Abstract
A critical feature of highly pathogenic avian influenza viruses (H5N1 and H7N7) is the efficient intracellular cleavage of the hemagglutinin (HA) protein. H7N7 viruses also exist in equine species, and a unique feature of the equine H7N7 HA is the presence of an eleven amino acid insertion directly N-terminal to a tetrabasic cleavage site. Here, we show that three histidine residues within the unique insertion of the equine H7N7 HA are essential for intracellular cleavage. An asparagine residue within the insertion-derived glycosylation site was also found to be essential for intracellular cleavage. The presence of the histidine residues also appear to be involved in triggering fusion, since mutation of the histidine residues resulted in a destabilizing effect. Importantly, the addition of a tetrabasic site and the eleven amino acid insertion conferred efficient intracellular cleavage to the HA of an H7N3 low pathogenicity avian influenza virus. Our studies show that acquisition of the eleven amino acid insertion offers an alternative mechanism for intracellular cleavage of influenza HA.
Keywords: Influenza, Hemagglutinin, Membrane fusion, Proteolytic, Cleavage, Histidine, Glycosylation
Introduction
Influenza virus is a member of the Orthomyxoviridae family and is classified into A, B, and C types (Knipe et al., 2007). Influenza A virus is subtyped by the distinct antigenicity of the hemagglutinin (HA) and neuraminidase (NA) surface proteins, resulting in 17 HA and 9 NA subtypes (Watanabe et al., 2012). The viral HA is further classified into two distinct structural groups, 1 and 2 (Air, 1981, Russell et al., 2004). All influenza A subtypes are thought to have originated from waterfowl, with some subtypes being transmitted to mammals such as horses, pigs, and humans (Horimoto and Kawaoka, 2005). In horses, the H3N8 and H7N7 influenza subtypes have been found to circulate widely. Currently it is the H3N8 virus that circulates in horses and is also associated with an epizootic and subsequent establishment of influenza H3N8 in dogs (Hayward et al., 2010). Influenza virus is highly contagious in horses, with infection generally limited to the respiratory tract and associated with fever, cough, and depression (Murcia et al., 2011, Timoney, 1996). The first report of H7N7 equine influenza occurred in 1956 (Sovinova and Tumova, 1958), and this virus circulated globally until the mid-1970 s. H7N7 is generally considered to no longer be present in the equine population, with the last viral isolate reported in 1980 (Daly et al., 1996). However, serology of the equine population suggests that the virus may still in fact be in circulation (Appleton and Gagliardo, 1992, Olusa et al., 2010).
H7 influenza viruses have historically been associated with many outbreaks of HPAI or “fowl plague” (Kaleta and Rülke, 2008). More recently, they have caused limited outbreaks in humans, where one of which resulted in a lethal infection (Bos et al., 2010). The equine H7N7 is considered to have arisen as a direct transmission event from birds (Baigent and McCauley, 2003). Interestingly, the equine H7N7 was found to be both highly pathogenic and neurovirulent in mice without adaptation (Kawaoka, 1991, Shinya et al., 2005, Shinya et al., 2007). Equine H7N7 viruses have other distinct properties, including that unlike HPAI, H7N7 it does not require coexpression of a functional M2 protein to maintain HA in its native conformation (Takeuchi et al., 1994).
The influenza HA is synthesized as a fusion inactive precursor (HA0) that must be cleaved into two functional subunits (HA1 and HA2) by host proteases, in order to gain the ability to fuse with the host endosome (Garten et al., 1981, Steinhauer, 1999, Taubenberger,1998). Cleavage of HA containing a monobasic cleavage site is most likely driven by extracellular or membrane-bound trypsin-like proteases, with the type II transmembrane serine protease (TMPRSS) family being the most recent examples (Böttcher et al., 2006, Bertram et al., 2010a, Chaipan et al, 2009, Hamilton et al., 2012). The HA cleavage sites of the low pathogenicity and highly pathogenic viruses are in general quite distinct, with the HA of low pathogenicity strains containing a monobasic cleavage site and highly pathogenic viruses containing a multibasic cleavage site. This difference in the cleavage site likely dictates which host proteases are able to cleave HA, and the cellular location where cleavage occurs. Cleavage of HA containing a monobasic cleavage site occurs extracellularly, most likely by trypsin-like proteases. In contrast, cleavage of HA containing a multibasic cleavage site occurs intracellularly by subtilisin-like proteases such as furin, which minimally recognizes a R–X–K/R–R motif (Bertram et al., 2010b, Zhang et al., 2003). This is best documented for the avian H5N1 and H7N7 viruses (HPAIs), which are associated with a high mortality rate in poultry, compared to LPAIs (Kaleta and Rülke, 2008). A large polybasic stretch of 6–7 residues is typically found in the cleavage site of most avian H5 and H7 HPAI subtypes (Fig. 1) (Senne et al., 1996). However, only four basic residues are found in the equine H7N7 HA cleavage site. This is intriguing, since intracellular cleavage was observed for the equine H7 HA (Gibson et al., 1992), (Takeuchi et al., 1994), but in contrast, was not observed by mutational analysis of other HA subtypes when the cleavage site contained only four basic residues (Gohrbandt et al., 2011, Walker and Kawaoka, 1993). A unique feature of the equine H7 HA cleavage site is the presence of an eleven amino acid insertion (consensus sequence=NSTHKQLTHHM) directly N-terminal to the tetrabasic cleavage site (RKKR) (Fig. 1). This insertion appears to have occurred coincident with the transmission of an avian H7N7 to horses (Gibson et al., 1992). It remains unclear whether there is a functional role of this unique eleven amino acid insertion, but it may play a role in HA stability or cleavage due to the presence of three ionizable histidine residues and the addition of a predicted glycosylation site within the insertion.
Fig. 1.

