and HIV-1-specific T cells in adult PBMC (top) or cord blood (bottom) were identified by tetramer staining followed by magnetic bead enrichment (left) and anti-CD45RO surface staining (right). Data is representative of two adults PBMCs and two cord bloods assayed in parallel (See also Fig. S4).
We also performed gene expression analysis for transcripts previously identified as being preferentially expressed in memory and naive human CD4+ T cell populations (Haining et al., 2008; Weng et al., 2012). Of the ten genes analyzed for differential gene expression, five (ANXA1, CCR6, S100A4, FAS, and SATB1) were statistically different between the CD45RO+ and CD45RO− HIV-1-specific lymphocytes (Figure 4B). Analysis of individual cells also showed that memory phenotype cells generally exhibited similar transcriptional patterns irrespective of tetramer staining (Figure S4A).
To examine TCR repertoire of memory phenotype T cells, we sorted CD45RO+ and CD45RO− HIV-1 tetramer labeled cells individually and performed TCR beta chain amplification and sequence analysis on each cell. A total of 112 HIV-1-specific CD45RO+ cells and 82 HIV-1-specific CD45RO− cells from 5 HIV negative individuals were analyzed. For HIV-1-specific memory cells, evidence of clonal expansion was observed in 4 out of 5 blood donors, with 15% to 50% of these cells from each donor sharing the exact same TCR sequence with that of another cell from the same person. This contrasts with the greater TCR sequence diversity observed in HIV-1-specific naïve T cells where each TCR sequence was unique. Similarly, T cell clonality was also observed for HSV-specific memory T cells for two out of four donors and in all 3 donors for HA-specific memory T cells (Fig. 4C). The extent of clonal expansion appeared less restricted for HIV-1-specific T cells, which had more diverse Vβ gene usage than cells that recognize HA (Fig. S4B).
Memory phenotype T cells are largely absent in umbilical cord blood
We also analyzed umbilical cord blood to examine whether these memory phenotype cells are present at birth. While the prevalence of a naïve phenotype in bulk cord blood T cells versus adults is well known (Cossarizza et al., 1996), the frequency and the phenotype of individual TCR specificities are not. Two HLA-DR4+ human umbilical cord blood samples were identified and the characteristics of HIV-1-specific T cells were analyzed and compared to that of adult blood. In these samples, we found no apparent difference in the frequencies of HIV-1-specific T cells, which were 3.6 and 8.2 per million in the cord blood, and are within one standard deviation from the average of 4.0 per million in adult samples (0 – 9.25) (Fig. 4D). Similarly, T cells recognizing the self-peptide from gp100 were also detectable in cord blood cells at comparable frequencies (Fig. S4C). In contrast, CD45RO positive cells were strikingly absent in the tetramer positive T cells in the cord blood specimens (Fig. 4D and S4C). These findings indicate that newborns possess a full complement of T cells that can recognize self- and foreign-antigens, but essentially all of the cells are naïve and therefore the memory phenotype lymphocytes described here are acquired sometime after birth and before adulthood.
T cells selected for HIV-1-specificity can recognize numerous microbial peptides
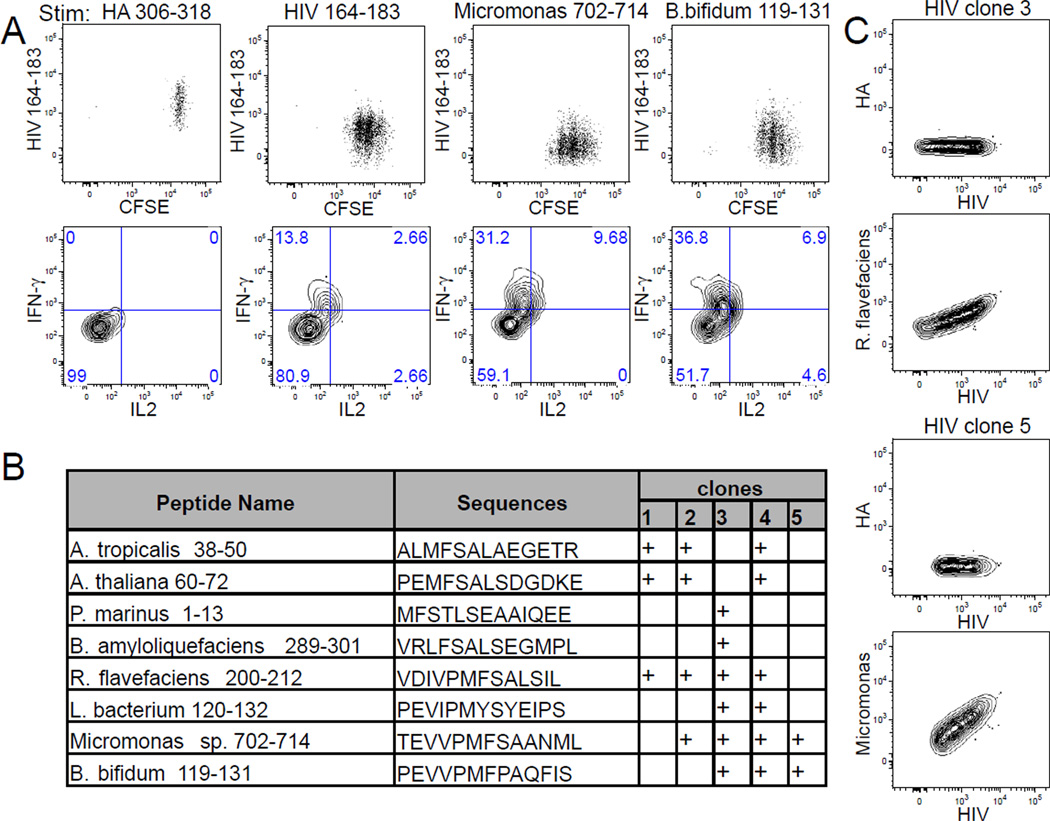
