Abstract
Genetic information in forensic studies is largely limited to CODIS data and the ability to match samples and assign them to an individual. However, there are circumstances, in which a given DNA sample does not match anyone in the CODIS database, and no other information about the donor is available. In this study, we determined 75 SNPs in 24 genes (previously implicated in human or animal pigmentation studies) for the analysis of single- and multi-locus associations with hair, skin, and eye color in 789 individuals of various ethnic backgrounds. Using multiple linear regression modeling, five SNPs in five genes were found to account for large proportions of pigmentation variation in hair, skin, and eyes in our across-population analyses. Thus, these models may be of predictive value to determine an individual’s pigmentation type from a forensic sample, independent of ethnic origin.
Keywords: forensic science, genetics, human, pigmentation, QTL, phenotype
FBI CODIS statistics showed that DNA forensic profiles increased exponentially from 2001–2006 (http://www.fbi.gov/hq/lab/codis/clickmap.htm). However, the DNA forensic profile hits increased linearly, suggesting that the discrepancy between unmatched DNA profiles and hits will continue to increase, as the CODIS DNA database increases. As a means of reducing the pool of suspects of an unmatched DNA profile, ancestry informative markers (AIMs) can indicate the ethnicity of an unknown sample. However, admixed samples are problematic, in that AIMs cannot categorize them into a particular ethnic group. Conceivably, an individual may have a majority of AIMs of one ethnic group, but depending on the amount of admixture, their physical appearance may be different than what might be expected.
While AIMs can enable inferences to be made about the ethnicity of an unknown sample, they do not enable identification of an individual based on physical characteristics within an ethnic group, hence as suggested by Tully (1), phenotype prediction based on genetic tests may be a useful tool in forensic analysis. In this study, we assayed genetic markers associated with pigmentation genes (some of which are also AIMs) to determine how much variation in human pigmentation they account for. These markers have been previously studied within specific ethnic groups. We demonstrate that in ethnically mixed populations, these same markers account for a significant fraction of the pigmentation phenotype.
There are two types of melanin, eumelanin (brown/black) and pheomelanin (yellow/red) that differ in sulfur content (2). Variation in human pigmentation results from differences in the type of melanin and amount of melanin synthesized in specialized vesicles (melanosomes) within pigment cells (melanocytes) and in the size, shape, and export of those melanosomes to the hair and skin (3). Although the rate of synthesis of melanin is much lower in the adult eye than in the skin and hair, additional background color is contained in the iris, making eye color a more complex trait. Genes previously implicated in mediating pigment variation include the melanocortin-1 receptor (MC1R) gene and a gene encoding its inhibitor, agouti signaling protein (ASIP); two genes associated with oculocutaneous albinism, P (OCA2) and SLC45A2 (OCA4, formerly named MATP); and most recently, SLC24A5, the human orthologue of the zebrafish golden gene.
Multiple polymorphisms in the MC1R gene have been linked to red hair and fair skin (4–8), and the ASIP gene, encoding an inhibitor of the MC1R ligand, α-MSH, has been linked to skin pigmentation (9–12). A wide range of OCA phenotypes has been noted for various mutations of OCA2 (13) and SLC45A2, formerly named MATP (14–16). Similarly, a wide range of coat color phenotypes is seen in mice with various mutations in their respective orthologous genes (p, [17,18] and Slc45a2, formerly named uw: [19]). These observations suggested that variations in these genes may be associated with variation in the normal range of human pigmentation. Indeed, gene(s) associated with brown eyes and brown hair were found to map to chromosome 15q, with the OCA2 gene as a prime candidate (20). Population studies have shown that specific polymorphisms in two genes associated with albinism, the OCA2 gene and the SLC45A2 gene, are strongly associated with variations in normal pigmentation of the hair (20–22), skin (22–25), and eyes (22,26–31).
In addition to its effects on pigmentation variation, the SLC45A2 polymorphism rs16891982 (L374F) is a useful marker of population origin (32,33). Another marker of population origin that plays an important role in pigmentation, SLC24A5 (or NCKX5, the human orthologue of the zebrafish golden gene), has been recently identified, with a coding polymorphism divergent between European/Caucasians and other human populations (32,34). SLC24A5 has been shown to biologically affect pigmentation in zebrafish, Danio rerio (34), murine, and cultured human epidermal melanocytes (35).
Alleles of other pigmentation genes such as tyrosinase-related protein 1 (TYRP1) and dopachrome tautomerase (DCT) have been statistically associated with human iris pigmentation (28). In association with certain alleles of other genes, specific alleles of agouti signaling protein have also been associated with human iris color (28) as well as skin color (9–11).
Previous studies have focused on the effects of a limited number of genes on hair or skin color (4,5,24,34). The most comprehensive studies to date have focused on genome-wide-association of SNPs with pigmentation within specific populations (8,12,36,37). In this report, we set out to determine the markers predictive for human pigmentation, independent of ethnic origin. We assayed 75 polymorphisms in 24 genes that were previously implicated in human or animal pigmentation studies for the analysis of single- and multi-locus associations with hair, skin, and eye color in 789 individuals of various ethnic backgrounds. Multiple linear regression (MLR) modeling revealed that a surprisingly small number of markers account for large proportions of pigmentation variation in hair, skin, and eyes in our across-population analyses.
Materials and Methods
Participants
Informed consent was obtained from 791 participants recruited at the University of Arizona between the ages of 18 and 40 with no gray hair and at least 1 inch (measured from the roots) of un-dyed scalp hair. Participants in this study roughly mirror the ethnic composition of the student population. Phenotype data, hair samples, and buccal cell samples were collected from each participant following an Institutional Review Board-approved protocol. Participant’s hair and eye color were independently scored by an investigator; participants also indicated other relevant information, such as tanning response and ethnicity. Buccal cell samples were collected using Catch-All Sample Collection Swabs (Epicentre, Madison, WI) and processed for DNA according to the manufacturer’s protocol.
Melanin Analysis
Approximately 300 scalp hairs (1 cm at the base) were collected from each subject. Hair samples from 186 randomly selected participants were analyzed for both total melanin (combined amount of eumelanin and pheomelanin) content and the two subtypes of melanin, eumelanin and pheomelanin, following a previously published protocol (38,39). As not all genotypes were determined for all SNPs for all individuals, subsets of the 186 samples (54–185) were used in generating the multiple linear regression models discussed later.