Multiple sequence alignment of the cleavage site region of HA from the H5 and H7 subtypes. Comparison of the consensus cleavage site region of the HPAI H5N1, HPAI H7N7 HA and LPAI H7N3 with the equine H7N7 HA sequence. Cleavage occurs at the C-terminal end of the Arg (R) bonded to the Gly (G) residue, which is the first amino acid of the fusion peptide. Representative sequences from the NCBI influenza resource (http://www.ncbi.nlm.nih.gov/genomes/FLU/FLU.html) were aligned with Clustal W. The A/chicken/Vietnam/2003 (H5N1) (accession # ABE97598), A/fowl/Weybridge/1934 (H7N7) (accession # AAA56803), A/Netherlands/2003 (H7N7) (accession # AAR02640), A/turkey/Italy/2002 (H7N3) (accession # AEK65956), and A/equine/Cornell/1974 (H7N7) (accession # ABY83099) were used as representative sequences.
In the present study, we have investigated the functional role of the eleven amino acid insertion of the equine H7 HA. Specifically, the focus of this study was to investigate both the role of the three histidine residues and the additional glycosylation site for the A/equine/Cornell/16/1974 strain of equine H7N7 influenza virus, which contains the consensus insert at the cleavage site position (NSTHKQLTHHMRKKR).
Results
The eleven amino acid insertion of the H7 HA insertion is essential for intracellular cleavage
As equine H7N7 viruses contain a unique eleven amino acid insertion adjacent to a minimal furin cleavage site, we first investigated the role of the unique insertion for HA cleavage. Specifically, we focused on the role of the three histidine residues and the additional glycosylation site, and generated the HA mutants summarized in Table 1. Both wild type and mutant HAs were analyzed for efficient cell surface expression (Supplementary Fig. 1). All mutants HAs showed >65% cell expression compared to wild type, with the exception of the H338A mutation.
Table 1.
Mutants of the influenza A/equine/Cornell/1974 (H7N7) HA generated in this study.
| H7 HA form | Sequence |
|---|---|
| EqHA wt | PENSTHKQLTHHMRKKRGLFG |
| EqHA H338A | PENSTAKQLTHHMRKKRGLFG |
| EqHA H343A/H344A | PENSTHKQLTAAMRKKRGLFG |
| EqHA H338A/H343A/H344A | PENSTAKQLTAAMRKKRGLFG |
| EqHA NST–AST | PEASTHKQLTHHMRKKRGLFG |
| EqHA NST–APA | PEAPAHKQLTHHMRKKRGLFG |
| EqHA Δ335–345 | PE - - - - - - - RKKRGLFG |
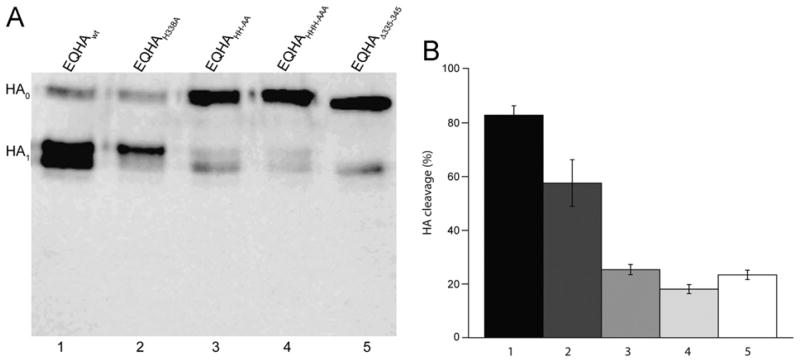
To determine whether the histidine residues of the H7 HA insertion are involved in intracellular cleavage, the wild type HA, individual and sequential mutations of the histidine residues, and the insert deletion mutant of the H7 HA were expressed in mammalian cells and assessed for intracellular cleavage by western blot analysis (Table 1). Efficient intracellular cleavage was observed for the wild type protein, with some reduction in cleavage for the HA containing a single histidine substitution (EQHAH338A) (Fig. 2A). However, intracellular cleavage was greatly reduced for mutants containing double and triple histidine substitutions (EQHAH343A/H344A and EQHAH338A/H343A/H344A), as well as with the eleven amino acid insert removed (EQHAΔ335–345) when compared to the wild type HA (Fig. 2A and B). The loss of cleavage was unlikely to be due to inefficient intracellular trafficking or cell expression, as mutants containing double and triple histidine substitutions, as well with the insert removed, showed efficient cell surface expression (see Supplementary Fig. 1). Overall, we show that sequential mutations of the histidine residues resulted in sequential loss of intracellular cleavage for equine H7N7 HA.
Fig. 2.
The three histidine residues of the insertion are essential for intracellular cleavage of the equine H7 HA. (A) Western blot analysis of the intracellular cleavage of EQHAwt (lane 1), EQHAH338A (lane 2), EQHAH343A/H344A(EQHAHH-AA) (lane 3), EQHAH338A/H343A/H344A(EQHAHHH-AAA) (lane 4), and EQHAΔ335-345 (lane 5) HA. (B) Quantification of the cleavage of each H7 HA form.
Since the eleven amino acid insertion of the A/Cornell/74(H7N7) HA includes a potential glycosylation site, and N-linked glycosylation near the cleavage site has been found to affect the cleavage efficiency of HA by proteases (Webster and Rott, 1987), the insertion glycosylation site was investigated to determine any effects to the cleavage of the H7 HA. Typically, the N-linked glycan resides near the cleavage site in the tertiary structure, but is distant in the primary structure. However, this is in contrast to the equine H7, where the N-linked glycan is adjacent to the cleavage site in the primary structure, and so may cause differing effects. Wild type H7N7 HA and well as the EQHANST–AST, and EQHANST–APA mutants were expressed in mammalian cells and first assessed for cell surface expression (Supplementary Fig. 1). The APA mutant (EQHANST–APA) was included as a subset of natural isolates of equine H7N7 (including the prototype virus A/equine/Prague/1956) that contain an APA sequence in place of NST. Cell surface expression of EQHANST–APA was relatively poor (approximately 25% of wild type HA), yet efficient intracellular cleavage was observed for the NST-APA mutant, by Western blot analysis, and was comparable to cleavage of wild type HA (Fig. 3A). However, near complete loss of both cell surface expression and intracellular cleavage was observed for the EQHANST–AST mutant HA. Furthermore, the EQHANST–AST mutant was insensitive to trypsin treatment, revealing inaccessibility of the cleavage site (Fig. 3A).
