Abstract
Background
Organ confined prostate cancer (PCa) can be cured by radical retropubic prostatectomy (RRP); however, some tumors will still recur. Current tools fail to identify patients at risk of recurrence. Glutathione-S-Transferases (GSTs) are involved in the metabolism of carcinogens, hormones and drugs. Thus, genetic polymorphisms that modify the GSTs activities may modify the risk of PCa recurrence.
Methods
We retrospectively recruited Argentine PCa patients treated with RRP to study the association between GSTs polymorphisms and PCa biochemical relapse after RRP. We genotyped germline DNA in 105 patients for: GSTP1 c.313 A>G (p.105 Ile>Val, rs1695) by PCR-RFLP; and GSTT1 null and GSTM1 null polymorphisms by multiplex-PCR. Kaplan-Meier curves and Cox proportional hazard models were used to evaluate these associations.
Results
Patients with GSTP1 c.313 GG genotype showed shorter biochemical relapse-free survival (BRFS) (p=0.003) and higher risk for recurrence in unadjusted (Hazard Ratio (HR)=3.16, 95% Confidence Interval (95% CI)=1.41–7.06, p=0.005) and multivariate models (HR=3.01, 95% CI=1.13–8.02, p=0.028). We did not find significant associations for GSTT1 and GSTM1 genotypes. In addition, we found shorter BRFS (p=0.010) and increased risk for recurrence for patients having 2 or more risk alleles when we combined the genotypes of the three GSTs in multivariate models (HR=3.06, 95% CI=1.20–7.80, p=0.019).
Conclusions
Our results give support to the implementation of GSTs genotyping for personalized therapies as a novel alternative for PCa management for patients who undergo RRP. This is the first study that examined GST polymorphisms in PCa progression in Argentine men. Replication of our findings in larger cohort is warranted.
Keywords: prostate cancer, biochemical relapse, GST, glutathione-S-transferase, polymorphism
Introduction
Prostate cancer (PCa) is the second most common type of cancer diagnosed in men and the sixth leading cause of cancer-related deaths worldwide. However, the incidence and mortality rates are highly variable between countries and ethnicities.1 In Argentina, PCa is the most frequent type of cancer and the second cause of death from cancer among Argentine men (http://www.msal.gov.ar/inc/equipos_analisis.php;).2
Early detection of PCa has resulted in more men being diagnosed with localized disease. Surgical resection of the entire gland, radical retropubic prostatectomy (RRP), is one therapeutic option with curative purposes for men with organ confined PCa. However, 30–40% of patients will have a biochemical relapse after RRP which may indicate clinical recurrence of aggressive disease. Current clinical indicators of PCa recurrence and mortality after RRP have limited sensitivity and specificity.3–5 Thus, the ability to establish the risk of progression after surgery is of high importance for patient management and to avoid overtreatment.
The Glutathione-S-Transferases (GSTs) are phase II enzymes involved in detoxification of reactive oxygen species, environmental carcinogens, metabolism of steroid hormones, and metabolism of chemotherapeutic agents.6 Different GST isoforms have diverse, but overlapping, substrate specificities and have been shown to be highly expressed in the prostate.7
Genetic polymorphisms that alter the activity of GSTs may affect the level of hormones and xenobiotics within the prostate which, in turn, may alter the risk of PCa development and progression.
Several studies have been conducted to determine the role of GSTs polymorphisms as risk factors for PCa development;8,9 but only few sought to analyze the effect of these polymorphisms on PCa progression, with inconclusive results.10–12 In addition, most studies that analyzed GST polymorphisms have been carried out in Caucasians, few in African-Americans and Asians, and only two were conducted in Hispanics to evaluate the effect of these polymorphisms on the risk PCa development.13,14 Therefore, the role of GSTs in PCa in the Hispanic population is understudied.
This study is the first to analyze genetic polymorphisms in Hispanic PCa patients from Argentina and to evaluate their effect of these variations on PCa progression. We examined the GSTT1 null, GSTM1 null, and GSTP1 c.313 A>G (p.105 Ile>Val) polymorphisms and their potential association with PCa biochemical relapse in a retrospective cohort of Argentine men who underwent RRP.
Materials and methods
Patients
We designed a hospital-based case-case study to determine the association between GSTs polymorphisms and PCa biochemical relapse after RRP. We retrospectively recruited 105 patients histologically diagnosed with PCa from August 2008 to November 2010 at the Hospital de Clínicas José de San Martín, Buenos Aires, Argentina. All patients underwent RRP as their primary therapeutic strategy (date of RRP from December 1998 to July 2010). Patient recruitment, follow-up and maintenance of updated medical records were performed by trained urologists and oncologists. All patients were Argentine citizens, and by definition Hispanics. Most of them had predominant Caucasian ancestry, although as reported for this population, some admixture of Amerindian and African ancestry is to be expected.15
The study protocol was approved by the Institutional Ethical Committee and followed the Ethical Principles enunciated by the Declaration of Helsinki. All patients who agreed to participate in the study signed a written informed consent.