Skin Reflectance
Skin reflectance was measured as others (40) have with a portable spectrophotometer (Mercury 1000, Datacolor International, Lawrenceville, NJ) fitted using a 15-mm aperture. This device measures in the visible light range of 400–700 nanometers, at intervals of 20 nm. Three independent reflectance measurements (measured as CIEL, L [lightness] scale of the International Committee on Illumination) of the inner aspect of the upper arm were recorded and averaged for each participant.
Eye Color
Eye color was measured by matching subjects’ eye color to the Kolberg Iris Color Chart® (ocularistsupplies.com) and recorded. Measurements were binned into six different color categories based on another study (41) that correlated pigmentation content to color. Categories were binned one through six (1 = blue, 2 = yellow brown, 3 = green, 4 = packets of brown + blue/green, 5 = brown, and 6 = dark brown/black), where bin 1 corresponded to the least amount of pigmentation, and 6 corresponded to the highest pigmentation.
Genotyping
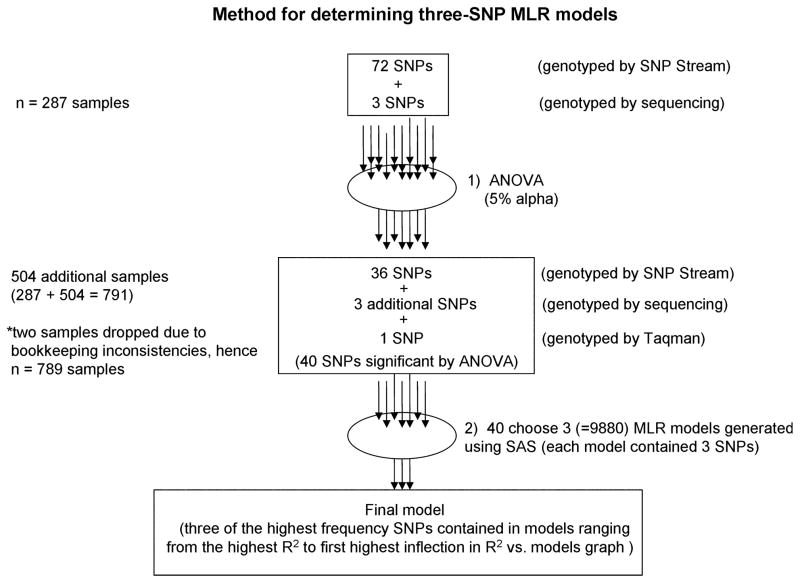
Some SNPs were determined by sequencing the PCR products (Fig. 1). In this case, each PCR consisted of the final concentrations/quantities of the following: 1 ×PCR buffer, 1.5 mM MgCl2, 10 pmol of each primer (forward and reverse; Table 1), 0.25 mM dNTPs, 1 U Taq, and ddH2O to 20 μL. PCR amplification was performed using a PTC-200 Thermal Cycler (MJ Research, Water-town, MA). Thermal cycle program was as follows: 3 min at 95°C, 34 cycles of 30 sec. at each temperature setting (95°C, 55°C, and 72°C), and a final extension of 5 min at 72°C.
FIG. 1.
Flow diagram of method for determining three-SNP multiple linear regression models.
TABLE 1.
Primers used to amplify OCA2 exons 9 and 13 and sequences containing SNPs rs1426654 and rs6058017.
| Primer Name | Primer Sequence | |
|---|---|---|
| rs1800401, and rs1800402 (OCA2) | ||
| forward | MHB581 | (5′-GGGCTGAATTGTTCCATTTG-3′) |
| reverse | MHB582 | (5′-TCTCACGGATCTCAAGCCTC-3′) |
| sequencing | MHB583 | (5′-TGGCTGATACAGAGGGAGGT-3′) |
| rs1800407 (OCA2) | ||
| Forward | MHB593 | (5′-GGCTCCCTGTTCTTAAAGTC-3′) |
| Reverse | MHB594 | (5′-TGAGAATGGAACCTGGAGCC-3′) |
| Sequencing | MHB595 | (5′-AAGGCTGCCTCTGTTCTACG-3′) |
| rs1426654 (SLC24A5) | ||
| Forward | MHB1755 | (5′-TCATAAAGAAGCAAAACATTGGA-3′) |
| Reverse | MHB1756 | (5′-AGCACAGATGCCAAGGAGAT-3′) |
| Sequencing | MHB1757 | (5′-TGCCAATATCTCCCTTTGTG-3′) |
| rs6058017 (ASIP) | ||
| Forward | MHB1836 | (5′-GGCTTCGATGAAGAAAGTGG-3′) |
| Reverse | MHB1837 | (5′-AGGGGATAGCCTCGTTCCTA-3′) |
| Sequencing | MHB1838 | (5′-ATGGGACTTCAGGGAGACCT-3′) |
Sequencing was performed by the Genomic Analysis and Technology Core at the University of Arizona on a 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA). In addition, the first 287 DNA samples collected were genotyped at 75 SNPs in 24 genes implicated in melanin biosynthesis by DNA Print Genomics (Sarasota, FL) following a previously published protocol (27). The remaining 504 individuals were also genotyped by DNA Print Genomics (Sarasota, FL) for 37 of the 75 SNPs that were statistically significant.
SNP rs12913832 (HERC2) was genotyped using TaqMan assay C__30724404 (Applied Biosystems).
Mathematical Modeling
Statistical analysis was performed using SAS (version 9.1) and JMP IN (release 5.1) statistical analysis software (SAS Institute, Cary, NC). The pool of SNPs was reduced from 75 (Fig. 1; Table 2) to 40 (within 15 genes) by choosing SNPs that were statistically significant (p < 0.05, Table 2) by ANOVA (n = 287 samples). Finally, rs1426654 (SLC24A5), rs6058017 (ASIP), and rs12913832 (HERC2) were genotyped for all individuals (n = 791 samples). Two samples were dropped from the study (sample 102 and 352) because of bookkeeping inconsistencies. In accordance with standard statistical procedure, the natural log of the ratio of eumelanin-to-pheomelanin was used to normalize the data.
TABLE 2.
