Abstract
Purpose
The ARMS2/HTRA1 genes at the 10q26 locus have been associated with risk of age-related macular degeneration (AMD), with the most significantly associated variants being A69S (rs10490924), del443ins54 (EU427539) and rs11200638. We wished to explore the association of the del443ins54 in two ethnically different populations from India and Australia.
Methods
The del443ins54 was screened in a large cohort of ~1500 subjects from these two populations by a combination of PCR-based agarose gel electrophoresis and validated by resequencing. Statistical analysis comprised the calculations of allele, genotype and haplotype frequencies along with their p values and corresponding odds ratios (OR), and 95% confidence intervals (95% CI) and measures of linkage disequilibrium (LD).
Results
The del443ins54 was significantly associated with AMD in both the Indian (p=1.74×10−13; OR=2.80, 95%CI, 2.12–3.70) and Australian cohorts (p=2.78×10−30; OR=3.15, 95%CI, 2.58–3.86). These associations were similar to those previously identified for the A69S and the rs11200638 variant in these populations that also exhibited high degrees of LD (D’ of 0.87-0.99). A major risk haplotype of “T-indel-A” (p=5.7×10−16; OR=3.16, 95%CI, 2.34–4.19 and p=6.33×10−30; OR=3.15, 95%CI, 2.57–3.85) and a protective haplotype of “G-wild type-G” (p=2.35×10−11; OR=0.39, 95%CI, 0.29–0.52 and p=1.02×10−30; OR=0.31, 95%CI, 0.25–0.38) were identified in the Indian and Australian cohorts, respectively.
Conclusions
These data provide an independent replication of the association of del443ins54 variant in two different ethnicities, despite differences in allele and haplotype frequencies between them. High levels of LD in both populations limit further genetic dissection of this region in AMD.
Introduction
Age-related macular degeneration (AMD) is a complex multifactorial disease and a leading cause of irreversible blindness in the world [1]. A major AMD susceptibility locus on 10q26 has been found to harbor risk associated variants in ARMS2 (rs10490924) and HTRA1 (rs11200638) in multiple populations worldwide [1-4]. An insertion-deletion (indel) polymorphism (EU427539) that affects the stability of ARMS2 mRNA by the removal of a polyadenylation signal (443 bases) and insertion of a 54bp AU rich element in the 3′-UTR (del443ins54), has also been identified in the 3′ end of the ARMS2 gene as increasing risk of AMD by several fold in both Caucasian [5] as well as Asian populations. Previous associations for this indel vary (from p=3.5×10−5 to p=8.4×10−34) in individuals of European origin [6-9], whereas, this has only been reported in two non-European cohorts consisting of Han Chinese [8] and Japanese populations [10].
We have previously reported a significant association of the A69S (rs10490924) and rs11200638 variants with AMD in both South Indian and Australian cohorts [11,12]. We now wished to determine the risk conferred by del443ins54 and its combined effect with both the A69S and rs11200638 variants with AMD susceptibility in two ethnically different cohorts from Southern India and Australia.
Methods
The study protocols adhered to the tenets of the Declaration of Helsinki and were approved by the Institutional Review Boards of the Royal Victorian Eye and Ear Hospital, Melbourne, Australia and L.V. Prasad Eye Institute, Hyderabad, India. The del443ins54 was screened in end stage AMD cases (mainly choroidal neovascular) and normal controls from cohorts in India (n=433) and Australia (n=1054). The detailed methods of clinical diagnosis along with the inclusion and exclusion criteria have been previously reported [11,12]. Amplification was performed using forward (5′-TCT GTG CAG CTG GTG AAA TC-3′) and reverse (5′-TCC AGG GTG GTG TAA TCC AT-3′) primers at an annealing temperature of 61 °C. Amplicons were visualized on a 2% agarose gel and genotypes directly scored from the gels. Subsets of samples were further validated by bi-directional sequencing on an automated DNA sequencer (ABI 3100), using the BigDye chemistry as per manufacturer’s guidelines (both from Applied Biosystems, Foster City, CA). Genotyping results were independently validated by a second investigator who was masked to the phenotype data.
Allele and genotype frequencies were determined by the gene counting method and estimates of Hardy–Weinberg equilibrium were assessed. Odds ratios (OR) and 95% confidence intervals (95%CI) were calculated to assess the risk conferred for each variant using the PLINK software [13]. Haplotypes were generated using various combinations of the A69S, the indel and rs11200638 variants and the estimated haplotype frequencies and linkage disequilibrium (LD) were assessed with the Haploview software (version 4.2) that uses the EM algorithm [14].