Genotyping
Germline DNA was extracted from peripheral blood. We genotyped three polymorphisms in three GST genes: GSTP1 c.313 A>G (NM_000852.3:c.313A>G; p.105 Ile>Val; rs1695), GSTT1 null, and GSTM1 null. The genotyping of GSTP1 c.313 A>G was performed by PCR-RFLP assay. GSTT1 null and GSTM1 null genotypes were assessed by multiplex-PCR reaction. This method allowed us to discriminate the null genotype (homozygote deletion), determined by the absence of band in the electrophoresis, from the heterozygote and homozygote present genotypes. We called the null genotype when there was an absence of a band for either GSTT1 or GSTM1 with at least one band for one of the two genes being present (internal PCR control). Samples that did not amplify for both genes were repeated twice or three times to discard a PCR failure. These samples were called null for both GSTT1 and GSTM1 only when the following criteria were met: i) all replications were concordant, ii) other samples within the same PCR reaction using the same PCR mix amplified (reaction control), and iii) PCR reactions for double-null samples showed the specific amplicon for other genes (DNA quality control). Details of methods are available online as supplemental information SI1. Genotyping call rates were 98% for GSTP1, 99% for GSTT1 and 98% for GSTM1. All PCR reactions were performed in a DNA Engine™ Thermocycler (Bio-rad, California, USA). PCR reactions and digested products were analyzed by 2% agarose (Genbiotech SRL, Buenos Aires, Argentina) gel electrophoresis in 1× TAE buffer (0.8 M Tris; 0.4 M sodium acetate; 0.04 M EDTA; pH 8.3) and dyed with ethidium bromide (Promega, Wisconsin, USA). Gels were photographed and analyzed with the G-Box system (Syngene, USA) and the Genesnap software (Syngene, USA). Samples that failed were repeated once or twice as needed. Genotyping outputs were read by 2 independent laboratory members, and 10–12% of blindly random selected samples were re-analyzed as quality control of the experiments. The results were considered for the final analyses when there was 100% agreement between the two independent readers, and when there was a 100% concordance between samples and blinded repeats.
Statistical analysis
Biochemical relapse was defined as a rise in serum PSA above 0.2 ng/ml after RRP. Univariate and multivariate analyses were conducted using Cox proportional hazard models to study the association between polymorphisms and PCa biochemical relapse and to estimate Hazard Ratios (HR) and 95% Confidence Intervals (95% CI). Multivariate models included margin involvement of the resected prostate, pathologic Gleason score, pathologic T stage, serum PSA level at diagnosis, family history of PCa in first-degree relatives, smoking status and age at diagnosis as covariates. Smoking status was stratified as follows: i) never smokers: patients that never smoke, ii) former smokers: patients that quit smoking at least 1 year prior PCa diagnosis, and iii) current smokers: patients that smoke at the time of PCa diagnosis or patients that quit smoking no more than 1 year prior PCa diagnosis. To study biochemical-relapse-free survival, time was calculated from date of RP to date of biochemical relapse or last follow up. Kaplan-Meier plots were used to evaluate the association between clinical variables or genotypes and biochemical relapse, and the comparison between groups was done using the log-rank test.
All statistical analyses were carried out using Stata/SE 11.2 (StataCorp. 2009. Stata Statistical Software: Release 11. College Station, TX: StataCorp LP).
Results
The clinico-pathological characteristics of the studied patients are shown in Table 1. Twelve (11.4%) patients were diagnosed with PCa with normal serum PSA levels (≤4 ng/ml). Nearly half presented with a Gleason score <7, and 66% of patients with a combined Gleason score ≥7 showed a 7(3+4) score. Most resected prostates showed tumor-free margins (77.5%) and 50.5% of patients were diagnosed with pT2-stage tumors. One-third of patients developed a relapse during follow-up. The median follow-up time for patients without relapse was 84 months (8–156), and 36 months (3–132) for patients who recurred.
Table 1.
Clinico-pathological characteristics of the study group.
| Variables | N | Percentage |
|---|---|---|
| Total cases | 105 | 100.0 |
| Age at diagnosis (years old) (median, range) | 65 | 49 – 74 |
| PSA at diagnosis (ng/ml) (median, range) | 6.87 | 0.77 – 28.90 |
| ≤4 | 12 | 11.4 |
| >4 – 10 | 63 | 60.0 |
| >10 | 30 | 28.6 |
| Smoking status | ||
| Never smoker | 29 | 27.6 |
| Former smoker | 56 | 53.3 |
| Current smoker | 20 | 19.1 |
| Family History of PCa | ||
| No | 88 | 83.8 |
| Yes | 17 | 16.2 |
| Pathological Gleason score | ||
| 5 | 6 | 5.7 |
| 6 | 46 | 43.8 |
| 7 (3+4) | 35 | 33.3 |
| 7 (4+3) | 13 | 12.4 |
| 8 | 5 | 4.8 |
| Pathological T stage | ||
| pT2 | 50 | 50.5 |
| pT3a | 47 | 47.5 |
| pT3b | 2 | 2.0 |
| missing | 6 | |
| Risk group for biochemical relapsea | ||
| Low | 26 | 26.3 |
| Intermediate | 21 | 21.2 |
| High | 52 | 52.5 |
| missing | 6 | |
| Weight of resected prostate (g) (median, range) | 45 | 10 – 143 |
| Missing data | 14 | |
| Tumor Volume (mm3) (median, range) | 1900 | 12 – 18000 |
| Missing data | 74 | |
| Margin involvement of the resected prostate | ||
| No | 79 | 77.5 |
| Yes | 23 | 22.5 |
| Missing data | 3 | |
| Biochemical Relapse | ||
| No | 70 | 66.7 |
| Yes | 35 | 33.3 |
risk groups were defined according to D’Amico as follows: low risk (PSA<10 ng/ml, pT2 stage and Gleason score ≤ 6), intermediate risk (PSA=10–20 ng/dL and pT2 stage and/or Gleason score 7), high risk (PSA>20 ng/dL or pT3 stage or Gleason score ≥ 8)3.