75 SNPs within 24 genes.
| Gene | SNP | Total Hair Melanin | Natural Log of Ratio of Melanins | Skin Reflectance (CIEL) | Eye Color | Gene | SNP | Total Hair Melanin | Natural Log of Ratio of Melanins | Skin Reflectance (CIEL) | Eye Color |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AP3D1 | rs2238600 | MC1R | rs3212353 | ||||||||
| AP3D1 | rs4806830 | MC1R | rs3212352 | ||||||||
| ASIP | rs2424984 | * | * | * | * | MC1R | rs3212351 | ||||
| ASIP | rs6058017 | * | * | * | * | MLPH | rs2292885 | * | * | ||
| ASIP | rs2296151 | MLPH | rs1343768 | ||||||||
| CYP2C8 | rs1341164 | MYO18A | rs11080078 | * | |||||||
| CYP2D6 | rs1058174 | MYO5A | rs1724630 | * | |||||||
| CYP4B1 | rs1572603 | * | MYO5A | rs752864 | * | ||||||
| DCT | rs1325611 | * | * | * | * | MYO5A | rs2290332 | * | * | * | |
| DCT | rs1407995 | * | * | * | * | MYO7A | rs2276289 | * | * | ||
| GPR143 | rs3044 | * | * | * | * | MYO7A | rs3737454 | * | * | ||
| GPR143 | rs3810741 | MYO7A | rs948962 | ||||||||
| GPR143 | rs2521667 | MYO7A | rs1320703 | ||||||||
| HERC2 | rs12913832 | * | * | * | * | MYO7A | rs2276288 | ||||
| HPS1 | rs1804689 | OCA2 | rs1037208 | * | |||||||
| HPS1 | rs2296430 | OCA2 | rs10852218 | * | * | ||||||
| HPS1 | rs3830015 | OCA2 | rs11638265 | * | * | * | |||||
| HPS3 | rs2689234 | OCA2 | rs1800404 | * | * | * | * | ||||
| HPS4 | rs1894704 | * | * | OCA2 | rs1800407 | * | * | ||||
| HPS4 | rs3752589 | * | OCA2 | rs1800410 | * | * | * | * | |||
| HPS4 | rs3752590 | * | OCA2 | rs1800411 | * | * | * | * | |||
| HPS4 | rs739289 | * | OCA2 | rs1800414 | * | ||||||
| HPS5 | rs2305564 | OCA2 | rs1900758 | * | * | * | * | ||||
| HPS6 | rs4917959 | OCA2 | rs749846 | * | * | * | |||||
| LYST | rs3768051 | OCA2 | rs1800402 | ||||||||
| MAOA | rs979605 | OCA2 | rs737051 | ||||||||
| MC1R | rs1805007 | * | * | OCA2 | rs1800415 | ||||||
| MC1R | rs1805008 | * | * | * | * | OCA2 | rs2305253 | ||||
| MC1R | rs3212346 | * | * | OCA2 | rs1800401 | ||||||
| MC1R | rs3212355 | * | OCA2 | rs2044627 | |||||||
| MC1R | rs3212357 | * | * | * | * | SLC24A5 | rs1426654 | * | * | * | * |
| MC1R | rs3212370 | SLC45A2 | rs16891982 | * | * | * | * | ||||
| MC1R | rs3212368 | SLC45A2 | rs2287949 | * | * | * | |||||
| MC1R | rs3212366 | SLC45A2 | rs26722 | * | * | * | * | ||||
| MC1R | rs3212363 | SLC45A2 | rs40132 | * | * | * | * | ||||
| MC1R | rs3212364 | TYRP1 | rs2733832 | * | * | * | * | ||||
| MC1R | rs2228479 | TYRP1 | rs683 | ||||||||
| MC1R | rs1805005 |
Significant SNPs by ANOVA (p<0.05) (40 SNPs within 16 genes) with respect to each of the four phenotypes for the first 287 samples. rs3212363 (MC1R) was genotyped for all individuals, it was not significant across all populations.
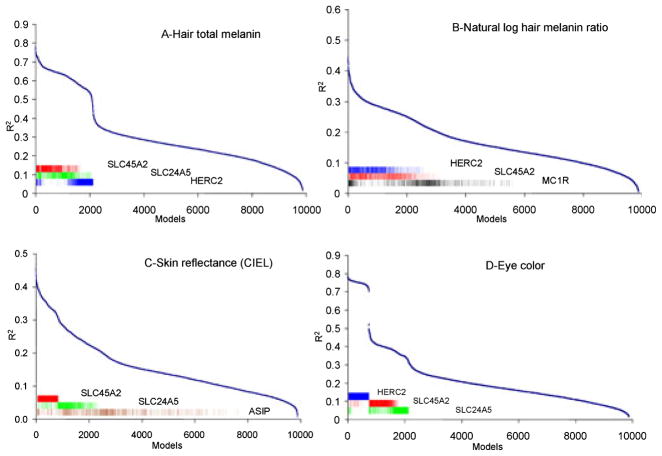
A total of 40 SNPs (Fig. 1; Table 2) were used to build MLR models of three SNPs. Initially, forward, reverse, and mixed stepwise regression methods were used to trim the number of SNPs. Different models were obtained based on the method used. To determine SNPs that were most significant in a three-SNP MLR model, we used SAS to generate all possible models of three SNPs from the pool of 40 significant SNPs. All models were then plotted using squared regression coefficient (R2) in descending order of value, and inflections in the resulting curves were noted. To find the basis of these inflections, we plotted histograms using JMP IN that contained all SNPs that comprised all models up to the R2 inflections (along the steepest initial slopes). In doing so, it became obvious which SNPs were predominantly responsible for the inflections. This method was performed in determining SNPs for each trait.
Results
Our primary aim was to develop a forensic DNA test predictive for pigmentation phenotype. Therefore, we analyzed the data across all populations of the study. The pool of SNPs was reduced from 75 to 40 SNPs by ANOVA (p < 0.05) (Fig. 1; Table 2). To determine the most likely SNPs for a given trait, all possible combinations of three-SNP models (40 choose 3) were generated, sorted by descending R2 value, and graphed. The three most frequent SNPs found in the range from the highest R2 model to the first inflection of the graph were used to construct an MLR model for each trait. These models accounted for significant variation in each of the four measured traits: scalp-hair total melanin (76.3%), natural log of the ratio of eumelanin-to-pheomelanin (43.2%), skin reflectance (45.7%), and eye color (74.8%) (Table 3).
TABLE 3.