Fig. 3.
The asparagine residue of the insertion is essential for intracellular cleavage of the H7N7 HA. (A) Western blot analysis of the intracellular cleavage of EQHAwt, EQHANST-AST, EQHANST-APA HA. (B) Western blot analysis of EQHAwt HA in the presence or absence of PNGase F. Glycosylated HA is indicated by (G) and deglycosylated HA is indicated by (DG).
Observation of the EQHAwt HA cleavage products revealed two forms of the HA1 subunit (Fig. 3A and B). However, cleavage of EQHANST–APA HA produced only a single HA1 band, which is typical amongst the HA subtypes (Fig. 3A). We therefore reasoned that the two HA1 forms observed for EQHAwt are due to incomplete glycosylation of the site found within the insertion, since mutation of the H7 HA from NST to APA eliminates the potential glycosylation site. The glycosidase, PNGase F cleaves N-linked glycans from proteins and so was incubated with EQHAwt and resulted in a collapse of the doublet HA1 band to a single product (Fig. 3B). Formation of the single HA1 product by PNGase F revealed incomplete glycosylation of the insertion site, where only ∼50% is found glycosylated.
The three histidine residues found in the H7 HA insertion affect the pH of fusion
Histidine residues often play important roles in regulating the activity of viral fusion proteins, being located in the vicinity of positively charged residues in the pre-fusion conformation, and subsequently forming salt bridges with negatively charged residues at low pH, in the post-fusion conformation (Chen et al., 1998, Stevens et al., 2004). For the influenza H5 virus, previous reports have demonstrated that histidine residues of the HA1 and HA2 subunits that are distal to the fusion peptide play a critical role in fusion. These histidine residues interact with a tryptophan residue found in the fusion peptide, and are thought to have a destabilizing effect once the pH has lowered to the point where the histidines become positively charged (Chen et al., 1998, Stevens et al., 2004). Interestingly, amino acid substitution of histidines that appear to be involved in triggering fusion resulted in both a stabilizing and destabilizing effect depending on which amino acid the histidine was substituted with, further demonstrating the importance of these residues in fusion (Daniels et al, 1985, Thoennes et al., 2008). Because of the known importance of histidines in regulating fusion activity, we examined the effects of the histidine residues within the unique H7 cleavage site insertion on the pH of fusion for A/equine/Cornell/16/1974 HA, by expressing HA and monitoring cell–cell fusion over a pH range. We examined wild type H7 HA, as well as mutant forms where the histidines were substituted with alanines. Fusion of the wild type or mutant equine H7 did not induce cell–cell fusion over the pH range of 5.4–5.8 for any of the H7 HA forms (data not shown) Therefore, we examined fusion of the wild type H7 HA, along with individual and sequential mutations of the histidine residues, and an insert deletion mutant of the H7 HA at a lower pH range of 4.8–5.2, under physiological salt conditions (Fig. 4).
Fig. 4.
The nine amino acid insertion of the equine H7 HA affects the pH of fusion. Immunofluorescence staining of EQHAwt, EQHAH338A, EQHAEQHAH343A/H344A (EQHAHH-AA), and EQHAEQHAH338A/H343A/H344A (EQHAHHH–AAA) HA over a pH range of 4.8–5.2 in three different salt buffers. The cell surface HA was stained with Alexa fluor 488 (green) and the nucleus is stained with DAPI (blue).
For the wild type H7 HA, we observed only a low level of syncytia at either pH 5.2 or pH 5.0. At pH 4.8 however, a marked increase in fusion efficiency was observed, as determined by visualization of syncytia formation (Fig. 4). As for the histidine mutants, efficient fusion was observed in each case over the pH range 4.8–5.2, under physiological salt conditions. In addition, the A/equine/Cornell/74 NA was co-expressed with the wild type HA to determine any effects on fusion, since previous reports demonstrated that fusion was enhanced in the presence of NA (Reed et al., 2010, Su et al., 2009). The fusion pH appeared to be unaffected by NA co-expression when compared to the fusion profile of HA alone over the pH range of 4.8–5.2 (data not shown). Thus, the histidine residues of the eleven amino acid insertion appear to be involved in triggering fusion, since a destabilizing effect was observed by substituting histidine with alanine.
We also examined fusion in a low salt buffer, since fusion can be affected by buffer conditions (Korte et al., 2007). In all cases, no fusion was observed in the low salt buffer at any pH tested (Fig. 4). However, fusion was recovered for each of the HA forms by the addition of calcium at each pH examined (Fig. 4).
Addition of the eleven amino acid insertion confers intracellular cleavage of a LPAI H7N3 HA
To determine whether the addition of both the polybasic site and insertion would promote intracellular cleavage of HA from a low pathogenicity influenza virus, the monobasic cleavage site of the avian H7N3 HA (A/turkey/Italy/2002) was replaced with a tetrabasic cleavage site, as well as a tetrabasic cleavage site followed by the addition of the eleven amino acid equine H7N7 insertion. Mutations are summarized in Table 2. Both mutant HAs showed efficient cell surface expression (approximately 80% of wild type; see Supplementary Fig. 1).
Table 2.