Association analyses between clinico-pathological variables and biochemical relapse
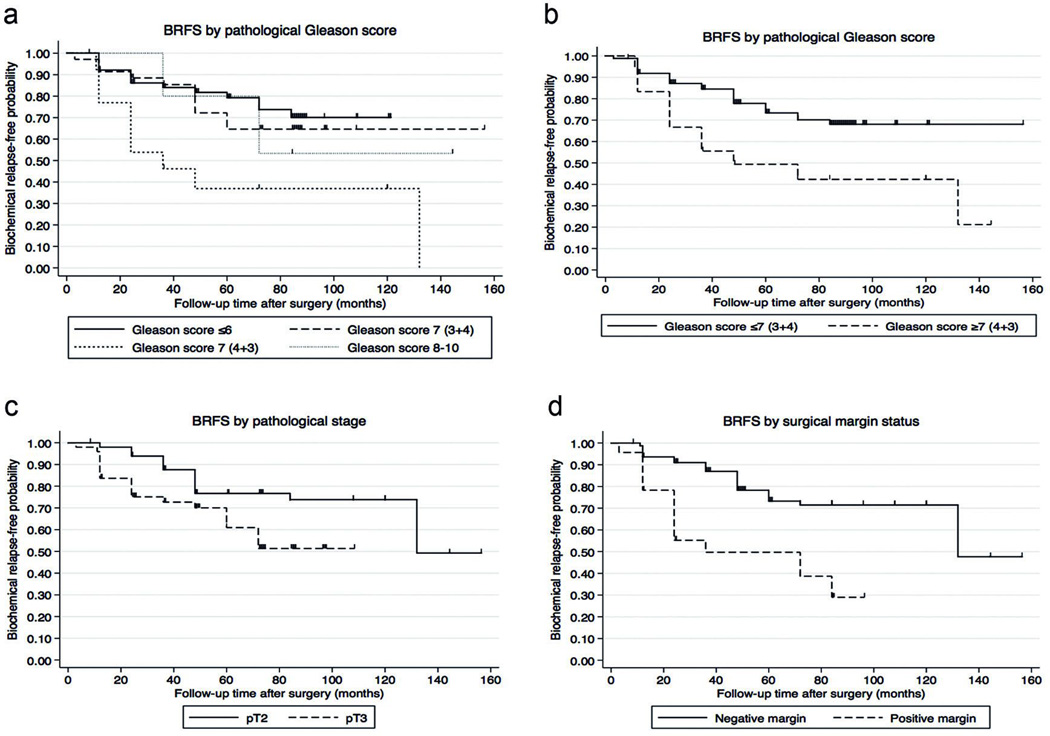
Kaplan-Meier curves were used to study biochemical recurrence-free survival (BRFS) across Gleason score categories, pathologic T stage, and margin involvement of the resected prostate, which are known risk factors for biochemical relapse. Gleason score was evaluated using the following categories: ≤6, 7 (3+4), 7 (4+3), and ≥8 (Figure 1A). Considering that the survival curve of patients with a Gleason score of 7 (3+4) is similar to the curve of patients with a score ≤6, we combined these two groups into a low-risk Gleason score. We also combined the Gleason scores 7 (4+3) and ≥8 into a high-risk Gleason score. Analysis of the dichotomized Gleason score showed a statistically significant difference in BRFS (Figure 1B) and was associated with a nearly 2.5-fold increased risk for biochemical relapse for patients with high-risk Gleason score (HR=2.45, 95% CI=1.18–5.09, p=0.016). Advanced tumors (pT3 stage) were also associated with higher risk for developing a biochemical relapse (HR=2.20, 95% CI=1.07–4.52, p=0.032) and shorter BRFS compared to patients with localized PCa (pT2) (Figure 1C). BRFS was also significantly different based on involvement of surgical margins (Figure 1D), and associated with more than 3-fold higher risk of biochemical relapse for positive surgical margins (HR=3.33, 95% CI=1.67–6.62, p=0.001).
Figure 1. Biochemical-relapse free survival analysis by different clinico-pathological characteristics.
To study biochemical-relapse free survival (BRFS), time was calculated from date of RRP to date of biochemical relapse or last follow up. The figure depicts the Kaplan-Meier survival analysis by: a) Gleason score (log-rank p=0.016), b) dichotomous Gleason score (log-rank p=0.011), c) pathological T stage (log-rank p=0.024), and d) margin status (log-rank p<0.001).
A recent study showed that margin involvement predicts biochemical relapse only in intermediate risk disease (PSA=10–20 ng/ml, stage pT2 and/or Gleason score=7).16 We found that high risk patients (PSA>20 ng/ml, stage pT3 or Gleason score ≥8; stratification according to D’Amico)3 with tumor-positive margins had shorter BRFS (log-rank p=0.026, data not shown). However, we observed no association with the other two groups, which might be due to small numbers.
Overall, the studied patients followed the current clinical criteria used to evaluate PCa biochemical relapse risk.
Analyses of genotypes and risk of biochemical relapse
Figure 2 shows one agarose gel electrophoresis for the multiplex PCR reaction. The genotypes distributions are shown in Table 2, and were similar to those reported for Caucasians and US Hispanics.
Figure 2. Agarose gel electrophoresis of the multiplex PCR reaction for GSTT1 and GSTM1 genotyping.