Because humans have copious exposure to environmental antigens, we sought to explore whether cross-reactivity to environmental antigens may have generated these memory phenotype T cells to novel antigens. To evaluate the extent of TCR cross-reactivity, twenty-four HIV-1-specific clones were generated using blood from three individuals by single cell cloning of tetramer positive cells as described previously in Fig. 1B. Responsiveness to the HIV-1 antigen was demonstrated by antigen stimulation with both the relevant peptide and the recombinant protein that it is derived from (Fig. S5A–B). Within the full-length HIV-1 peptide, there are two potential registers: HIV-1 167–179 was previously shown to bind a similar DR allele (Zavala-Ruiz et al., 2004), and HIV-1 171–181 contains the predicted anchor residues for HLA-DR4 (Hammer et al., 1994; Sette et al., 1993; Southwood et al., 1998). We first showed that both peptides stimulated different HIV-1 tetramer labeled clones (Figure S5C) then used these to identify homologous sequences by searching the NCBI-blast non-redundant protein sequence database. In total, 24 homologous sequences were examined for cross-reactivity. Whether HIV-1-reactive clones become stimulated by these peptides was determined by IFN-γ and interleukin-2 (IL2) cytokine production and validated by the induction of cellular proliferation using carboxyfluorescein diacetate succinimidyl ester (CFSE). We found TCR reactivity was quite common and that 21% of the clones (5/24) responded to at least two of these homologous ligands (Fig. 5A–B, Fig. S5D). The sources of peptides included bacteria from the gut and soil, other bacteria, as well as ocean algae and plants. Notably, these peptides not only stimulated HIV-1 reactive clones to produce cytokine and proliferate, but some likely also had high TCR avidity and could bind by tetramers (Fig. 5C). Together, these data show that homologous peptides in microbial genomes can trigger HIV-1-specific responses.
Figure 5. HIV-1-specific T cells exhibit extensive TCR cross-reactivity.
(A) FACS plots of CFSE staining and cytokine production by a representative HIV-1-reactive clone (HIV-1 clone 5). T cell proliferation and cytokine production was observed in response to HIV-1, Micromonas, and B. bifidum derived peptides but not to an irrelevant HA peptide. (B) Table summarizing peptide cross-reactivity for five HIV-1-specific clones that recognize alternate peptide sequences. Response of each clone to peptide stimulation was repeated at least 3 times. (C) Contour plots showing two cross-reactive clones that bind HIV-1 and R. flavefaciens tetramers (HIV-1 clone 3) or HIV-1 and Micromonas tetramers (HIV-1 clone 5), but not HA tetramers. Data is representative of 3 independent experiments (See also Fig. S5).
Flu-specific T cells recognize other microbial peptides
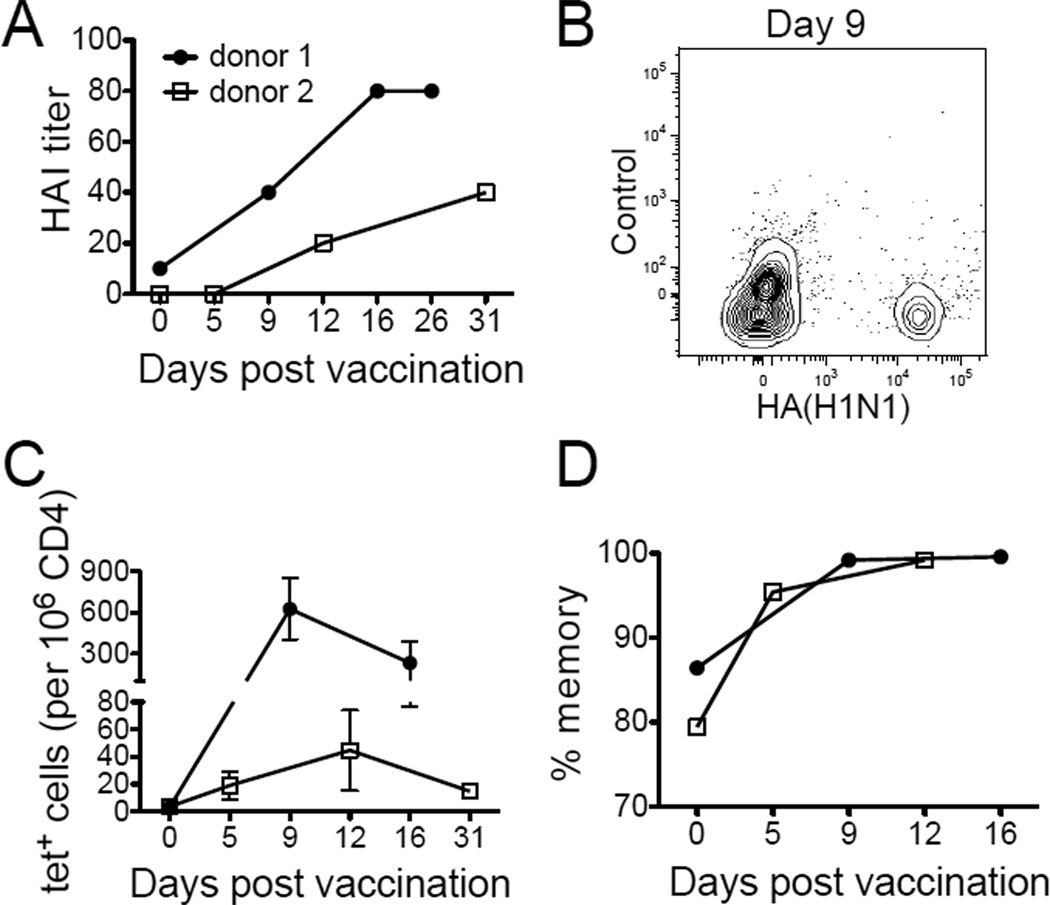
In order to study cross-reactive T cell response to a known antigen exposure, we recruited two individuals who have not had flu vaccines for at least 5 years to receive the 2011 seasonal influenza vaccine containing A/California/7/2009 (H1N1). Response to vaccination was validated by an increase in anti-influenza antibody to a protective titer of at least 1:40 (Fig. 6A). To analyze the corresponding T cell response, we examined 10 HA specificities and found cells recognizing HA amino acids 391–410 to be highly expanded following immunization. The T cell response to this epitope was tracked by HLA-DR4 tetramers loaded with HA 391–410 peptide (HA(H1N1)) and appeared as early as day 5 in the second donor when his HAI titer was still undetectable (Fig. 6B–C). In both individuals a dominant population of memory phenotype T cells specific for HA 391–410 was already present prior to vaccination consistent with conservation of this peptide in other influenza strains (Fig. 6D).