Mutants of the influenza A/Turkey/Italy/2002 (H7N3) HA generated in this study.
| H7 HA form | Sequence |
|---|---|
| H7N3 wt | PE - - - - - - - - - - RGLFG |
| H7N3 MF | PE - - - - - - - RKKRGLFG |
| H7N3 MF+I | PENSTHKQLTHHMRKKRGLFG |
The H7N3wt HA was completely trypsin dependent, where no intracellular cleavage was observed (Fig. 5A). In the case of H7N3MF HA, only slight cleavage was observed by replacement of the monobasic cleavage site with a tetrabasic site (Fig. 5A). However, in the case of H7N3MF+I HA, replacement of the monobasic cleavage site with a tetrabasic site coupled with the addition of the insertion resulted in efficient intracellular cleavage (Fig. 5A). A similar effect was observed in the cell–cell fusion assay, where a high degree of syncytia formation was only observed for H7N3MF+I in the absence of exogenous proteases, and furthermore, demonstrated that cleavage produces a fusogenic HA (Fig. 5B). These results would suggest that intracellular cleavage caused by the insertion is not unique to the equine H7 HA, where efficient intracellular cleavage of other HA molecules can be achieved by acquiring the insertion.
Fig. 5.
Addition of a tetrabasic site and the nine amino acid insertion confers intracellular cleavage of the LPAI, H7N3 HA. (A) Western blot analysis of the cleavage of H7N3wt, H7N3MF, and H7N3MF+I HA. (B) Immunofluorescence staining of each form examined in (A). The surface HA was stained with Alexa fluor 488 (green) and the nuclei were stained with DAPI (blue). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Discussion
HA cleavage activation is a critical step in the influenza replication cycle that is driven by host cell proteases. While mutations in several viral genes (e.g. polymerase), as well at the host response, affect viral pathogenicity, a localized infection is generally associated with the influenza virus subtypes containing a monobasic site, since cleavage activation occurs only in a limited number of organs (Böttcher et al., 2009). However, infection by subtypes containing a polybasic cleavage site are not limited to particular organs due to the ubiquitous nature of the enzyme involved in cleavage, and therefore, have the potential to be highly pathogenic (Peiris et al., 2007). Acquisition of a polybasic site is essential for high pathogenicity in poultry. Furthermore, a high occurrence of avian viruses that have directly transmitted to humans and are associated with a high mortality rate contain the polybasic cleavage site (Klenk et al., 2011). Considering the HPAI H7N7 and H5N1 outbreaks (Chan, 2002, Koopmans et al., 2004), it is of the utmost importance to have a complete understanding of the molecular determinants required for efficient intracellular cleavage. Of note, recent cases of H7 influenza in Mexico (i.e. A/chicken/Jalisco/CPA1/2012 (H7N3)) contain a cleavage site with a relatively low potential to be cleaved by furin, but have a peptide insert in the cleavage site that contains a histidine residue (PENPKDRKSRRHRRTR-GLFG; GenBank accession #AFN85519). We consider that equine H7N7 influenza can serve as a model to help determine the factors involved in generation of a highly pathogenic influenza virus in mammals, since it has already established itself in the mammalian population and contains at least one of the requirements of a highly pathogenic influenza virus, i.e. being cleaved intracellularly.
The histidine residues located in the unique, eleven amino insertion of the equine H7N7 HA were investigated to determine if there is a functional role in the pH of fusion, since previous results have demonstrated that histidine residues affect HA fusion. When fusion was examined in a physiological salt buffer, the pH of fusion was decreased when compared to that of a typical HA fusion pH range. However, with even a single His mutation, the pH of fusion increased that resembled the HA fusion profile more typical for influenza HA, e.g. H3N2. Interestingly, no fusion was observed for each of the H7 HA forms examined in a low salt buffer. This result is intriguing since Korte et al. demonstrated that fusion by the H3N2 HA appeared to be enhanced in a low salt buffer, when compared to a physiological salt buffer (Korte et al., 2007). Another interesting observation is that fusion in a low salt buffer was recovered upon the addition of calcium for all HA forms. Previous reports have demonstrated that calcium is involved in membrane fusion, and appears to affect fusion of the H7 HA (Friedrich et al., 2008) (Collins et al., 2012). Considering the previous analysis of the His residues role in fusion (Chen et al., 1998, Stevens et al., 2004), a plausible explanation for this observation is that the three His residues do in fact form a tight interaction with the fusion peptide, and the pKa of these histidine is decreased by their local environment when compared to histidine pockets from other HA subtypes. This scenario will require additional experimentation and would be aided by a crystal structure for equine H7, which is currently lacking. Additionally, both the degree of HA cleavage and the pH at which fusion occurs appear to affect the pathogenicity of the virus (DuBois et al., 2011, Horimoto and Kawaoka, 1997, Reed et al., 2010). Therefore, one other unresolved question is whether this lowered pH of fusion has any impact to the virulence of the virus, albeit a positive or negative effect.
The three histidine residues of the insertion were also examined to determine if they play a role in intracellular cleavage. Sequential mutations of the histidine residues resulted in a sequential loss of intracellular cleavage, where these histidines appear to play an essential role in cleavage. This result in combination with low pH of fusion gives the appearance that the histidine residues have a dual purpose to the activation of HA fusion. However, it is possible that the histidine residues are optimal for either cleavage or fusion and coincidentally have an effect on the other process. We reasoned that the most plausible explanation to the effect on cleavage is that histidine residues interact with the fusion peptide in both the pre-cleavage and post-cleavage states, and by doing so alter the conformation of the cleavage site region, allowing for accessibility by subtilisin-like proteases. Ultimately, obtaining a crystal structure would shed light on the role of the histidines in both fusion and cleavage, and determine if all of the histidine residues are present post-cleavage and not removed by a carboxypeptidase known to trim back the new C-terminus of HA1 (Garten and Klenk, 1983). Determining the structure of the equine, H7 HA will be a future aim of the research.