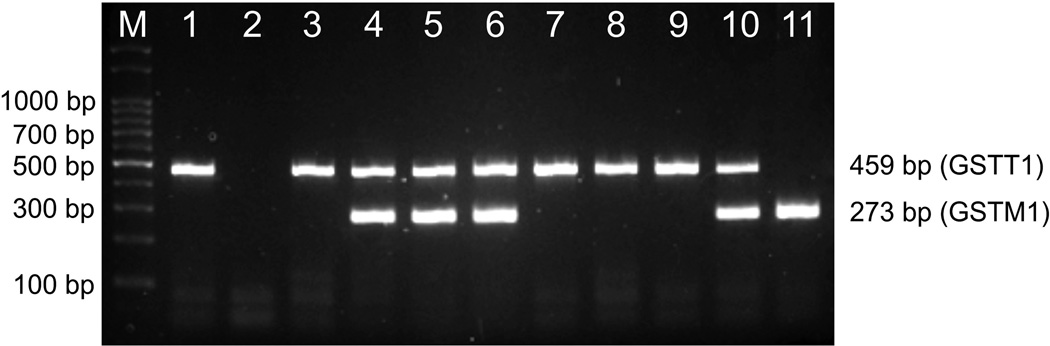
The figure depicts an example of one 2% agarose gel electrophoresis dyed with ethidium bromide used to determine the GSTT1 and GSTM1 genotypes by multiplex PCR. Lanes 1, 3, 7, 8 and 9: GSTT1 present/GSTM1 null; lane 2: GSTT1 null/GSTM1 null; lanes 4, 5, 6 and 10: GSTT1 present/GSTM1 present; lane 11: GSTT1 null/GSTM1 present; M: 100 bp marker (Productos Bio-Lógicos, Buenos Aires, Argentina). Samples that did not amplify for both genes were repeated twice or three times to discard a PCR failure.
Table 2.
Genotypic distribution and comparison with other populations.
| Genotype | N | Percent | Reported percentage in US Hispanicsa |
Reported percentage in Hispanics from Chileb |
Reported percentage in Caucasiansc |
|---|---|---|---|---|---|
| Total cases | 105 | 100.0 | - | - | - |
| GSTT1 null | |||||
| Present | 82 | 78.8 | 86 | 89–94 | 71–85 |
| Null | 22 | 21.2 | 14 | 6–11 | 15–29 |
| Missing data | 1 | ||||
| GSTM1 null | |||||
| Present | 57 | 55.3 | 57 | 64–77 | 52–55 |
| Null | 46 | 44.7 | 43 | 23–36 | 45–48 |
| Missing data | 2 | ||||
| GSTP1 c.313 A>G (p.105 Ile>Val) | |||||
| AA (Ile/Ile) | 51 | 49.5 | 52 | NA | 48–50 |
| AG (Ile/Val) | 41 | 39.8 | 35 | NA | 30–42 |
| GG (Val/Val) | 11 | 10.7 | 13 | NA | 10–22 |
| A (Ile) | 143 | 69.4 | NA | NA | NA |
| G (Val) | 63 | 30.6 | NA | NA | NA |
| Missing data | 2 | ||||
| GSTT1 null + GSTM1 null + GSTP1 c.313 A>G d | |||||
| 0 risk allele | 42 | 42.0 | NA | NA | NA |
| 1 risk allele | 40 | 40.0 | NA | NA | NA |
| 2 risk alleles | 17 | 17.0 | NA | NA | NA |
| 3 risk alleles | 1 | 1.0 | NA | NA | NA |
| Missing data | 5 | ||||
Hispanic genotype frequency ranges were obtained from dbSNP (accessed in August 2012; http://www.ncbi.nlm.nih.gov/snp) for HISP1 population and a report by Block et al..19
Genotype frequency ranges for Caucasians with Spanish and Italian ancestry were obtained from dbSNP (accessed in August 2012; http://www.ncbi.nlm.nih.gov/snp) for CAUC1 and AFD_EUR_PANEL populations, and reports by To-Figueras et al.,20 Ladero et al.,21 Raimondi et al.,22 and Agudo et al..23
for GSTP1 c.313 A>G the recessive model was considered
Abbreviations: NA, not available
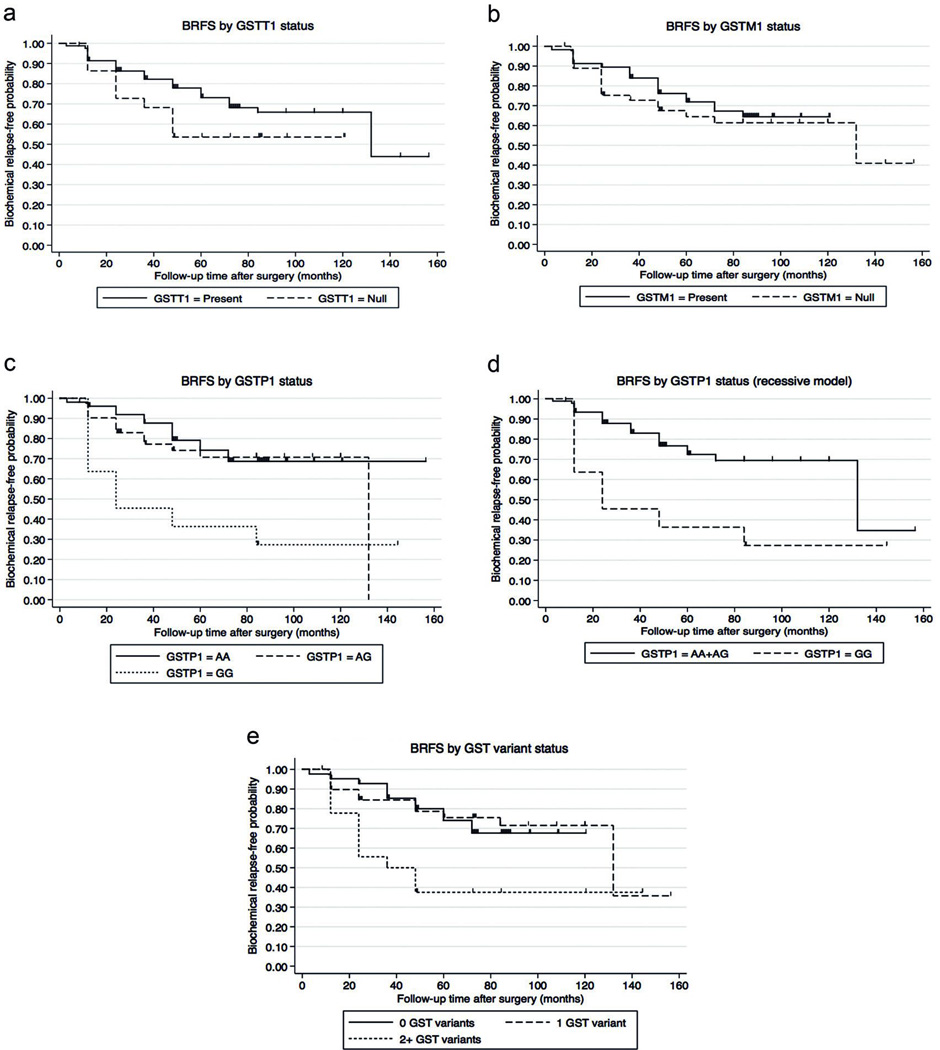
We found that carriers of the null genotypes had slightly shorter BRFS than carriers of the non-null genotypes, albeit these differences were not statistically significant (Figures 3A and 3B for GSTT1 and GSTM1, respectively). Non-statistically significant associations were found between these polymorphisms and biochemical relapse risk in the unadjusted and multivariate Cox models (Table 3 and supplemental information SI2).