Figure 6. Antigen-specific T cell response to the 2009 H1N1 vaccine.
(A) Antibody response to A/California/7/2009 (H1N1) virus by HAI titer at various time points before and after influenza vaccination. (B) Contour plot showing expansion of HA(H1N1)-specific T cells of donor 1 nine days after vaccination. (C) The kinetics of HA(H1N1)-specific T cells. Error bars represent SEM. Experiments from each donor at each time point were performed 1–4 times, depending on sample availability. (D) The percentage of HA(H1N1)-specific T cells that are CD45RO+.
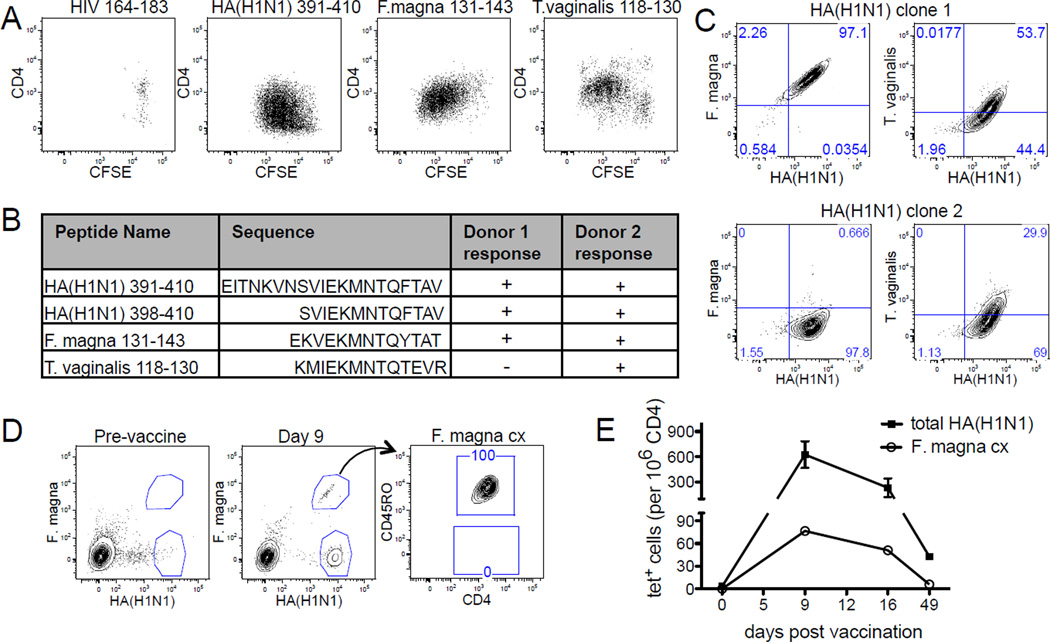
To evaluate T cell cross-reactivity, HA(H1N1)-specific T cells were cloned from post-vaccination blood samples and assayed for their response to other microbial peptides identified using a similar approach as for HIV-1-specific T cells. T cell stimulation by these peptides demonstrated that HA(H1N1)-specific T cell clones responded specifically to the HA 391–410 peptide sequence by which they were isolated, and to peptides from F. magna (1352 hypothetical protein 131–143) and T. vaginalis (TVAG_489980 hypothetical protein 118–130), but not to an irrelevant HIV-1 peptide (Fig. 7A). Although the cross-reactive peptides are quite similar to the HA core sequence (HA 398–410) and also to each other, these peptides differed from HA 398–410 in their ability to stimulate HA-reactive clones and elicited distinct responses in the two donors (Fig. 7B, Fig. S6). Differences in the fine specificities to these peptides were further evident by tetramer labeling, which showed F. magna and T. vaginalis peptides bound different HA(H1N1)-specific T cell clones with distinct avidity (Fig. 7C).
Figure 7. T cells responsive to flu vaccination also cross-recognized microbial peptides.
(A) CFSE staining of a representative HA(H1N1)-specific clones that proliferated in response to HA 391–410 peptide and also to a F. magna and T. vaginalis derived peptides but not to a control HIV-1 peptide. T cell stimulation with each peptide was repeated at least 3 times. (B) Table showing the sequences of full length HA 391–410 peptide, HA 398–410 core sequence, F. magna peptide, and T. vaginalis peptide. Twenty-one clones from donor 1 and twelve clones from donor 2 were combined and assayed for response to each peptide. Data is representative of two independent experiments. (C) Tetramer labeling of two HA(H1N1)-reactive clones by HA(H1N1), F. magna, and T. vaginalis tetramers. Experiment was repeated two times. (D) Direct ex vivo analysis of HA(H1N1) and F. magna tetramer positive cells. A population of influenza and F. magna cross-reactive cells (F. magna cx) became detectable on day 9 and exhibited 100% memory phenotype. (E) Frequencies of the total HA(H1N1) tetramer positive cells and cross-reactive T cells co-labeled with F. magna tetramers (F. magna cx). Error bars represent SEM. Experiments at each time point were performed 1–3 times, depending on sample availability (See also Fig. S6 and S7).