Across populations R2 values of individual SNPs, interaction terms, full model, and sample size of each ethnic group (self-reported).
| Individual R2 Values % (sample size; p value) | Caucasian | African-American | Hispanic | South Asian | East Asian | Native American | Admixture | Not Listed | |
|---|---|---|---|---|---|---|---|---|---|
| Scalp-hair total melanin | |||||||||
| rs16891982 (SLC45A2) | 63.7 (n = 172, p =7.2 × 10–38) | 102 | 18 | 14 | 4 | 10 | 5 | 19 | |
| rs1426654 (SLC24A5) | 62.3 (n = 166, p = 2.76 × 10–35) | 101 | 18 | 11 | 2 | 8 | 5 | 21 | |
| rs12913832 (HERC2) | 54.0 (n = 176, p = 6.69 × 10–30) | 108 | 16 | 13 | 4 | 10 | 5 | 20 | |
| Model | 76.3 (n = 143) | 86 | 14 | 10 | 2 | 8 | 5 | 18 | |
| ln(eumelanin/pheomelanin) | |||||||||
| rs16891982 (SLC45A2) | 24.7 (n = 172, 3.88 × 10–11) | 102 | 18 | 14 | 4 | 10 | 5 | 19 | |
| rs12913832 (HERC2) | 26.5 (n = 176, p = 2.79 × 10–12) | 108 | 16 | 13 | 4 | 10 | 5 | 20 | |
| rs1805007 (MC1R) | 14.0 (n = 186, p = 1.41 × 10–7) | 113 | 19 | 14 | 4 | 10 | 5 | 21 | |
| Model | 43.2 (n = 162) | 97 | 15 | 13 | 4 | 10 | 5 | 18 | |
| Average skin reflectance | |||||||||
| rs16891982 (SLC45A2) | 32.1 (n = 487, p = 2.28 × 10–41) | 267 | 19 | 83 | 23 | 42 | 14 | 30 | 9 |
| rs1426654 (SLC24A5) | 23.9 (n = 528, p = 7.09 × 10–32) | 296 | 20 | 88 | 22 | 41 | 15 | 33 | 13 |
| rs2424984 (ASIP) | 15.6 (n = 538, p = 2.18 × 10–20) | 300 | 20 | 89 | 25 | 44 | 15 | 32 | 13 |
| Model | 45.7 (n = 447) | 244 | 19 | 74 | 20 | 39 | 14 | 28 | 9 |
| Model+rs16891982* rs2424984 | 49.6 (n = 447) | ||||||||
| Eye color | |||||||||
| rs12913832 (HERC2) | 74.8 (n = 484, p = 1.09 × 10–144) | 253 | 19 | 70 | 39 | 48 | 15 | 28 | 12 |
| rs16891982 (SLC45A2) | 38.4 (n = 397, p = 3.55 × 10–42) | 191 | 21 | 56 | 35 | 47 | 13 | 24 | 10 |
| rs1426654 (SLC24A5) | 34.3 (n = 490, p = 3.64 × 10–45) | 260 | 21 | 69 | 36 | 46 | 15 | 30 | 13 |
| Model | 76.4 (n = 353) | 171 | 16 | 49 | 32 | 43 | 13 | 20 | 9 |
Scalp-Hair Total Melanin
The R2 curve generated from the three-SNP models showed SNPs rs16891982 (SLC45A2), rs1426654 (SLC24A5), and rs12913832 (HERC2) to be the most frequent SNPs (310 or 3.1%) above the first inflection of the R2 curve (Fig. 2A). Together these three SNPs yielded an R2 of 76.3% (n = 143 samples).
FIG. 2.
Three-SNP multiple linear regression (MLR) models for four pigmentation traits across populations. In each panel, the horizontal-axis depicts all 9880 combinations of significant SNPs in a three-SNP MLR model (i.e., 40 choose 3). The vertical-axis is the R2 value for each model. The three SNPs that occurred most frequently in the models with the highest R2 values are indicated by the colored bar insets. Each model contained rs16891982 (SLC45A2) in red. (A) Hair total melanin’s second and third SNPs were rs1426654 (SLC24A5) in green, and rs12913832 (HERC2) in blue, model R2 value of 76.3%. (B) Natural log of hair melanin ratio’s second and third SNPs were rs1426654 (SLC24A5) and rs1805007 (MC1R) in black, model R2 value of 43.2%. (C) Skin reflectance’s second and third SNPs were rs1426654 (SLC24A5) and rs2424984 (ASIP) in brown, model R2 value of 45.7%. (D) Eye color’s SNP was rs12913832 (HERC2) in blue, model R2 value of 74.8%. The second and third SNPs rs16891982 (SLC45A2) and rs1426654 (SLC24A5) only marginally increased the model R2 value to 76.4%.
Natural Log of the Ratio of Eumelanin-to-Pheomelanin
The R2 curve generated from the three-SNP models showed SNPs rs16891982 (SLC45A2), rs1426654 (SLC24A5), and rs1805007 (MC1R) to be the most frequent SNPs (430 or 4.4%) above the first inflection of the R2 curve (Fig. 2B). Together these three SNPs yielded an R2 of 43.2% (n = 162 samples).
Skin Reflectance
The R2 curve generated from the three-SNP models showed SNPs rs16891982 (SLC45A2), rs1426654 (SLC24A5), and rs2424984 (ASIP) to be the most frequent SNPs (750 or 7.6%) above the first inflection of the R2 curve (Fig. 2C). Together these three SNPs yielded an R2 value of 45.7% (n = 447 samples). An interaction term of rs2424984 (ASIP) and rs16891982 (SLC45A2) increased the R2 value of the model by approximately 4% to 49.6% (n = 447 samples).
Eye Color
The R2 curve generated from the three-SNP models showed SNP rs12913832 (HERC2) to be the most frequent SNP (710 or 7.2%) above the first inflection of the R2 curve (Fig. 2D). rs12913832 yielded an R2 value of 74.8% (n = 484 samples). The next two inflection points showed SNPs rs12913832 (HERC2), rs16891982 (SLC45A2), and rs1426654 (SLC24A5) to be the most frequent SNPs. Together these three SNPs yielded an R2 value of 76.4% (n = 353 samples).
Discussion
Using MLR, we have shown that a large proportion of variation in hair, skin, and eye color across diverse human populations can be accounted for by a small number of SNPs. Starting from an initial candidate group of 75 SNPs, we found by ANOVA that 40 of these SNPs were significantly (p < 0.05) associated with hair pigmentation, skin reflectance, or eye color phenotype. Initially, we trimmed the number of SNPs by forward, backward, and/or a combination of both step-wise regression methods. However, each method yielded a different model. In an attempt to circumvent this problem, we modeled all possible combinations of significant SNPs. The R2 values of the top models differed on average by <3–10 thousandths of a percent or smaller. The question arose as to which was the best model. To answer this question, we chose SNPs that were most frequent in the highest R2 three-SNP models. This method ultimately trimmed the SNPs to five SNPs (three coding and two noncoding) in five genes that accounted for most of the variance (76.3% for hair total melanin; 43.2% for hair eumelanin-to-pheomelanin ratio; 45.7% for skin CIEL; and 74.8% for eye color).