The glycosylation site asparagine located in the insertion was also found to be essential for intracellular cleavage, and furthermore, was completely insensitive to trypsin treatment when mutated. Intracellular cleavage of the equine HA does not appear, however, to be solely dependent on the presence of a N-linked glycan, since both the glycosylated and non-glycosylated forms of the insertion glycosylation site were efficiently cleaved. Furthermore, in place of the NST sequence, isolates of this virus were found to contain either an APA or NSI sequence, which deviates from the glycosylation site consensus sequence (N–X–S/T). One possibility is that the glycosylation increases cleavage site accessiblity by altering the local conformation, and when absent, the asparagine residue causes this same change in local conformation by hydrogen bonding with neighboring residues, causing a structural turn. In the case of A/equine/Prague/1956, it is possible that the known propensity of proline to kink alpha-helices (Branden and Tooze, 1999) results in the APA sequence conferring a similar structural turn adjacent to the cleavage site. It is also possible that mutation of the asparagine residue affects the proper folding of HA in the endoplasmic reticulum (ER), resulting in insensitivity to proteolytic cleavage. We consider that the additional glycan in A/equine/Cornell/1974 (as compared to A/equine/Prague/1956) arose as the result of altered antigenicity as the virus evolved in horses.
Both a tetrabasic site and the eleven amino acid insertion were added to the LPAI, H7N3 HA to determine if intracellular cleavage can be gained, or that the insertion-dependent, intracellular cleavage is unique to the equine H7 HA. Efficient intracellular cleavage of the LPAI, H7N3 HA was only observed by the addition of a tetrabasic site and insertion, demonstrating that the insertion-dependent, intracellular cleavage is not limited to the equine HA. Khatchikian et al. demonstrated that extensive passages of a LPAI H7N3 in the absence of trypsin resulted in acquisition of an 18-residue insertion at the cleavage site by recombination with ribosomal RNA (Khatchikian et al., 1989). Other events, such as recombination with the nucleoprotein (NP) or matrix (M) proteins, or polymerase slippage have occurred that resulted in the addition of amino acids adjacent to the HA cleavage site (Pasick et al., 2005, Perdue, 2001) (García et al., 1996). The equine H7 naturally acquired an insertion, most likely by recombination, and appears to have done so in horses. However, the origin of the recombinant insert remains a mystery. Previous analysis of either sense of the nucleotide sequence in the nucleotide databases failed to identify significant homology (Gibson et al., 1992), and re-analysis by our laboratory has confirmed a lack of homology with currently available database sequences. Therefore, it is possible that emergence of a HPAI, at least in part, can occur by novel insertions in the cleavage site and should be considered in surveillance studies.
To conclude, uncertainty still remains in the underlying factors that contribute to the conversion of a LPAI to a highly pathogenic strain. One factor that is well understood is that acquisition of a polybasic cleavage site in the HA molecule is essential for conversion to a HPAI. Additional factors influencing HA cleavage activation are: (1) distal glycosylation sites that influence protease accessibility to the cleavage site, and (2) distal histidine residues that modulate conformational changes post-cleavage. The number of basic residues found within the polybasic site also affects the efficiency of intracellular cleavage, where a tetrabasic site offers only limited cleavage. We show here that acquisition of a single unique insertion by the equine H7 HA, containing a glycosylation site and three histidine residues, has allowed for efficient intracellular cleavage activation despite having only a tetrabasic cleavage site. While not necessarily impacting the tropism or pathogenesis of equine H7N7 in its natural host, the results described here represent a new alternative mechanism by which an H7N7 influenza virus can acquire efficient intracellular cleavage by including a distinct regulatory module adjacent to the cleavage site, and thus increase its potential to become highly pathogenic.
Materials and methods
Strains and culture media
Vero cells (American Type Culture Collection) were maintained in Dulbecco modified Eagle medium (DMEM) (Cellgro) supplemented with 10% fetal bovine serum (Gibco), 100 units/mL penicillin (Cellgro), and 10 units/mL streptomycin (Cellgro). Plasmids encoding the A/equine/Cornell/1974 (H7N7) HA and A/turkey/Italy/2002 (H7N3) HA were synthesized by GeneArt and subcloned into a pEF4 plasmid. Trypsin (TPCK-treated) from bovine pancreas was obtained from Pierce.
Mutagenesis
The gene encoding for both the A/equine/Cornell/1974 (H7N7) HA and A/turkey/Italy/2002 (H7N3) HA were mutated by site-directed mutagenesis. Successful mutation of the desired residues and the absence of undesired mutations were assessed by sequencing the entire gene of each HA.
Cell–cell fusion assay
Vero cells were grown in 24 well plates containing a glass cover slip. The cells were then transfected with 0.8 μg of A/equine/Cornell/1974 HA, mutants thereof, A/turkey/Italy HA, mutants thereof, and the A/Aichi/68 HA-expressing plasmids using Lipofectamine 2000 and incubated for 12 h at 37 °C. Cells were also transfected with 0.4 μg of A/equine/Cornell/1974 HA and 0.4 μg of A/equine/Cornell/1974 NA using Lipofectamine 2000 and incubated for 12 h at 37 °C for co-expression of influenza surface proteins. The cells were then washed with phosphate buffered saline (PBS) and treated with 6 μg/mL of trypsin for 10 min at 37 °C. The cells were then washed with PBS and incubated in a low salt, calcium buffer (50 mM MES, 10 mM CaCl2), low salt buffer (10 mM K2PO4), and physiological salt buffer (5 mM HEPES, 5 mM MES, 5 mM succinate, 150, mM NaCl) in 0.2 unit increments of a pH range of 4.8–5.8 for 3 min at 37 °C. The cells were then washed with PBS and incubated for 1 h in DMEM at 37 °C. Cell–cell fusion was then analyzed by immunofluorescence staining using anti-A/equine/Prague/56 (H7N7) HA antibody (NIAID Biodefense & Emerging Infections Research Resource Repository) along with Alexa Fluor 488 anti-goat antibody (Invitrogen). The nuclei were stained with Hoechst 33258 (Invitrogen). The percent syncytia formation was determined by dividing the total number of transfected cells involved in syncytia formation by the total number of cells in a specified region.