Figure 3. Biochemical-relapse free survival analysis by GST genotype.
To study biochemical-relapse free survival (BRFS), time was calculated from date of RRP to date of biochemical relapse or last follow up. The figure depicts the Kaplan-Meier survival analysis by genotypes: a) GSTT1 (log-rank p=0.1480), b) GSTM1 (log-rank p=0.4901), c) GSTP1 co-dominant model (log-rank p<0.010), d) GSTP1 recessive model (log-rank p=0.003), and e) GST combined genotype using the GSTP1 recessive model (log-rank p=0.010).
Table 3.
Unadjusted and multivariate Cox proportional hazard models for biochemical relapse by GST genotypes.
| Genotypes | HR (95% CI) | HRadja (95% CI) |
|---|---|---|
| GSTT1 | ||
| Present | 1.00 (reference) | 1.00 (reference) |
| Null | 1.69 (0.81–3.53) | 2.05 (0.92–4.54) |
| p-value | 0.164 | 0.078 |
| GSTM1 | ||
| Present | 1.00 (reference) | 1.00 (reference) |
| Null | 1.26 (0.64–2.47) | 0.97 (0.47–2.01) |
| p-value | 0.503 | 0.937 |
| GSTP1 c.313 A>G | ||
| AA+AG | 1.00 (reference) | 1.00 (reference) |
| GG | 3.16 (1.41–7.06) | 3.01 (1.13–8.02) |
| p-value | 0.005 | 0.028 |
| GSTP1 c.313 A>G + GSTT1 null + GSTM1 nullb | ||
| 0 risk allele | 1.00 (reference) | 1.00 (reference) |
| 1 risk allele | 0.97 (0.43–2.21) | 0.74 (0.30–1.84) |
| p-value (1 vs 0) | 0.994 | 0.512 |
| 2+ risk alleles | 2.82 (1.23–6.49) | 3.06 (1.20–7.80) |
| p-value (2+ vs 0) | 0.015 | 0.019 |
Statistical significant associations are bolded.
adjusted for margin, Gleason score (low-risk vs high-risk), pathological T stage (pT2 vs pT3), PSA level at diagnosis (≤4 vs >4–10 vs >10), family history of PCa, smoking status (never vs former vs current) and age at diagnosis (continuous variable)
for GSTP1 c.313 A>G the recessive model was considered
A Supplementary Table SI2 is available online showing the results of the other models tested.
Figure 4 shows one agarose gel electrophoresis for the enzymatic digestion of the GSTP1 amplicon. The genotype distribution and allelic frequencies are shown in Table 2, and were similar to those reported for the Caucasian and US Hispanic populations.
Figure 4. Agarose gel electrophoresis of GSTP1 amplicons digested with Alw26I restriction enzyme.
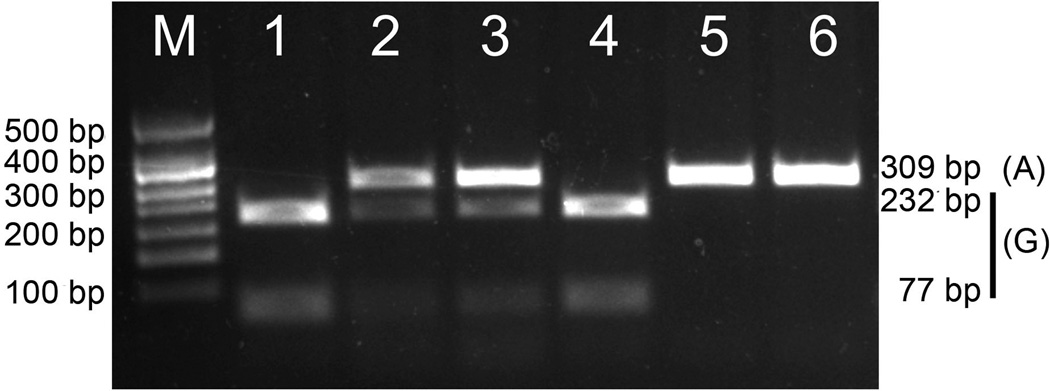
The figure shows an example of one 2% agarose gel electrophoresis dyed with ethidium bromide used to determine the GSTP1 genotype by PCR-RFLP. Lanes 1 and 4: c.313 GG; lanes 2 and 3: c.313 AG; lanes 5 and 6: c.313 AA; M: 50 bp marker (Genbiotech, Buenos Aires, Argentina).