Total Hair Melanin and Eumelanin-to-Pheomelanin Ratio
Previous studies have examined the overall color of hair in relationship to various genetic markers. Overall hair color is the result of at least two parameters: total melanin and the ratio of eumelanin-to-pheomelanin. These can be measured objectively by chemical analysis (2). We found that although both parameters are associated with SNPs from SLC45A2 and HERC2, they differ in the third most significant genetic contributor (SLC24A5 for total hair melanin and MC1R for the ratio of eumelanin-to-pheomelanin).
A high proportion of phenotypic variance of total hair melanin (76.3%) can be accounted for by three SNPs: rs1426654 (SLC24A5), rs16891982 (SLC45A2), and rs12913832 (HERC2) (Fig. 2A). While SLC24A5 is an AIM, it plays an important role in pigmentation (34). And in studies carefully controlled for population stratification, the coding SNP rs1426654 is indeed a determinant for normal human pigmentation variation (42). Similarly, rs16891982 (SLC45A2) has been shown to be an AIM and has been shown to be associated with pigmentation variation in mice (19) and humans (22,32) in studies controlled for population stratification (42).
For the analysis of total hair melanin, SNP rs12913832 (HERC2) was the third most significant contributor in a three-SNP model. Although this SNP lies within the HERC2 gene, it may be part of a promoter region for the adjacent OCA2 gene (37,43–46). OCA2 has been associated with albinism in diverse populations (13) and has also been associated with hair color by linkage analysis (20).
For the natural log of the ratio of eumelanin-to-pheomelanin, a high proportion of phenotypic variance (43.2%) can be accounted for by three SNPs. Two of these are in common with total hair melanin: rs1426654 (SLC24A5) and rs16891982 (SLC45A2) (Fig. 2B). SNP rs1805007 (MC1R) was the third most significant contributor in a three-SNP model. MC1R has been shown to be a major determinant in whether eumelanin or pheomelanin is produced (21,47). MC1R variants that decrease the protein’s functionality have been shown to be associated with an increased incidence of red hair color in humans (4,5,48). Chemically, this translates to increased pheomelanin relative to eumelanin production.
Different studies have analyzed MC1R in different ways. Many analyzed specific populations to determine which SNPs were associated with variation in hair color, other studies focused on specific populations and on red hair. Moreover, the statistical analyses employed were different, and different variants have been studied. In this study, we analyzed for significance by (i) one-way ANOVA and (ii) a three-SNP model based on the frequency of contributors in the models with the highest R2 values (Table 4).
TABLE 4.
ANOVA results for MC1R SNPs with respect to hair total melanin and natural log of eumelanin-to-pheomelanin.
| MC1R SNP | Total Melanin (From First 287 Individuals) | n | Ratio | Total Melanin (From Full Data Set) | n | Ratio |
|---|---|---|---|---|---|---|
| rs3212357 | † | 141 | † | † | 219 | † |
| rs3212346 | * | 138 | † | † | 191 | † |
| rs1805008 | * | 141 | † | § | 218 | † |
| rs1805007 | § | 141 | † | † | 219 | † |
| rs3212355 | § | 137 | § | † | 214 | § |
| rs3212370 | ‡ | 141 | ‡ | |||
| rs3212368 | ‡ | 141 | ‡ | |||
| rs3212364 | ‡ | 141 | ‡ | |||
| rs3212352 | ‡ | 141 | ‡ | |||
| rs3212351 | ‡ | 141 | ‡ | |||
| rs3212366 | § | 137 | § | |||
| rs3212353 | § | 136 | § | |||
| rs1805005 | § | 124 | § | |||
| rs2228479 | § | 122 | § |
p < 0.05.
p < 0.01.
No variation.
p > 0.05.
Although we did not examine all known SNPs of MC1R, some of the SNPs we examined have been studied in relationship to red hair color. Valverde et al. (1995) found rs2228479 (V60L) in combination with other nonsynonymous MC1R SNPs in British and Irish to be associated with red hair color. In contrast, two studies (5,7) did not find an association of rs2228479 with red hair. Looking across populations of all hair colors, we did not find rs2228479 to be significant by ANOVA. In addition, Flanagan et al. (2000) and Branicki et al. (2007) found that rs1805007 (R151G) in combination with rs1805008 (R160W) to be associated with red hair color. Moreover, Sulem et al. (2007) found both of these SNPs to be associated with hair color in Icelandic and Dutch populations. In our three-SNP MLR model, rs1805007 (R151G) was the third most important genetic contributor for the ratio of pigmentation in hair color. We note that our analysis of the ratio of hair pigmentation did not focus on any particular hair color.
Skin Reflectance (CIEL)
A high proportion of phenotypic variance of skin reflectance (45.7%) can be accounted for by three SNPs: rs1426654 (SLC24A5), rs16891982 (SLC45A2), and rs2424984 (ASIP) (Fig. 2C). Studies have examined SNPs rs1426654 (SLC24A5) and rs16891982 (SLC45A2) and found both to be associated with normal human pigmentation within various ethnic groups and allele frequencies across various ethnic groups (24,32,34,42). Thus, rs1426654 (SLC24A5) and rs16891982 (SLC45A2) mediate pigmentation variation, and they are also AIMs. Lamason et al. (34) found SNP rs1426654 to be an AIM in determining European vs. non-European ethnic origin. Stokowski et al. (2007) controlled for population stratification and showed rs1426654 (SLC24A5) to be significantly associated with a dichotomously defined skin reflectance in South Asians. Although it is an AIM, it does not clearly distinguish between Europeans and Sri Lankans (32). In contrast, SNP rs16891982 (SLC45A2) does distinguish between Europeans and Sri Lankans (32).
The third most significant genetic contributor was SNP rs2424984 (ASIP). ASIP has been shown to be associated with skin pigmentation (12), namely for rs6058017 (9,10). Although we found rs6058017 to be significant by ANOVA, we did not find it to be a better predictor in skin reflectance than rs2424984. For both a single SNP and a three-SNP model, rs2424984 was a better predictor for skin reflectance.
Other studies have found MC1R (21,25,43,47), OCA2, and DCT (49) to be associated with normal skin pigmentation. Similarly, we found SNPs within the above genes to be associated by ANOVA with normal skin pigmentation. However, none were found to be significant contributors in a three-SNP MLR model. This does not imply that these genes are not important in pigmentation, it simply means that our method was unable to detect their significance. This may be attributed to a variety of factors such as sampling error because of small sample size, different populations studied, and/or different methods of measuring skin reflectance.