Analysis of HA cleavage
Vero cells were grown in 12 well plates and transfected with 1 μg of HA-expressing plasmids using Lipofectamine 2000 and incubated for 12 h at 37 °C. The cells were then washed with PBS and incubated with 9 μg/mL of trypsin (when indicated) for 10 min at 37 °C. The cells were processed by cell surface biotinylation as described in Sun et al. (Sun et al., 2010). Briefly, the free lysines of cell surface proteins were biotinylated using NHS-biotin (Pierce) and isolated by overnight incubation with streptavidin agarose resin (Pierce). The streptavidin resin containing the cell surface proteins was heated in protein-loading dye at 95 °C for 5 min, and the resin was removed by centrifugation. HA cleavage was than analyzed by western blot using the antibodies described above coupled with an anti-goat HRP antibody (Invitrogen). To determine if the wild type, HA1 cleavage products were a mixture of glycosylated and non-glycosylated protein, HA was heated for 10 min at 80 °C in buffer A (1% SDS, 50 mM TRIS, pH 7.5) and both 1% NP-40 and 10 U/mL of PNGase F were subsequently added. The reaction was incubated at 37 °C for 4 h and assessed by western blot using the antibodies described above. The percent HA cleavage and cell surface expression was determined by densitometry.
Supplementary Material
Acknowledgments
The authors thank Ruth Collins, Susan Daniel, Brian Crane and all members of the Whittaker Lab for helpful discussions. We would also like to thank Judy Appleton for advice, and provision of antibodies and viruses. These studies were funded by the United States Department of Health and Human Services contract HHSN266200700008C (NIAID Centers of Excellence for Influenza Research and Surveillance). Work in the author's laboratory was also supported by a research grant from the Harry M. Zweig Memorial Fund for Equine Research.
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.virol.2012.09.004.
References
- Air GM. Sequence relationships among the hemagglutinin genes of 12 subtypes of influenza A virus. Proc Natl Acad Sci. 1981;78:7639–7643. doi: 10.1073/pnas.78.12.7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleton JA, Gagliardo LF. Diversity of the antibody responses produced in ponies and mice against the equine influenza A virus H7 haemagglutinin. J Gen Virol. 1992;73:1569–1573. doi: 10.1099/0022-1317-73-6-1569. [DOI] [PubMed] [Google Scholar]
- Böttcher E, Freuer C, Steinmetzer T, Klenk HD, Garten W. MDCK cells that express proteases TMPRSS2 and HAT provide a cell system to propagate influenza viruses in the absence of trypsin and to study cleavage of HA and its inhibition. Vaccine. 2009;27:6324–6329. doi: 10.1016/j.vaccine.2009.03.029. [DOI] [PubMed] [Google Scholar]
- Böttcher E, Matrosovich T, Beyerle M, Klenk HD, Garten W, Matrosovich M. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J Virol. 2006;80:9896–9898. doi: 10.1128/JVI.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baigent SJ, McCauley JW. Influenza type A in humans, mammals and birds: determinants of virus virulence, host-range and interspecies transmission. BioEssays. 2003;25:657–671. doi: 10.1002/bies.10303. [DOI] [PubMed] [Google Scholar]
- Bertram S, Glowacka I, Blazejewska P, Soilleux E, Allen P, Danisch S, Steffen I, Choi SY, Park Y, Schneider H, et al. TMPRSS2 and TMPRSS4 facilitate trypsin-independent spread of influenza virus in Caco-2 cells. J Virol. 2010a;84:10016–10025. doi: 10.1128/JVI.00239-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S, Glowacka I, Steffen I, Kühl A, Pöhlmann S. Novel insights into proteolytic cleavage of influenza virus hemagglutinin. Rev Med Virol. 2010b;20:298–310. doi: 10.1002/rmv.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos MEH, te Beest DE, van Boven M, van Beest Holle MRDR, Meijer A, Bosman A, Mulder YM, Koopmans MPG, Stegeman A. High probability of Avian influenza virus (H7N7) transmission from poultry to humans active in disease control on infected farms. J Infect Dis. 2010;201:1390–1396. doi: 10.1086/651663. [DOI] [PubMed] [Google Scholar]
- Branden CL, Tooze J. Introduction to Protein Structure. 2. Garland; New York: 1999. [Google Scholar]
- Chaipan C, Kobasa D, Bertram S, Glowacka I, Steffen I, Solomon Tsegaye T, Takeda M, Bugge TH, Kim S, Park Y, et al. Proteolytic activation of the 1918 influenza virus hemagglutinin. J Virol. 2009;83:3200–3211. doi: 10.1128/JVI.02205-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PKS. Outbreak of Avian influenza A(H5N1) virus infection in Hong Kong in 1997. Clin Infect Dis. 2002;34:S58–S64. doi: 10.1086/338820. [DOI] [PubMed] [Google Scholar]
- Chen J, Lee KH, Steinhauer DA, Stevens DJ, Skehel JJ, Wiley DC. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell. 1998;95:409–417. doi: 10.1016/s0092-8674(00)81771-7. [DOI] [PubMed] [Google Scholar]
- Collins RN, Holtz RW, Zimmerberg J. The biophysics of membrane fusion. Compr Biophys. 2012;5:244–260. [Google Scholar]
- Daly JM, Lai ACK, Binns MM, Chambers TM, Barrandeguy M, Mumford JA. Antigenic and genetic evolution of equine H3N8 influenza A viruses. J Gen Virol. 1996;77:661–671. doi: 10.1099/0022-1317-77-4-661. [DOI] [PubMed] [Google Scholar]