For the analysis of the GSTP1 SNP, we first considered an additive model with the c.313 AA genotype (p.105 Ile/Ile, highest enzymatic activity) as reference, the heterozygote genotype as intermediate activity, and the c.313 GG genotype (p.105 Val/Val) as the lowest activity. We observed that carriers of the GSTP1 c.313 GG genotype (p.105 Val/Val) had shorter BRFS when compared to patients with the GSTP1 c.313 AA (p.105 Ile/Ile) and c.313 AG (p.105 Ile/Val, intermediate enzymatic activity) genotypes (Figure 3C). Given that we did not observe meaningful differences between survival of patients with the GSTP1 c.313 AA and c.313 AG genotypes, we considered a recessive model in which a modification of the enzymatic activity and phenotype is only obtainable when two G alleles are present. We found that homozygote G patients had shorter BRFS when compared to patients with the AA or AG combined genotypes (Figure 3D) and was associated with a 3-fold higher risk for recurrence in the unadjusted model (Table 3). This association remained statistically significant when the model was further adjusted for margin status, Gleason score, pT stage, PSA level at diagnosis, family history of PCa, smoking status and age at diagnosis (Table 3 and supplemental information SI2). The estimates did not significantly change when further adjusting for GSTT1 and GSTM1 genotypes (supplemental information SI2).
Because GSTs are enzymes that typically participate in the same metabolism pathways with overlapping substrate specificity, we considered an additive combined score that captures information on the genotypes of the three GSTs. We stratified the genotypes as follows: 0-risk-allele genotype (GSTT1 present, GSTM1 present, and GSTP1 c.313 AA+AG), 1-risk-allele genotype (GSTT1 null, GSTM1 present, and GSTP1 c.313 AA+AG; or GSTT1 present, GSTM1 null, and GSTP1 c.313 AA+AG; or GSTT1 present, GSTM1 present, and GSTP1 c.313 GG), 2-risk-allele genotype (GSTT1 null, GSTM1 null, and GSTP1 c.313 AA+AG; or GSTT1 null, GSTM1 present, and GSTP1 c.313 GG; or GSTT1 present, GSTM1 null, and GSTP1 c.313 GG), and 3-risk-allele genotype (GSTT1 null, GSTM1 null, and GSTP1 c.313 GG). The genotype distribution is shown in Table 2. Only one patient presented the 3-risk-allele genotype; therefore, he was pooled with the 2-risk-allele group. We found that patients who carried 2 or more (2+) GST risk alleles had shorter BRFS compared to patients with 0 or 1 risk allele (Figure 3E). The unadjusted proportional hazard model showed that patients with 2+ risk alleles had a nearly 3-fold increased risk for biochemical relapse compared to patients with 0 risk alleles (Table 3). This association remained statistically significant after adjustment for other potential risk factors in multivariate models (Table 3 and supplemental information SI2).
Discussion
The clinical course of localized PCa is difficult to predict given that men with similar tumor features can experience strikingly diverse outcomes. Clinicians face the struggle of efficiently identifying high-risk patients given the limited accuracy of currently available staging tools for defining patients at risk of progression to lethal disease. Our study is the first study conducted among Argentine patients to examine the association between GSTs polymorphisms and the risk of PCa biochemical relapse. We found that GSTP1 c.313 A>G polymorphism and the combined genotype of three GSTs are associated with the risk of recurrence and the time to recurrence in Argentine men with localized PCa.
Whereas many studies have investigated the role of GSTs polymorphisms as risk factors for PCa development,8,9,13,14 fewer have analyzed the potential role of these polymorphisms on PCa recurrence, progression and PCa-specific death.10–12 In particular, none of these studies were conducted among Hispanics. Agalliu et al. found an increased risk for PCa-specific mortality among Caucasians who have the GSTM1 null genotype after adjustment for potential confounders (HR=3.76, 95% CI=1.59–8.91); however, the small number of PCa-specific deaths limited the power of the study.11 No associations with recurrence and disease progression were found.11 Another study failed to find a statistically significant increased risk for recurrence in Caucasian men for individual polymorphisms (GSTT1 null, GSTM1 null and GSTP1 c.313 A>G) and for the combined genotypes.10 However, an increased risk for biochemical relapse was found for the GSTM1 null genotype among patients with high-grade (Gleason score ≥8) or high stage (stage ≥T3a) tumors.10 In our patient cohort we had few men with Gleason score ≥8; therefore, we were unable to do analyses among high-grade patients. However, overall, our findings are in agreement with the published data given that we found a positive non-significant association between the GSTM1 and GSTT1 null genotypes and risk for recurrence and shorter BRFS. This lack of significance might be partly due to the modest number of patients enrolled in our study.