Eye Color
A high proportion of phenotypic variance of eye color (74.8%) can be accounted for by one SNP rs12913832 (HERC2) (Fig. 2D). Previous studies have examined eye color in relationship to various genetic markers. OCA2 (8,20,26,28,29), SLC45A2 (22), and MC1R (8,50) have been statistically associated with variation in eye color.
Duffy et al. (2007) found three SNPs within intron 1 of OCA2 that when considered as a haplotype–diplotype explained about 74% of eye color variation. However, there are significant differences between our results and theirs. Among the differences were populations studied, SNPs genotyped, and binning of eye colors. The population they studied was Northern European, whereas we analyzed across various ethnic populations, and we genotyped additional SNPs within genes other than OCA2.
Most recently, genome-wide-association studies have shown that intronic SNPs of a gene 5′ to OCA2, HERC2, have given the highest association with eye color (37,43,45). Studies suggest that HERC2 contains a promoter region for OCA2 (37,43,45,46,51). To date, SNP rs12913832 (HERC2) in intron 86 was shown to have the highest association and most likely causative SNP in determining eye color (45), explaining 68% of the variance between blue eye and brown eye color in an Anglo-Celtic population. Therefore, rs12913832 is likely the causative SNP for European/Caucasians. We note that rs12913832 varies in Caucasian/European populations (where eye color is varied), but only one allele is found in non-Caucasian/European populations (where most non-Caucasian/Europeans have brown/dark eye color). Regardless of whether rs12913832 is causative or an AIM, it is extremely predictive for eye color both within and across populations.
A three-SNP model of rs12913832 (HERC2), rs16891982 (SLC45A2), rs1426654 (SLC24A5) is only marginally better at explaining the variance (76%) than rs12913832 (HERC2) alone. Like, rs12913832 (HERC2), SNPs rs16891982 (SLC45A2), rs1426654 (SLC24A5) are AIMs that distinguish European/Caucasians from non-European/Caucasians. However, individually rs16891982 (SLC45A2) accounts for 38% of eye color variance, and rs1426654 (SLC24A5) accounts for 34% of eye color variance, significantly less than rs12913832 (HERC2) that accounts for 74% of the variance.
Statistical Interactions
Statistical interaction of SNPs rs12910433 (OCA2) and rs2228479 (MC1R) has been associated with skin reflectance in a Tibetan population (40). We did not find this interaction; however, we did find an interaction between SNPs rs16891982 (SLC45A2) and rs2424984 (ASIP) for the across-population skin reflectance MLR model. These differences may reflect our use of an ethnically diverse sample; thus, the statistical product of these two genes might be more predictive of skin reflectance across populations. Other interactions, such as between rs12913832 (HERC2) and rs1800407 (OCA2), may exist in explaining variation in eye color (45), but we cannot confirm this from our data.
Conclusion
We found five SNPs in five genes that were informative for normal human pigmentation (SLC45A2, SLC24A5, MC1R, HERC2, and ASIP). Three of these (SLC45A2, SLC24A5, and MC1R) were coding. Clearly, variations within these proteins can result in functional variation that contributes to the pigmentation variation. In contrast, the most significant SNPs found in OCA2-HERC2 and ASIP were noncoding. We genotyped most of all known coding SNPs within these genes that showed allele frequency differences across populations, and none were as significantly correlated with pigmentation as the informative noncoding SNPs. This suggests that the regulation of the expression of OCA2 and ASIP may underlie pigmentation variation. Although our results demonstrate that relatively few SNPs in a relatively few genes control a significant proportion of normal human pigmentation variation, it is certain that additional polymorphisms in these and other genes account for the remaining variation.
The polymorphisms that we report are in genes known to regulate pigmentation. Some or all of the variance in pigmentation explained by them may be because they are also AIMs. Nevertheless, they are predictive markers for normal human pigmentation variation across various ethnic backgrounds. We note that these models of human constitutive pigmentation phenotype have significant implications for forensic science. These results suggest that assays can be developed, independent of ethnicity, to predict hair, skin, and eye color from DNA samples. Preliminary analysis of an independent sample set (n = 261 samples) has validated the predictive utility of these models (Valenzuela et al., in preparation). In addition, recent genome-wide-association studies have uncovered a variety of other candidate SNPs that contribute to phenotypic variation in pigmentation. These include SNPs rs12896399 (SLC24A4), rs12203592 (IRF4), rs1540771 (6p25.3), and rs35264875 (TPCN2) (12,43,46). Future studies are needed to evaluate these and other SNPs as they are uncovered, to further refine these predictive models such that they can accurately predict the pigmentation phenotype of the donors of otherwise unknown forensic samples.
Acknowledgments
We thank Domonique Smith and Benjamin Metelits for their technical help with this project.
Footnotes
Funded by the National Institute of Justice (2002-1J-CX-K010).
Conflict of Interest Statement
Dr. Tony Frudakis is and Dr. Matthew Thomas was an employee and shareholder of a company, DNAPrint, which performs forensic DNA analysis.