- Daniels RS, Downie JC, Hay AJ, Knossow M, Skehel JJ, Wang ML, Wiley DC. Fusion mutants of the influenza virus hemagglutinin glycoprotein. Cell. 1985;40:431–439. doi: 10.1016/0092-8674(85)90157-6. [DOI] [PubMed] [Google Scholar]
- DuBois RM, Zaraket H, Reddivari M, Heath RJ, White SW, Russell CJ. Acid stability of the hemagglutinin protein regulates H5N1 influenza virus pathogenicity. PLoS Pathog. 2011;7:e1002398. doi: 10.1371/journal.ppat.1002398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich R, Groffen AJ, Connell E, van Weering JRT, Gutman O, Henis YI, Davletov B, Ashery U. DOC2B acts as a calcium switch and enhances vesicle fusion. J Neurosci. 2008;28:6794–6806. doi: 10.1523/JNEUROSCI.0538-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García M, Crawford JM, Latimer JW, Rivera-Cruz E, Perdue ML. Heterogeneity in the haemagglutinin gene and emergence of the highly pathogenic phenotype among recent H5N2 avian influenza viruses from Mexico. J Gen Virol. 1996;77:1493–1504. doi: 10.1099/0022-1317-77-7-1493. [DOI] [PubMed] [Google Scholar]
- Garten W, Bosch FX, Linder D, Rott R, Klenk HD. Proteolytic activation of the influenza virus hemagglutinin: the structure of the cleavage site and the enzymes involved in cleavage. Virology. 1981;115:361–374. doi: 10.1016/0042-6822(81)90117-3. [DOI] [PubMed] [Google Scholar]
- Garten W, Klenk HD. Characterization of the carboxypeptidase involved in the proteolytic cleavage of the influenza haemagglutinin. J Gen Virol. 1983;64:2127–2137. doi: 10.1099/0022-1317-64-10-2127. [DOI] [PubMed] [Google Scholar]
- Gibson CA, Daniels RS, Oxford JS, McCauley JW. Sequence analysis of the equine H7 influenza virus haemagglutinin gene. Virus Res. 1992;22:93–106. doi: 10.1016/0168-1702(92)90037-a. [DOI] [PubMed] [Google Scholar]
- Gohrbandt S, Veits J, Breithaupt A, Hundt J, Teifke JP, Stech O, Mettenleiter TC, StechJr TC. H9 avian influenza reassortant with engineered polybasic cleavage site displays a highly pathogenic phenotype in chicken. J Gen Virol. 2011;92:1843–1853. doi: 10.1099/vir.0.031591-0. [DOI] [PubMed] [Google Scholar]
- Hamilton BS, Gludish DWJ, Whittaker GR. Cleavage activation of the human-adapted influenza virus subtypes by matriptase reveals both subtype-and strain-specificity. J Virol. 2012;86:10579–10586. doi: 10.1128/JVI.00306-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward JJ, Dubovi EJ, Scarlett JM, Janeczko S, Holmes EC, Parrish CR. Microevolution of Canine influenza virus in shelters and its molecular epidemiology in the United States. J Virol. 2010;84:12636–12645. doi: 10.1128/JVI.01350-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horimoto T, Kawaoka Y. Biologic effects of introducing additional basic amino acid residues into the hemagglutinin cleavage site of a virulent avian influenza virus. Virus Res. 1997;50:35–40. doi: 10.1016/s0168-1702(97)00050-6. [DOI] [PubMed] [Google Scholar]
- Horimoto T, Kawaoka Y. Influenza: lessons from past pandemics, warnings from current incidents. Nat Rev Micro. 2005;3:591–600. doi: 10.1038/nrmicro1208. [DOI] [PubMed] [Google Scholar]
- Kaleta EF, Rülke CPA. Avian Influenza. Blackwell Publishing Ltd; 2008. The Beginning and Spread of Fowl Plague (H7 High Pathogenicity Avian Influenza) across Europe and Asia (1878-1955) pp. 145–189. [Google Scholar]
- Kawaoka Y. Equine H7N7 influenza A viruses are highly pathogenic in mice without adaptation: potential use as an animal model. J Virol. 1991;65:3891–3894. doi: 10.1128/jvi.65.7.3891-3894.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatchikian D, Orlich M, Rott R. Increased viral pathogenicity after insertion of a 28S ribosomal RNA sequence into the haemagglutinin gene of an influenza virus. Nature. 1989;340:156–157. doi: 10.1038/340156a0. [DOI] [PubMed] [Google Scholar]
- Klenk HD, Garten W, Matrosovich M. Molecular mechanisms of inter species transmission and pathogenicity of influenza viruses: lessons from the 2009 pandemic. BioEssays. 2011;33:180–188. doi: 10.1002/bies.201000118. [DOI] [PubMed] [Google Scholar]
- Knipe D, Howley P, Griffin D, Lamb R, Martin M, Roizman B, Straus S. Fields Virology. 5th. Philidelphia: Lippincott Williams & Wilkins; 2007. [Google Scholar]
- Koopmans M, Wilbrink B, Conyn M, Natrop G, van der Nat H, Vennema H, Meijer A, van Steenbergen J, Fouchier R, Osterhaus A, et al. Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in the Netherlands. The Lancet. 2004;363:587–593. doi: 10.1016/S0140-6736(04)15589-X. [DOI] [PubMed] [Google Scholar]
- Korte T, Ludwig K, Huang Q, Rachakonda P, Herrmann A. Conformational change of influenza virus hemagglutinin is sensitive to ionic concentration. Eur Biophys J. 2007;36:327–335. doi: 10.1007/s00249-006-0116-0. [DOI] [PubMed] [Google Scholar]
- Murcia PR, Wood JLN, Holmes EC. Genome-scale evolution and phylodynamics of Equine H3N8 influenza A virus. J Virol. 2011;85:5312–5322. doi: 10.1128/JVI.02619-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olusa TAO, Adegunwa AK, Aderonmu AA, Adeyefa CAO. Serologic evidence of Equine H7 influenza in polo horses in Nigeria. Sci World J. 2010;5:17–19. [Google Scholar]