The GSTP1 c.313 A>G (p.105 Ile>Val) single nucleotide polymorphism (SNP) alters the substrate specificity, activity and thermostability of the GSTpi enzyme.17 Our results also showed that the GSTP1 c.313 A>G (p.105 Ile>Val) SNP is associated with the risk of biochemical relapse, and patients carrying the GSTP1 c.313 GG genotype were at higher risk of recurrence. Two other studies that investigated this polymorphism did not find statistically significant associations with PCa recurrence.10,11 These discrepancies might be partially due to the fact that the latter studies included Caucasian men, whereas our patients are Hispanic. Our results showed that the genotype frequencies among these Argentine men are more similar to Caucasians from Spain and Italy than to US Hispanics and Hispanics from Chile, in agreement with the large proportion of Spanish and Italian ancestry among Argentines from Buenos Aires, from where patients in our study came from. However, Argentines, as all Hispanics, have an admixed genetic background that includes Europeans, Amerindians, and African ancestries.15 Our study did not include genetic ancestry estimation using Ancestry Informative Markers (AIMs); therefore, we were unable to adjust for potential confounding by population admixture. Our study did not include genetic ancestry estimation using ancestry informative markers (AIMs); therefore, we were unable to adjust for potential confounding by population admixture.
We also found that patients with more than two risk alleles of the combined GSTT1, GSTM1 and GSTP1 polymorphisms were at higher risk for developing a biochemical relapse. These results agree with the results reported by Nock et al. who found that Caucasian men with more than two GST risk alleles and more aggressive tumors (Gleason score ≥8 or pT stage ≥3a) had an increased risk for recurrence.10
The primary strength of our study is the inclusion of patients who underwent RRP without neoadjuvant therapy, which avoided the potential confounding effect of other types of treatment. Key limitations are the modest number of patients included, the possible long-survivorship bias due to the retrospective study design, and that eight men were only followed for periods shorter than the median time to relapse (36 months) and might develop a biochemical relapse later on. Moreover, we did not evaluate GSTP1 promoter methylation, which is a frequent epigenetic event in PCa and is suggested to be a predictor for PCa recurrence.18 It was reported that serum GSTP1 hypermethylation was detected in 15% of the patients who recurred.18 Therefore, it might be challenging to clearly determine the role of the GSTP1 SNP in biochemical relapse without considering methylation status.
GSTT1 null, GSTM1 null and GSTP1 c.313 A>G polymorphisms might be relevant predictors of biochemical relapse of PCa in the Argentine population. However, these results need to be validated with a larger patient cohort. Our results give support to the inclusion of molecular markers into clinical practice in an effort to better classify men with localized PCa according to their risk of progression, which may lead to tailored post-surgery therapies and, therefore, will avoid overtreatment.
Supplementary Material
SI1. This file contains detailed information on the genotyping methods, such as, primer sequences, PCR conditions, and enzyme digestion settings (.pdf).
SI2. This file shows the results obtained for all statistical models used to study the association between the polymorphisms and PCa biochemical relapse (.pdf).
Acknowledgments
The authors acknowledge Diego Fernandez-Ortiz for his help with the clinical records. Financial support: Agencia Nacional de Promoción Científica y Tecnológica: PICT 2006-0228 and PICT-RAICES 2006-0367. National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institute of Health (NIH), through Grant Award Number TL1RR031992.
Footnotes
Conflict of interest
The authors declare that they have no competing interests.
Supplementary Information accompanies the paper on the Prostate Cancer and Prostatic Diseases website (http://www.nature.com/pcan).
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Matos EL, Loria DI, Zengarini N, Fernandez MM, Guevel CG, Marconi E, et al. Atlas de mortalidad por cáncer en Argentina 1997–2001. Asociación Argentina de Cáncer: Buenos Aires. 2003 [Google Scholar]
- 3.D'Amico AV, Whittington R, Malkowicz SB, Schultz D, Blank K, Broderick GA, et al. Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. JAMA. 1998;280(11):969–974. doi: 10.1001/jama.280.11.969. [DOI] [PubMed] [Google Scholar]
- 4.Kattan MW, Wheeler TM, Scardino PT. Postoperative nomogram for disease recurrence after radical prostatectomy for prostate cancer. J Clin Oncol. 1999;17(5):1499–1507. doi: 10.1200/JCO.1999.17.5.1499. [DOI] [PubMed] [Google Scholar]
- 5.Ramos CG, Roehl KA, Antenor JA, Humphrey PA, Catalona WJ. Percent carcinoma in prostatectomy specimen is associated with risk of recurrence after radical prostatectomy in patients with pathologically organ confined prostate cancer. The Journal of urology. 2004;172(1):137–140. doi: 10.1097/01.ju.0000132139.40964.75. [DOI] [PubMed] [Google Scholar]