References
- 1.Tully G. Genotype versus phenotype: human pigmentation. Forensic Sci Int Genet. 2007;1:105–10. doi: 10.1016/j.fsigen.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Wakamatsu K, Ito S. Advanced chemical methods in melanin determination. Pigment Cell Res. 2002;15:174–83. doi: 10.1034/j.1600-0749.2002.02017.x. [DOI] [PubMed] [Google Scholar]
- 3.Nordlund JJ, Ortonne J. The normal color of human skin. In: Nordlund JJ, Boissy RE, Hearing VJ, King RA, Oetting WS, Ortonne J, editors. The pigmentary system. 2. Malden, MA: Blackwell Publishing; 2006. pp. 504–20. [Google Scholar]
- 4.Valverde P, Healy E, Jackson I, Rees JL, Thody AJ. Variants of the melanocyte-stimulating hormone receptor gene are associated with red hair and fair skin in humans. Nat Genet. 1995;11:328–30. doi: 10.1038/ng1195-328. [DOI] [PubMed] [Google Scholar]
- 5.Flanagan N, Healy E, Ray A, Philips S, Todd C, Jackson IJ, et al. Pleiotropic effects of the melanocortin 1 receptor (MC1R) gene on human pigmentation. Hum Mol Genet. 2000;9:2531–7. doi: 10.1093/hmg/9.17.2531. [DOI] [PubMed] [Google Scholar]
- 6.Rees JL. The genetics of sun sensitivity in humans. Am J Hum Genet. 2004;75:739–51. doi: 10.1086/425285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Branicki W, Brudnik U, Kupiec T, Wolanska-Nowak P, Wojas-Pelc A. Determination of phenotype associated SNPs in the MC1R gene. J Forensic Sci. 2007;52:349–54. doi: 10.1111/j.1556-4029.2006.00361.x. [DOI] [PubMed] [Google Scholar]
- 8.Sulem P, Gudbjartsson DF, Stacey SN, Helgason A, Rafnar T, Magnusson KP, et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat Genet. 2007;39(12):1443–52. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- 9.Bonilla C, Boxill LA, Donald SA, Williams T, Sylvester N, Parra EJ, et al. The 8818G allele of the agouti signaling protein (ASIP) gene is ancestral and is associated with darker skin color in African Americans. Hum Genet. 2005;116:402–6. doi: 10.1007/s00439-004-1251-2. [DOI] [PubMed] [Google Scholar]
- 10.Kanetsky PA, Swoyer J, Panossian S, Holmes R, Guerry D, Rebbeck TR. A polymorphism in the agouti signaling protein gene is associated with human pigmentation. Am J Hum Genet. 2002;70:770–5. doi: 10.1086/339076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voisey J, del Gomez-Cabrera MC, Smit DJ, Leonard JH, Sturm RA, van Daal A. A polymorphism in the agouti signalling protein (ASIP) is associated with decreased levels of mRNA. Pigment Cell Res. 2006;19:226–31. doi: 10.1111/j.1600-0749.2006.00301.x. [DOI] [PubMed] [Google Scholar]
- 12.Sulem P, Gudbjartsson DF, Stacey SN, Helgason A, Rafnar T, Jakobsdottir M, et al. Two newly identified genetic determinants of pigmentation in Europeans. Nat Genet. 2008;40(7):835–7. doi: 10.1038/ng.160. [DOI] [PubMed] [Google Scholar]
- 13.Oetting WS, King RA. Molecular basis of albinism: mutations and polymorphisms of pigmentation genes associated with albinism. Hum Mutat. 1999;13:99–115. doi: 10.1002/(SICI)1098-1004(1999)13:2<99::AID-HUMU2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 14.Inagaki K, Suzuki T, Shimizu H, Ishii N, Umezawa Y, Tada J, et al. Oculocutaneous albinism type 4 is one of the most common types of albinism in Japan. Am J Hum Genet. 2004;74:466–71. doi: 10.1086/382195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newton JM, Cohen-Barak O, Hagiwara N, Gardner JM, Davisson MT, King RA, et al. Mutations in the human orthologue of the mouse under-white gene (uw) underlie a new form of oculocutaneous albinism, OCA4. Am J Hum Genet. 2001;69:981–8. doi: 10.1086/324340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rundshagen U, Zuhlke C, Opitz S, Schwinger E, Kasmann-Kellner B. Mutations in the MATP gene in five German patients affected by oculocutaneous albinism type 4. Hum Mutat. 2004;23:106–10. doi: 10.1002/humu.10311. [DOI] [PubMed] [Google Scholar]
- 17.Johnson DK, Stubbs LJ, Culiat CT, Montgomery CS, Russell LB, Rinchik EM. Molecular analysis of 36 mutations at the mouse pink-eyed dilution (p) locus. Genetics. 1995;141:1563–71. doi: 10.1093/genetics/141.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lyon MF, King TR, Gondo Y, Gardner JM, Nakatsu Y, Eicher EM, et al. Genetic and molecular analysis of recessive alleles at the pink-eyed dilution (p) locus of the mouse. Proc Natl Acad Sci USA. 1992;89:6968–72. doi: 10.1073/pnas.89.15.6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sweet HO, Brilliant MH, Cook SA, Johnson KR, Davisson MT. A new allelic series for the underwhite gene on mouse chromosome 15. J Hered. 1998;89:546–51. doi: 10.1093/jhered/89.6.546. [DOI] [PubMed] [Google Scholar]
- 20.Eiberg H, Mohr J. Assignment of genes coding for brown eye colour (BEY2) and brown hair colour (HCL3) on chromosome 15q. Eur J Hum Genet. 1996;4:237–41. doi: 10.1159/000472205. [DOI] [PubMed] [Google Scholar]
- 21.Sturm RA, Teasdale RD, Box NF. Human pigmentation genes: identification, structure and consequences of polymorphic variation. Gene. 2001;277:49–62. doi: 10.1016/s0378-1119(01)00694-1. [DOI] [PubMed] [Google Scholar]
- 22.Graf J, Hodgson R, van Daal A. Single nucleotide polymorphisms in the MATP gene are associated with normal human pigmentation variation. Hum Mutat. 2005;25:278–84. doi: 10.1002/humu.20143. [DOI] [PubMed] [Google Scholar]
- 23.Duffy DL, Box NF, Chen W, Palmer JS, Montgomery GW, James MR, et al. Interactive effects of MC1R and OCA2 on melanoma risk phenotypes. Hum Mol Genet. 2004;13:447–61. doi: 10.1093/hmg/ddh043. [DOI] [PubMed] [Google Scholar]
- 24.Nakayama K, Fukamachi S, Kimura H, Koda Y, Soemantri A, Ishida T. Distinctive distribution of AIM1 polymorphism among major human populations with different skin color. J Hum Genet. 2002;47:92–4. doi: 10.1007/s100380200007. [DOI] [PubMed] [Google Scholar]
- 25.Shriver MD, Parra EJ, Dios S, Bonilla C, Norton H, Jovel C, et al. Skin pigmentation, biogeographical ancestry and admixture mapping. Hum Genet. 2003;112:387–99. doi: 10.1007/s00439-002-0896-y. [DOI] [PubMed] [Google Scholar]
- 26.Duffy DL, Montgomery GW, Chen W, Zhao ZZ, Le L, James MR, et al. A three-single-nucleotide polymorphism haplotype in intron 1 of OCA2 explains most human eye-color variation. Am J Hum Genet. 2007;80:241–52. doi: 10.1086/510885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frudakis T, Terravainen T, Thomas M. Multilocus OCA2 genotypes specify human iris colors. Hum Genet. 2007;122(3–4):311–26. doi: 10.1007/s00439-007-0401-8. [DOI] [PubMed] [Google Scholar]