We further demonstrated that flu vaccination induced expansion of cross-reactive T cells in vivo. A previously undetectable population of cells labeled with both influenza and F. magna tetramers and consisted entirely of memory phenotype cells were observed post-vaccination (Fig. 7D). The extent of this cross-reactive T cell expansion is transient and followed a very similar trajectory as the HA(H1N1)-specific population (Fig. 7E). By day 49, the frequency of F. magna cross-reactive T cells was down to a range previously considered to be for naïve T cells at 5.9 cells/million. Expansion of cross-reactive T cells was also observed in the second donor (Fig S7A–B), with the fraction of cross-reactive T cells accounting for 14% and 4% of total HA(H1N1) reactive T cells in donors 1 and 2, respectively (Fig S7C). Together, these experiments show directly that immunization with one microbe can give rise to T cells specific for another cross-reactive specificity.
Discussion
The work of Jenkins and colleagues has shown that, by using a combination of peptide-MHC tetramer staining and enrichment, it is possible to directly study the pre-immune repertoire of αβ T cells in mice (Moon et al., 2007). Here we have extended these studies to the analysis of CD4+ T cells in human peripheral blood to characterize the T cell precursor frequency and phenotype. Examination of a variety of human class II MHC restricted self and non-self epitopes revealed that most specificities range in frequency from 1 to 10 per million CD4+ T cells. Very recent work by Kwok et al., measuring the frequency of precursor cells recognizing Bacillus anthracis finds a similar frequency range (Kwok et al., 2012). The data described here represents a comprehensive analysis of the human CD4+ T cell repertoire and includes multiple self and microbial antigens from different organisms in adults and compares the frequency and phenotype of some of these in newborns. It shows that despite humans having ~1000x greater number of the T cells than mice, their ligand repertoire for CD4+ T cells is very similar to that of mice. This larger human TCR repertoire relative to mice suggests that humans may have a greater degree of redundancy in the numbers of TCRs that can recognize the same antigen. In agreement with this conclusion, we found here that TCRs specific to a particular HIV-1 peptide/HLA-DR4 ligand encompass a large number of different TCR sequences. In addition to the repertoire composition, analysis of the antigen-specific T cells by peptide-MHC tetramer enrichment revealed an intriguing 1.7-fold increase in autoantigen-specific T cells in females compared to males. While speculative, a gender disparity in the number of autoreactive T cells may help to explain why autoimmunity is often more prevalent in females.
But arguably the most intriguing finding from this characterization of human T cell precursors is the abundance of memory phenotype T cells in healthy adults for foreign antigens that those individuals have never encountered. These CD45RO+ CD4+ T cells exhibited rapid IFN-γ production, evidence of clonal expansion, and the preferential expression of memory T cell-associated transcripts. Previous studies in murine CD8+ T cells have also found a memory phenotype population in precursor T cells (Sprent and Surh, 2011). Because memory phenotype T cells have also been found in germ free mice, they are thought to have arisen through homeostatic proliferation driven by self-antigen interactions (Haluszczak et al., 2009). Antigen-specific precursor memory cells have subsequently been found in other mouse studies and shown to be generated in the periphery during neonatal lymphopenia, and thereafter are stably maintained, increasing with age (Akue et al., 2012; Decman et al., 2012; Rudd et al., 2011). For CD4+ T cells, homeostatic proliferation also induces memory phenotype T cells in lymphopenic animals (Min et al., 2004), but CD4+ T cells appear to be less sensitive to homeostatic signals and differ from CD8+ T cells in their TCR-dependent lipid raft regulation and response to IL-7 signaling (Cho et al., 2010; Guimond et al., 2009). Studies on precursor CD4+ T cells in immunocompetent mice have generally found these cells to have a naïve phenotype (Chu et al., 2010; Moon et al., 2007). In humans, homeostatic proliferation maintains the peripheral naïve T cell pool in adults (den Braber et al., 2012), but whether or not a portion of the T cells undergoing homeostatic cell turnover can develop a memory phenotype in a healthy human is not known.
Given these caveats on how homeostatic proliferation may pertain to CD4+ T cells in healthy adults and also because humans are exposed to copious environmental antigens, we sought to investigate an alternate mechanism whereby these memory phenotype cells are generated by cross-reactivity with environmental antigens. Unlike CD8+ T cells that interact mostly with endogenous antigens, CD4+ T cells, by virtue of being restricted to professional antigen presenting cells that engulf and sample external antigens, are intrinsically predisposed to respond to changes in the environment. Moreover, there is a clear tendency on the part of at least some TCRs to recognize distinctly different peptide-MHC combinations, thereby enabling the same T cell to be activated by different ligands. Cross-reactive TCRs have been implicated in the pathogenesis of a number of autoimmune diseases (Benoist and Mathis, 2001) and have been proposed to explain why sequential infections in mice result in differences in immune pathology and the hierarchy of immunodominance (Welsh et al., 2010). In humans, there is also a growing recognition that vaccinations and their sequence of exposure can have a more general impact on morbidity and mortality beyond the expected benefit in preventing the targeted disease (Shann et al., 2010). For example, several large retrospective and longitudinal studies on young children have found a survival benefit from measles vaccines even when death from measles was excluded (Kristensen et al., 2000; Veirum et al., 2005). Consistent with these observations is the result of a randomized control trial involving over 6000 children, which also showed a vaccine-dependent mortality reduction that is independent of measles related death (Aaby et al., 2010). The dose and sequence of immunization appear to matter, and children given diphtheria-tetanus-pertussis (DTP) vaccine after being immunized for measles, particularly if high titer measles vaccine was used, have an increased risk of death and hospitalization (Aaby et al., 2003). In light of the data presented here it seems possible that these broader vaccine effects may be mediated by cross-reactive T cell memory. The paucity of these cross-reactive cells could also help to explain why very young children are more susceptible to infectious diseases than older ones (Casanova and Abel, 2005; Moore et al., 1999) and whether these responses are generated could mean the difference between survival and death.