- 6.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 7.Di Paolo OA, Teitel CH, Nowell S, Coles BF, Kadlubar FF. Expression of cytochromes P450 and glutathione S-transferases in human prostate, and the potential for activation of heterocyclic amine carcinogens via acetyl-coA-, PAPS- and ATP-dependent pathways. Int J Cancer. 2005;117(1):8–13. doi: 10.1002/ijc.21152. [DOI] [PubMed] [Google Scholar]
- 8.Mo Z, Gao Y, Cao Y, Gao F, Jian L. An updating meta-analysis of the GSTM1, GSTT1, and GSTP1 polymorphisms and prostate cancer: a HuGE review. Prostate. 2009;69(6):662–688. doi: 10.1002/pros.20907. [DOI] [PubMed] [Google Scholar]
- 9.Ntais C, Polycarpou A, Ioannidis JP. Association of GSTM1, GSTT1, and GSTP1 gene polymorphisms with the risk of prostate cancer: a meta-analysis. Cancer Epidemiol Biomarkers Prev. 2005;14(1):176–181. [PubMed] [Google Scholar]
- 10.Nock NL, Bock C, Neslund-Dudas C, Beebe-Dimmer J, Rundle A, Tang D, et al. Polymorphisms in glutathione S-transferase genes increase risk of prostate cancer biochemical recurrence differentially by ethnicity and disease severity. Cancer Causes Control. 2009;20(10):1915–1926. doi: 10.1007/s10552-009-9385-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agalliu I, Lin DW, Salinas CA, Feng Z, Stanford JL. Polymorphisms in the glutathione Stransferase M1, T1, and P1 genes and prostate cancer prognosis. Prostate. 2006;66(14):1535–1541. doi: 10.1002/pros.20491. [DOI] [PubMed] [Google Scholar]
- 12.Norskov MS, Frikke-Schmidt R, Bojesen SE, Nordestgaard BG, Loft S, Tybjaerg-Hansen A. Copy number variation in glutathione-S-transferase T1 and M1 predicts incidence and 5-year survival from prostate and bladder cancer, and incidence of corpus uteri cancer in the general population. Pharmacogenomics J. 2011;11(4):292–299. doi: 10.1038/tpj.2010.38. [DOI] [PubMed] [Google Scholar]
- 13.Acevedo C, Opazo JL, Huidobro C, Cabezas J, Iturrieta J, Quinones Sepulveda L. Positive correlation between single or combined genotypes of CYP1A1 and GSTM1 in relation to prostate cancer in Chilean people. Prostate. 2003;57(2):111–117. doi: 10.1002/pros.10274. [DOI] [PubMed] [Google Scholar]
- 14.Caceres DD, Iturrieta J, Acevedo C, Huidobro C, Varela N, Quinones L. Relationship among metabolizing genes, smoking and alcohol used as modifier factors on prostate cancer risk: exploring some gene-gene and gene-environment interactions. Eur J Epidemiol. 2005;20(1):79–88. doi: 10.1007/s10654-004-1632-9. [DOI] [PubMed] [Google Scholar]
- 15.Avena S, Via M, Ziv E, Perez-Stable EJ, Gignoux CR, Dejean C, et al. Heterogeneity in genetic admixture across different regions of Argentina. PLoS One. 2012;7(4):e34695. doi: 10.1371/journal.pone.0034695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corcoran NM, Hovens CM, Metcalfe C, Hong MK, Pedersen J, Casey RG, et al. Positive surgical margins are a risk factor for significant biochemical recurrence only in intermediate-risk disease. BJU Int. 2012 doi: 10.1111/j.1464-410X.2011.10868.x. [DOI] [PubMed] [Google Scholar]
- 17.Srivastava SK, Singhal SS, Hu X, Awasthi YC, Zimniak P, Singh SV. Differential catalytic efficiency of allelic variants of human glutathione S-transferase Pi in catalyzing the glutathione conjugation of thiotepa. Arch Biochem Biophys. 1999;366(1):89–94. doi: 10.1006/abbi.1999.1217. [DOI] [PubMed] [Google Scholar]
- 18.Bastian PJ, Palapattu GS, Lin X, Yegnasubramanian S, Mangold LA, Trock B, et al. Preoperative serum DNA GSTP1 CpG island hypermethylation and the risk of early prostate-specific antigen recurrence following radical prostatectomy. Clin Cancer Res. 2005;11(11):4037–4043. doi: 10.1158/1078-0432.CCR-04-2446. [DOI] [PubMed] [Google Scholar]
- 19.Block G, Shaikh N, Jensen CD, Volberg V, Holland N. Serum vitamin C and other biomarkers differ by genotype of phase 2 enzyme genes GSTM1 and GSTT1. Am J Clin Nutr. 2011;94(3):929–937. doi: 10.3945/ajcn.111.011460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.To-Figueras J, Gene M, Gomez-Catalan J, Pique E, Borrego N, Carrasco JL, et al. Genetic polymorphism of glutathione S-transferase P1 gene and lung cancer risk. Cancer Causes Control. 1999;10(1):65–70. doi: 10.1023/a:1008811824890. [DOI] [PubMed] [Google Scholar]
- 21.Ladero JM, Martinez C, Garcia-Martin E, Ropero P, Briceno O, Villegas A, et al. Glutathione S-transferase M1 and T1 genetic polymorphisms are not related to the risk of hepatocellular carcinoma: a study in the Spanish population. Eur J Cancer. 2006;42(1):73–77. doi: 10.1016/j.ejca.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 22.Raimondi S, Paracchini V, Autrup H, Barros-Dios JM, Benhamou S, Boffetta P, et al. Meta- and pooled analysis of GSTT1 and lung cancer: a HuGE-GSEC review. Am J Epidemiol. 2006;164(11):1027–1042. doi: 10.1093/aje/kwj321. [DOI] [PubMed] [Google Scholar]
- 23.Agudo A, Peluso M, Sala N, Capella G, Munnia A, Piro S, et al. Aromatic DNA adducts and polymorphisms in metabolic genes in healthy adults: findings from the EPIC-Spain cohort. Carcinogenesis. 2009;30(6):968–976. doi: 10.1093/carcin/bgp062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SI1. This file contains detailed information on the genotyping methods, such as, primer sequences, PCR conditions, and enzyme digestion settings (.pdf).
SI2. This file shows the results obtained for all statistical models used to study the association between the polymorphisms and PCa biochemical relapse (.pdf).