- 28.Frudakis T, Thomas M, Gaskin Z, Venkateswarlu K, Chandra KS, Ginjupalli S, et al. Sequences associated with human iris pigmentation. Genetics. 2003;165:2071–83. doi: 10.1093/genetics/165.4.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rebbeck TR, Kanetsky PA, Walker AH, Holmes R, Halpern AC, Schuchter LM, et al. P gene as an inherited biomarker of human eye color. Cancer Epidemiol Biomarkers Prev. 2002;11:782–4. [PubMed] [Google Scholar]
- 30.Sturm RA, Frudakis TN. Eye colour: portals into pigmentation genes and ancestry. Trends Genet. 2004;20:327–32. doi: 10.1016/j.tig.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 31.Zhu G, Evans DM, Duffy DL, Montgomery GW, Medland SE, Gillespie NA, et al. A genome scan for eye color in 502 twin families: most variation is due to a QTL on chromosome 15q. Twin Res. 2004;7:197–210. doi: 10.1375/136905204323016186. [DOI] [PubMed] [Google Scholar]
- 32.Soejima M, Koda Y. Population differences of two coding SNPs in pigmentation-related genes SLC24A5 and SLC45A2. Int J Legal Med. 2007;121:36–9. doi: 10.1007/s00414-006-0112-z. [DOI] [PubMed] [Google Scholar]
- 33.Yuasa I, Umetsu K, Harihara S, Kido A, Miyoshi A, Saitou N, et al. Distribution of the F374 allele of the SLC45A2 (MATP) gene and founder-haplotype analysis. Ann Hum Genet. 2006;70:802–11. doi: 10.1111/j.1469-1809.2006.00261.x. [DOI] [PubMed] [Google Scholar]
- 34.Lamason RL, Mohideen MA, Mest JR, Wong AC, Norton HL, Aros MC, et al. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 2005;310:1782–6. doi: 10.1126/science.1116238. [DOI] [PubMed] [Google Scholar]
- 35.Ginger RS, Askew SE, Ogborne RM, Wilson S, Ferdinando D, Dadd T, et al. SLC24A5 encodes a trans-Golgi network protein with potassium-dependent sodium-calcium exchange activity that regulates human epidermal melanogenesis. J Biol Chem. 2008;283:5486–95. doi: 10.1074/jbc.M707521200. [DOI] [PubMed] [Google Scholar]
- 36.Stokowski RP, Krishna Pant PV, Tony D, Fereday A, Hinds DA, Jarman C, et al. A genomewide association study of skin pigmentation in a South Asian population. Am J Hum Genet. 2007;81(6):1119–32. doi: 10.1086/522235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kayser M, Liu F, Janssens AC, Rivadeneira F, Lao O, van Duijn K, et al. Three genome-wide association studies and a linkage analysis identify HERC2 as a human iris color gene. Am J Hum Genet. 2008;82:411–23. doi: 10.1016/j.ajhg.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ito S, Wakamatsu K. An improved modification of permanganate oxidation of eumelanin that gives a constant yield of pyrrole-2,3,5-tricarbox-ylic acid. Pigment Cell Res. 1994;7:141–4. doi: 10.1111/j.1600-0749.1994.tb00041.x. [DOI] [PubMed] [Google Scholar]
- 39.Wakamatsu K, Ito S, Rees JL. The usefulness of 4-amino-3-hydroxyphe-nylalanine as a specific marker of pheomelanin. Pigment Cell Res. 2002;15:225–32. doi: 10.1034/j.1600-0749.2002.02009.x. [DOI] [PubMed] [Google Scholar]
- 40.Akey JM, Wang H, Xiong M, Wu H, Liu W, Shriver MD, et al. Interaction between the melanocortin-1 receptor and P genes contributes to inter-individual variation in skin pigmentation phenotypes in a Tibetan population. Hum Genet. 2001;108:516–20. doi: 10.1007/s004390100524. [DOI] [PubMed] [Google Scholar]
- 41.Wakamatsu K, Hu DN, McCormick SA, Ito S. Characterization of melanin in human iridal and choroidal melanocytes from eyes with various colored irides. Pigment Cell Melanoma Res. 2008;21:97–105. doi: 10.1111/j.1755-148X.2007.00415.x. [DOI] [PubMed] [Google Scholar]
- 42.Stokowski RP, Pant PV, Dadd T, Fereday A, Hinds DA, Jarman C, et al. A genomewide association study of skin pigmentation in a South Asian population. Am J Hum Genet. 2007;81:1119–32. doi: 10.1086/522235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sulem P, Gudbjartsson DF, Stacey SN, Helgason A, Rafnar T, Magnusson KP, et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat Genet. 2007;39:1443–52. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- 44.Lehman AL, Silvers WK, Puri N, Wakamatsu K, Ito S, Brilliant MH. The underwhite (uw) locus acts autonomously and reduces the production of melanin. J Invest Dermatol. 2000;115:601–6. doi: 10.1046/j.1523-1747.2000.00107.x. [DOI] [PubMed] [Google Scholar]
- 45.Sturm RA, Duffy DL, Zhao ZZ, Leite FP, Stark MS, Hayward NK, et al. A single SNP in an evolutionary conserved region within intron 86 of the HERC2 gene determines human blue-brown eye color. Am J Hum Genet. 2008;82:424–31. doi: 10.1016/j.ajhg.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han J, Kraft P, Nan H, Guo Q, Chen C, Qureshi A, et al. A genome-wide association study identifies novel alleles associated with hair color and skin pigmentation. PLoS Genet. 2008;4:e1000074. doi: 10.1371/journal.pgen.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rees JL. Genetics of hair and skin color. Annu Rev Genet. 2003;37:67–90. doi: 10.1146/annurev.genet.37.110801.143233. [DOI] [PubMed] [Google Scholar]
- 48.Beaumont KA, Shekar SN, Cook AL, Duffy DL, Sturm RA. Red hair is the null phenotype of MC1R. Hum Mutat. 2008;29(8):E88–94. doi: 10.1002/humu.20788. [DOI] [PubMed] [Google Scholar]
- 49.Lao O, de Gruijter JM, van Duijn K, Navarro A, Kayser M. Signatures of positive selection in genes associated with human skin pigmentation as revealed from analyses of single nucleotide polymorphisms. Ann Hum Genet. 2007;71:354–69. doi: 10.1111/j.1469-1809.2006.00341.x. [DOI] [PubMed] [Google Scholar]
- 50.Kanetsky PA, Ge F, Najarian D, Swoyer J, Panossian S, Schuchter L, et al. Assessment of polymorphic variants in the melanocortin-1 receptor gene with cutaneous pigmentation using an evolutionary approach. Cancer Epidemiol Biomarkers Prev. 2004;13:808–19. [PubMed] [Google Scholar]
- 51.Lehman AL, Nakatsu Y, Ching A, Bronson RT, Oakey RJ, Keiper-Hrynko N, et al. A very large protein with diverse functional motifs is deficient in rjs (runty, jerky, sterile) mice. Proc Natl Acad Sci USA. 1998;95:9436–41. doi: 10.1073/pnas.95.16.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]