- Pasick J, Handel K, Robinson J, Copps J, Ridd D, Hills K, Kehler H, Cottam-Birt C, Neufeld J, Berhane Y, et al. Intersegmental recombination between the haemagglutinin and matrix genes was responsible for the emergence of a highly pathogenic H7N3 avian influenza virus in British Columbia. J Gen Virol. 2005;86:727–731. doi: 10.1099/vir.0.80478-0. [DOI] [PubMed] [Google Scholar]
- Peiris JSM, de Jong MD, Guan Y. Avian influenza virus (H5N1): a threat to human health. Clin Microbiol Rev. 2007;20:243–267. doi: 10.1128/CMR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdue ML. Avian Influenza Viruses. eLS: John Wiley & Sons, Ltd; 2001. [Google Scholar]
- Reed ML, Bridges OA, Seiler P, Kim JK, Yen HL, Salomon R, Govorkova EA, Webster RG, Russell CJ. The pH of activation of the hemagglutinin protein regulates H5N1 influenza virus pathogenicity and transmissibility in ducks. J Virol. 2010;84:1527–1535. doi: 10.1128/JVI.02069-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RJ, Gamblin SJ, Haire LF, Stevens DJ, Xiao B, Ha Y, Skehel JJ. H1 and H7 influenza haemagglutinin structures extend a structural classification of haemagglutinin subtypes. Virology. 2004;325:287–296. doi: 10.1016/j.virol.2004.04.040. [DOI] [PubMed] [Google Scholar]
- Senne DA, Panigrahy B, Kawaoka Y, Pearson JE, Süss J, Lipkind M, Kida H, Webster RG. Survey of the hemagglutinin (HA) cleavage site sequence of H5 and H7 Avian influenza viruses: amino acid sequence at the HA cleavage site as a marker of pathogenicity potential. Avian Dis. 1996;40:425–437. [PubMed] [Google Scholar]
- Shinya K, Suto A, Kawakami M, Sakamoto H, Umemura T, Kawaoka Y, Kasai N, Ito T. Neurovirulence of H7N7 influenza A virus: brain stem encephalitis accompanied with aspiration pneumonia in mice. Arch Virol. 2005;150:1653–1660. doi: 10.1007/s00705-005-0539-4. [DOI] [PubMed] [Google Scholar]
- Shinya K, Watanabe S, Ito T, Kasai N, Kawaoka Y. Adaptation of an H7N7 equine influenza A virus in mice. J Gen Virol. 2007;88:547–553. doi: 10.1099/vir.0.82411-0. [DOI] [PubMed] [Google Scholar]
- Sovinova O, Tumova B. Isolation of a virus causing respiratory disease in horses. Acta Virol. 1958;2:52–61. [PubMed] [Google Scholar]
- Steinhauer DA. Role of Hemagglutinin cleavage for the pathogenicity of influenza virus. Virology. 1999;258:1–20. doi: 10.1006/viro.1999.9716. [DOI] [PubMed] [Google Scholar]
- Stevens J, Corper AL, Basler CF, Taubenberger JK, Palese P, Wilson IA. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science. 2004;303:1866–1870. doi: 10.1126/science.1093373. [DOI] [PubMed] [Google Scholar]
- Su B, Wurtzer Sb, Rameix-Welti MA, Dwyer D, van der Werf S, Naffakh N, Clavel Fo, Labrosse Ba. Enhancement of the influenza A hemagglutinin (HA)-mediated cell–cell fusion and virus entry by the viral neuraminidase (NA) PLoS ONE. 2009;4:e8495. doi: 10.1371/journal.pone.0008495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Tse LV, Ferguson AD, Whittaker GR. Modifications to the hemagglutinin cleavage site control the virulence of a neurotropic H1N1 influenza virus. J Virol. 2010;84:8683–8690. doi: 10.1128/JVI.00797-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi K, Shaughnessy MA, Lamb RA. Influenza virus M2 protein ion channel activity is not required to maintain the Equine-1 hemagglutinin in its native form in infected cells. Virology. 1994;202:1007–1011. doi: 10.1006/viro.1994.1428. [DOI] [PubMed] [Google Scholar]
- Taubenberger JK. Influenza virus hemagglutinin cleavage into HA1, HA2: no laughing matter. Proc Natl Acad Sci. 1998;95:9713–9715. doi: 10.1073/pnas.95.17.9713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoennes S, Li ZN, Lee BJ, Langley WA, Skehel JJ, Russell RJ, Steinhauer DA. Analysis of residues near the fusion peptide in the influenza hemagglutinin structure for roles in triggering membrane fusion. Virology. 2008;370:403–414. doi: 10.1016/j.virol.2007.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timoney PJ. Equine influenza. Comp Immunol Microbiol Infect Dis. 1996;19:205–211. doi: 10.1016/0147-9571(96)00006-9. [DOI] [PubMed] [Google Scholar]
- Walker JA, Kawaoka Y. Importance of conserved amino acids at the cleavage site of the haemagglutinin of a virulent avian influenza A virus. J Gen Virol. 1993;74:311–314. doi: 10.1099/0022-1317-74-2-311. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Ibrahim MS, Suzuki Y, Ikuta K. The changing nature of Avian influenza A virus (H5N1) Trends Microbiol. 2012;20:11–20. doi: 10.1016/j.tim.2011.10.003. [DOI] [PubMed] [Google Scholar]
- Webster RG, Rott R. Influenza virus a pathogenicity: the pivotal role of hemagglutinin. Cell. 1987;50:665–666. doi: 10.1016/0092-8674(87)90321-7. [DOI] [PubMed] [Google Scholar]
- Zhang X, Fugére M, Day R, Kielian M. Furin processing and proteolytic activation of semliki forest virus. J Virol. 2003;77:2981–2989. doi: 10.1128/JVI.77.5.2981-2989.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.