In agreement with the relevance of cross-reactivity, we have shown here that this is not a rare phenomenon. Of the twenty-four HIV-1-reactive T cell clones analyzed, 21% had the ability to recognize multiple alternate ligands from environmental sources. While it is also possible that our data reflect a potential bias of repertoires selected on HLA-DR4 allele, this is still almost certainly an underestimate of the extent of cross-reactivity, since the propensity of T cells to cross-react with very different peptide sequences is well known and probably is directly enabled by the flexibility of the TCR binding site (Newell et al., 2011; Reinherz et al., 1999; Reiser et al., 2003). To directly test the hypothesis that CD4+ T cells can become activated and acquire a memory phenotype through cross-reactive interactions, we vaccinated two individuals with the seasonal flu vaccine containing the influenza virus, A/California/7/2009 (H1N1). We showed that T cells specific for both an HA epitope and a related sequence from F. magna, a human commensal bacteria (Levy et al., 2009), are both stimulated to proliferate and have the typical rise and fall of tetramer+ T cells post vaccination. We also detected donor-specific differences with one donor’s cells responding to both F. magna and a second cross-reactive epitope from T. vaginalis, a common sexually transmitted infection (Johnston and Mabey, 2008). This shows directly that exposure to one microbe is able to stimulate immune cells specific for cross-reactive peptides from very different microbes. We postulate that differences in microbial exposure and subsequent T cell cross-reactivity may help to explain some of the large variation in how individuals respond to infectious diseases. Our findings complement the existing data on homeostatic mechanisms of memory induction and provide a basis for further investigations on how environmental exposure may shape one’s immune repertoire.
In summary, our analysis of the human CD4+ T cell repertoire has uncovered a number of novel findings, but especially that healthy adults have numerous memory cells for foreign antigens to which they have never been exposed. Our data argues for TCR cross-reactivity in the acquisition of these cells and demonstrates inter-individual differences in this response. Regardless of the mechanism, these memory phenotype T cells specific for novel pathogens may confer an advantage for survival against infections. Individual differences in cross-reactive T cell memory may further explain the heterogeneity of response to infections in humans.
Material and Methods
Human Subjects
PBMCs were obtained from the Stanford Blood Bank. Cells in de-identified leukoreduction chambers from healthy platelet donors were processed as soon as possible and no later than 18 hours after plateletpheresis. For analysis of precursor T cells, an experiment uses at least 30 million CD4+ T cells, and typically between 50 – 150 million CD4+ T cells. For flu vaccination study, two healthy adults were recruited to receive the 2011 seasonal influenza vaccination. Details on the donor viral testing and vaccine study recruitment and sample processing are described in supplemental methods. Study subject recruitment was conducted in accordance with the rules and regulations of the Stanford institutional review board.
Identification and Analysis of Antigen-Specific T Cells
PBMCs were enriched for CD4+ cells by Rosettasep and ficoll centrifugation (Stemcell Tech). Tetramer staining was carried out at room temperature for 1 hour using 10 ug/ml of tetramers in the presence of purified anti-CD16, anti-CD32, and non-CD4 exclusion markers. Cells were incubated for an additional 10 minutes at room temperature with Agua live-dead dye, anti-CD4, anti-CD45RO, and anti-CCR7 antibodies for memory phenotyping. Tetramer tagged cells were enriched by adding anti-PE and/or anti-APC magnetic beads, and passing the mixture through a magnetized column (Miltenyi). Samples were analyzed by flow cytometry using LSRII (Becton Dickinson) and FlowJo software (Tree Star). Cells were stimulated immediately after isolation with PMA and ionomycin for cytokine analysis, sorted according to CD45RO expression for gene expression and TCR sequencing, or cultured individually to generate antigen-specific clones. Briefly, for cytokine analysis cells were stimulated with PMA (150ng/ml) and ionomycin (1mM) for 3 hours and assayed for intracellular IL2 and IFN-γ production in the presence of protein transport inhibitor. Multiplex real-time PCR and gene expression analysis was performed with TagMan® Gene Expression primers (Applied Biosystems) using BioMark system (Fluidgm). TCR sequences from single cells were obtained by a series of three nested PCR reactions. To generate antigen-specific clones, tetramer labeled cells were sorted individually and expanded by PHA and IL2. See supplemental methods for the list of antibodies and tetramers and also for detailed descriptions of frequency and sensitivity calculation, gene expression analysis, TCR sequencing, and cloning procedure.
Statistics
For analysis of experiments comparing a single variable, statistical significance was analyzed using student’s t-test. Spearman rank correlation and least squares fit regression were applied to measure the degree of association. A p-value of <0.05 was used as a cutoff for statistical significance. Differential gene expression was analyzed by a two-sided Mann-Whitney test and corrected for multiple hypothesis testing using false discovery rates (Benjamini and Hochberg). Genes were considered to be significantly differentially expressed between groups if they had p-values and false discovery rates < 0.05. All data analysis was performed using R statistical package v. 2.13 and GraphPad Prism.
Supplementary Material
Highlights.
Healthy adults have memory cells to unexposed foreign antigens
Pre-existing memory phenotype cells present in adults are absent in newborns
HIV-1 and influenza-specific T cells cross-react with microbial peptides
Influenza vaccination induces cross-reactive T cell expansion
Acknowledgements
We thank Dr. Kai Wucherpfennig for providing the HLA-DR4 stable transfectant cell line for the production of HLA-DR monomers. Thanks also to Yueh-hsiu Chien, John Imboden, Jonathan Graf, Elizabeth Mellins, Peter Ebert, Marc Jenkins and Wong Yu for helpful discussions. We thank Bithi Chatterjee for helpful advice on deriving dendritic cells, Andrew Prentice for helpful suggestions and Cristina Tato for manuscript edits.
L.F.S was supported by a Daland fellowship from the American Philosophical Foundation, and an ACR REF Rheumatology Scientist Development Award, and is currently supported by a K08 award from National Institute of Health grant (K08 AR059760). A.H. is supported by NIH training grant 5 T32 DK007056-36. Other research support came from grants from the National Institutes of Health (U19 AI057229 and U19AI090019) and the Howard Hughes Medical Institute to M.M.D.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contribution: L.F.S and M.M.D were responsible for design of the research. L.F.S performed experiments and analyzed data. A.H. designed the TCR sequencing primers and protocol. B.A.K. provided statistical expertise and analyzed gene expression data. J.J.K. assisted with HA epitope identification and performed the HAI assays. L.F.S wrote the paper. The manuscript was edited by M.M.D, A.H., B.A.K, and J.J.K.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
References
- Aaby P, Jensen H, Samb B, Cisse B, Sodemann M, Jakobsen M, Poulsen A, Rodrigues A, Lisse IM, Simondon F, Whittle H. Differences in female-male mortality after high-titre measles vaccine and association with subsequent vaccination with diphtheria-tetanus-pertussis and inactivated poliovirus: reanalysis of West African studies. Lancet. 2003;361:2183–2188. doi: 10.1016/S0140-6736(03)13771-3. [DOI] [PubMed] [Google Scholar]
- Aaby P, Martins CL, Garly ML, Bale C, Andersen A, Rodrigues A, Ravn H, Lisse IM, Benn CS, Whittle HC. Non-specific effects of standard measles vaccine at 4.5 and 9 months of age on childhood mortality: randomised controlled trial. BMJ. 2010;341:c6495. doi: 10.1136/bmj.c6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akue AD, Lee JY, Jameson SC. Derivation and maintenance of virtual memory CD8 T cells. J Immunol. 2012;188:2516–2523. doi: 10.4049/jimmunol.1102213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander J, Bilsel P, del Guercio MF, Stewart S, Marinkovic-Petrovic A, Southwood S, Crimi C, Vang L, Walker L, Ishioka G, et al. Universal influenza DNA vaccine encoding conserved CD4+ T cell epitopes protects against lethal viral challenge in HLA-DR transgenic mice. Vaccine. 2010;28:664–672. doi: 10.1016/j.vaccine.2009.10.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appay V, van Lier RA, Sallusto F, Roederer M. Phenotype and function of human T lymphocyte subsets: consensus and issues. Cytometry A. 2008;73:975–983. doi: 10.1002/cyto.a.20643. [DOI] [PubMed] [Google Scholar]
- Arif S, Tree TI, Astill TP, Tremble JM, Bishop AJ, Dayan CM, Roep BO, Peakman M. Autoreactive T cell responses show proinflammatory polarization in diabetes but a regulatory phenotype in health. J Clin Invest. 2004;113:451–463. doi: 10.1172/JCI19585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auger I, Sebbag M, Vincent C, Balandraud N, Guis S, Nogueira L, Svensson B, Cantagrel A, Serre G, Roudier J. Influence of HLA-DR genes on the production of rheumatoid arthritis-specific autoantibodies to citrullinated fibrinogen. Arthritis Rheum. 2005;52:3424–3432. doi: 10.1002/art.21391. [DOI] [PubMed] [Google Scholar]
- Benoist C, Mathis D. Autoimmunity provoked by infection: how good is the case for T cell epitope mimicry? Nat Immunol. 2001;2:797–801. doi: 10.1038/ni0901-797. [DOI] [PubMed] [Google Scholar]
- Bronke C, Palmer NM, Westerlaken GH, Toebes M, van Schijndel GM, Purwaha V, van Meijgaarden KE, Schumacher TN, van Baarle D, Tesselaar K, Geluk A. Direct ex vivo detection of HLA-DR3-restricted cytomegalovirus-and Mycobacterium tuberculosis-specific CD4+ T cells. Hum Immunol. 2005;66:950–961. doi: 10.1016/j.humimm.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Casanova JL, Abel L. Inborn errors of immunity to infection: the rule rather than the exception. J Exp Med. 2005;202:197–201. doi: 10.1084/jem.20050854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JH, Kim HO, Surh CD, Sprent J. T cell receptor-dependent regulation of lipid rafts controls naive CD8+ T cell homeostasis. Immunity. 2010;32:214–226. doi: 10.1016/j.immuni.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu HH, Moon JJ, Kruse AC, Pepper M, Jenkins MK. Negative selection and peptide chemistry determine the size of naive foreign peptide-MHC class II-specific CD4+ T cell populations. J Immunol. 2010;185:4705–4713. doi: 10.4049/jimmunol.1002276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossarizza A, Ortolani C, Paganelli R, Barbieri D, Monti D, Sansoni P, Fagiolo U, Castellani G, Bersani F, Londei M, Franceschi C. CD45 isoforms expression on CD4+ and CD8+ T cells throughout life, from newborns to centenarians: implications for T cell memory. Mech Ageing Dev. 1996;86:173–195. doi: 10.1016/0047-6374(95)01691-0. [DOI] [PubMed] [Google Scholar]
- Day CL, Seth NP, Lucas M, Appel H, Gauthier L, Lauer GM, Robbins GK, Szczepiorkowski ZM, Casson DR, Chung RT, et al. Ex vivo analysis of human memory CD4 T cells specific for hepatitis C virus using MHC class II tetramers. J Clin Invest. 2003;112:831–842. doi: 10.1172/JCI18509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decman V, Laidlaw BJ, Doering TA, Leng J, Ertl HC, Goldstein DR, Wherry EJ. Defective CD8 T cell responses in aged mice are due to quantitative and qualitative changes in virus-specific precursors. J Immunol. 2012;188:1933–1941. doi: 10.4049/jimmunol.1101098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Braber I, Mugwagwa T, Vrisekoop N, Westera L, Mogling R, de Boer AB, Willems N, Schrijver EH, Spierenburg G, Gaiser K, et al. Maintenance of peripheral naive T cells is sustained by thymus output in mice but not humans. Immunity. 2012;36:288–297. doi: 10.1016/j.immuni.2012.02.006. [DOI] [PubMed] [Google Scholar]
- Diethelm-Okita BM, Raju R, Okita DK, Conti-Fine BM. Epitope repertoire of human CD4+ T cells on tetanus toxin: identification of immunodominant sequence segments. J Infect Dis. 1997;175:382–391. doi: 10.1093/infdis/175.2.382. [DOI] [PubMed] [Google Scholar]
- Flynn KJ, Belz GT, Altman JD, Ahmed R, Woodland DL, Doherty PC. Virus-specific CD8+ T cells in primary and secondary influenza pneumonia. Immunity. 1998;8:683–691. doi: 10.1016/s1074-7613(00)80573-7. [DOI] [PubMed] [Google Scholar]
- Guimond M, Veenstra RG, Grindler DJ, Zhang H, Cui Y, Murphy RD, Kim SY, Na R, Hennighausen L, Kurtulus S, et al. Interleukin 7 signaling in dendritic cells regulates the homeostatic proliferation and niche size of CD4+ T cells. Nat Immunol. 2009;10:149–157. doi: 10.1038/ni.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haining WN, Angelosanto J, Brosnahan K, Ross K, Hahn C, Russell K, Drury L, Norton S, Nadler L, Stegmaier K. High-throughput gene expression profiling of memory differentiation in primary human T cells. BMC Immunol. 2008;9:44. doi: 10.1186/1471-2172-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haluszczak C, Akue AD, Hamilton SE, Johnson LD, Pujanauski L, Teodorovic L, Jameson SC, Kedl RM. The antigen-specific CD8+ T cell repertoire in unimmunized mice includes memory phenotype cells bearing markers of homeostatic expansion. J Exp Med. 2009;206:435–448. doi: 10.1084/jem.20081829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer J, Belunis C, Bolin D, Papadopoulos J, Walsky R, Higelin J, Danho W, Sinigaglia F, Nagy ZA. High-affinity binding of short peptides to major histocompatibility complex class II molecules by anchor combinations. Proc Natl Acad Sci U S A. 1994;91:4456–4460. doi: 10.1073/pnas.91.10.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson SC, Masopust D. Diversity in T cell memory: an embarrassment of riches. Immunity. 2009;31:859–871. doi: 10.1016/j.immuni.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston VJ, Mabey DC. Global epidemiology and control of Trichomonas vaginalis. Curr Opin Infect Dis. 2008;21:56–64. doi: 10.1097/QCO.0b013e3282f3d999. [DOI] [PubMed] [Google Scholar]
- Kristensen I, Aaby P, Jensen H. Routine vaccinations and child survival: follow up study in Guinea-Bissau, West Africa. BMJ. 2000;321:1435–1438. doi: 10.1136/bmj.321.7274.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok WW, Tan V, Gillette L, Littell CT, Soltis MA, LaFond RB, Yang J, James EA, DeLong JH. Frequency of epitope-specific naive CD4(+) T cells correlates with immunodominance in the human memory repertoire. J Immunol. 2012;188:2537–2544. doi: 10.4049/jimmunol.1102190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackritz EM, Satten GA, Aberle-Grasse J, Dodd RY, Raimondi VP, Janssen RS, Lewis WF, Notari EPt, Petersen LR. Estimated risk of transmission of the human immunodeficiency virus by screened blood in the United States. N Engl J Med. 1995;333:1721–1725. doi: 10.1056/NEJM199512283332601. [DOI] [PubMed] [Google Scholar]
- Lamb JR, Eckels DD, Lake P, Woody JN, Green N. Human T-cell clones recognize chemically synthesized peptides of influenza haemagglutinin. Nature. 1982;300:66–69. doi: 10.1038/300066a0. [DOI] [PubMed] [Google Scholar]
- Levy PY, Fenollar F, Stein A, Borrione F, Raoult D. Finegoldia magna: a forgotten pathogen in prosthetic joint infection rediscovered by molecular biology. Clin Infect Dis. 2009;49:1244–1247. doi: 10.1086/605672. [DOI] [PubMed] [Google Scholar]
- Mascher B, Schlenke P, Seyfarth M. Expression and kinetics of cytokines determined by intracellular staining using flow cytometry. J Immunol Methods. 1999;223:115–121. doi: 10.1016/s0022-1759(98)00200-2. [DOI] [PubMed] [Google Scholar]
- Min B, Foucras G, Meier-Schellersheim M, Paul WE. Spontaneous proliferation, a response of naive CD4 T cells determined by the diversity of the memory cell repertoire. Proc Natl Acad Sci U S A. 2004;101:3874–3879. doi: 10.1073/pnas.0400606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon JJ, Chu HH, Pepper M, McSorley SJ, Jameson SC, Kedl RM, Jenkins MK. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 2007;27:203–213. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore SE, Cole TJ, Collinson AC, Poskitt EM, McGregor IA, Prentice AM. Prenatal or early postnatal events predict infectious deaths in young adulthood in rural Africa. Int J Epidemiol. 1999;28:1088–1095. doi: 10.1093/ije/28.6.1088. [DOI] [PubMed] [Google Scholar]
- Newell EW, Ely LK, Kruse AC, Reay PA, Rodriguez SN, Lin AE, Kuhns MS, Garcia KC, Davis MM. Structural basis of specificity and cross-reactivity in T cell receptors specific for cytochrome c-I-E(k) J Immunol. 2011;186:5823–5832. doi: 10.4049/jimmunol.1100197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielen MM, van der Horst AR, van Schaardenburg D, van der Horst-Bruinsma IE, van de Stadt RJ, Aarden L, Dijkmans BA, Hamann D. Antibodies to citrullinated human fibrinogen (ACF) have diagnostic and prognostic value in early arthritis. Ann Rheum Dis. 2005;64:1199–1204. doi: 10.1136/ard.2004.029389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris PJ, Moffett HF, Brander C, Allen TM, O'Sullivan KM, Cosimi LA, Kaufmann DE, Walker BD, Rosenberg ES. Fine specificity and cross-clade reactivity of HIV type 1 Gag-specific CD4+ T cells. AIDS Res Hum Retroviruses. 2004;20:315–325. doi: 10.1089/088922204322996554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak EJ, Liu AW, Gebe JA, Falk BA, Nepom GT, Koelle DM, Kwok WW. Tetramer-guided epitope mapping: rapid identification and characterization of immunodominant CD4+ T cell epitopes from complex antigens. J Immunol. 2001;166:6665–6670. doi: 10.4049/jimmunol.166.11.6665. [DOI] [PubMed] [Google Scholar]
- Obar JJ, Khanna KM, Lefrancois L. Endogenous naive CD8+ T cell precursor frequency regulates primary and memory responses to infection. Immunity. 2008;28:859–869. doi: 10.1016/j.immuni.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan GQ, Touloukian CE, Yang JC, Restifo NP, Sherry RM, Hwu P, Topalian SL, Schwartzentruber DJ, Seipp CA, Freezer LJ, et al. Immunization of patients with metastatic melanoma using both class I- and class II-restricted peptides from melanoma-associated antigens. J Immunother. 2003;26:349–356. doi: 10.1097/00002371-200307000-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinherz EL, Tan K, Tang L, Kern P, Liu J, Xiong Y, Hussey RE, Smolyar A, Hare B, Zhang R, et al. The crystal structure of a T cell receptor in complex with peptide and MHC class II. Science. 1999;286:1913–1921. doi: 10.1126/science.286.5446.1913. [DOI] [PubMed] [Google Scholar]
- Reiser JB, Darnault C, Gregoire C, Mosser T, Mazza G, Kearney A, van der Merwe PA, Fontecilla-Camps JC, Housset D, Malissen B. CDR3 loop flexibility contributes to the degeneracy of TCR recognition. Nat Immunol. 2003;4:241–247. doi: 10.1038/ni891. [DOI] [PubMed] [Google Scholar]
- Rudd BD, Venturi V, Li G, Samadder P, Ertelt JM, Way SS, Davenport MP, Nikolich-Zugich J. Nonrandom attrition of the naive CD8+ T-cell pool with aging governed by T-cell receptor:pMHC interactions. Proc Natl Acad Sci U S A. 2011;108:13694–13699. doi: 10.1073/pnas.1107594108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A, Sidney J, Oseroff C, del Guercio MF, Southwood S, Arrhenius T, Powell MF, Colon SM, Gaeta FC, Grey HM. HLA DR4w4-binding motifs illustrate the biochemical basis of degeneracy and specificity in peptide-DR interactions. J Immunol. 1993;151:3163–3170. [PubMed] [Google Scholar]
- Shann F, Nohynek H, Scott JA, Hesseling A, Flanagan KL. Randomized trials to study the nonspecific effects of vaccines in children in low-income countries. Pediatr Infect Dis J. 2010;29:457–461. doi: 10.1097/INF.0b013e3181c91361. [DOI] [PubMed] [Google Scholar]
- Southwood S, Sidney J, Kondo A, del Guercio MF, Appella E, Hoffman S, Kubo RT, Chesnut RW, Grey HM, Sette A. Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol. 1998;160:3363–3373. [PubMed] [Google Scholar]
- Sprent J, Surh CD. Normal T cell homeostasis: the conversion of naive cells into memory-phenotype cells. Nat Immunol. 2011;12:478–484. doi: 10.1038/ni.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veirum JE, Sodemann M, Biai S, Jakobsen M, Garly ML, Hedegaard K, Jensen H, Aaby P. Routine vaccinations associated with divergent effects on female and male mortality at the paediatric ward in Bissau, Guinea-Bissau. Vaccine. 2005;23:1197–1204. doi: 10.1016/j.vaccine.2004.02.053. [DOI] [PubMed] [Google Scholar]
- Welsh RM, Che JW, Brehm MA, Selin LK. Heterologous immunity between viruses. Immunol Rev. 2010;235:244–266. doi: 10.1111/j.0105-2896.2010.00897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng NP, Araki Y, Subedi K. The molecular basis of the memory T cell response: differential gene expression and its epigenetic regulation. Nat Rev Immunol. 2012;12:306–315. doi: 10.1038/nri3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Ku GY, Gallardo HF, Orlandi F, Manukian G, Rasalan TS, Xu Y, Li H, Vyas S, Mu Z, et al. Safety and immunogenicity of a human and mouse gp100 DNA vaccine in a phase I trial of patients with melanoma. Cancer Immun. 2009;9:5. [PMC free article] [PubMed] [Google Scholar]
- Zavala-Ruiz Z, Strug I, Walker BD, Norris PJ, Stern LJ. A hairpin turn in a class II MHC-bound peptide orients residues outside the binding groove for T cell recognition. Proc Natl Acad Sci U S A. 2004;101:13279–13284. doi: 10.1073/pnas.0403371101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Nakayama M, Eisenbarth GS. Insulin as an autoantigen in NOD/human diabetes. Curr Opin Immunol. 2008;20:111–118. doi: 10.1016/j.coi.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.