Results
All statistical analyses were based on samples where genotyping was successful across all three A69S, del443ins54 and rs11200638 genetic variants. Each variant was in Hardy–Weinberg equilibrium in both the Indian and Australian cohorts (p>0.05). Allele frequencies for each of the risk variants (T allele of A69S, presence of the indel in del443ins54 and the A allele of rs11200638) exhibited a relatively higher frequency (0.60–0.63) in the Indian cohort compared to the European cohorts (0.36–0.53) but lesser than that reported in Han Chinese (0.73–0.77) or the in the Japanese (0.86–0.88) populations (Table 1). The allele frequencies in the Australian cohort were similar (0.44) to those previously reported for other European cohorts (Table 1).
Table 1. Risk allele frequencies of the ARMS2 (A69S and del443ins54) and HTRA1 SNPs in different populations.
| Population (N=Cases, Controls) |
rs10490924 (A69S; “T” risk allele) |
del443ins54 (Indel) |
rs11200638 (“A” risk allele) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | P value | OR (95%CI) | Cases | Controls | P value | OR(95%CI) | Cases | Controls | P value | OR (95%CI) | |
| German (760, 549)5 |
0.424 |
0.193 |
2.8 x10-29 |
2.86(2.38-3.44) |
0.424 |
0.193 |
4.1 x10-29 |
2.85(2.37-3.43) |
0.426 |
0.199 |
6.9 x 10-29 |
2.85(2.37-3.42) |
| Caucasian (819, 329)6 |
0.412 |
0.248 |
1.89 x 10-13 |
2.13 (1.74-2.61) |
0.409 |
0.248 |
3.62 x 10-13 |
2.1 (1.71-2.57) |
NA |
NA |
NA |
NA |
| Caucasian (291, 191)7 |
0.36 |
0.23 |
3.31 x 10-5 |
1.86 |
0.36 |
0.23 |
3.46 x 10-5 |
1.85 |
0.36 |
0.24 |
6.41 x 10-5 |
1.8 |
| Utah (705, 650)8 |
0.38 |
0.2 |
8.61 x 10-26 |
NA |
0.39 |
0.2 |
1.9 x 10-26 |
NA |
0.41 |
0.22 |
3.64 x 10-26 |
NA |
| Northern European (442, 434)8 |
0.52 |
0.24 |
4.87 x 10-34 |
NA |
0.53 |
0.25 |
8.35x10-34 |
NA |
0.53 |
0.25 |
2.52 x 10-34 |
NA |
| Italian (159, 286)9 |
NA |
NA |
NA |
NA |
0.51 |
0.24 |
2.7x10-15 |
3.25 (2.36-4.41) |
NA |
NA |
NA |
NA |
| Han Chinese (138, 591)8 |
0.74 |
0.49 |
1.15 x 10-13 |
NA |
0.73 |
0.49 |
6.03 x 10-13 |
NA |
0.77 |
0.52 |
5.10 x 10-13 |
NA |
| Japanese (56, 77)10 |
0.86 |
0.62 |
NA |
NA |
0.875 |
0.66 |
NA |
NA |
NA |
NA |
NA |
NA |
| Australian (624,430)* |
0.445 |
0.202 |
1.97x10-30 |
3.14(2.58-3.86) |
0.446 |
0.199 |
2.78x10-30 |
3.15 (2.58-3.86) |
0.441 |
0.202 |
1.43x10-29 |
3.11(2.54-3.80) |
| South Indian (227, 206)* | 0.63 | 0.36 | 1.85x10-15 | 3.06 (2.31-4.04) | 0.63 | 0.38 | 1.74x10-13 | 2.8 (2.12-3.70) | 0.6 | 0.35 | 9.11x10-11 | 2.76 (2.02-3.77) |
*Data from the current study ; NA = Data not available
The frequency of the del443ins54 variant was significantly higher among AMD cases than controls in both the Indian (p=1.74×10−13; OR=2.80, 95%CI, 2.12–3.70) and Australian (p=2.78×10−30; OR=3.15, 95%CI, 2.58–3.86), cohorts (Table 1). This increased risk was also observed with respect to the del443ins54 genotypes. These findings were similar for the A69S (ARMS2) and the rs11200638 (HTRA1) variants in both the Indian and Australian cohorts, respectively (Table 2).
Table 2. Genotype counts of the ARMS2 and HTRA1 SNPs in the Indian and Australian cohorts.
| Population | SNP (gene) | Genotypes | Genotype counts |
P value | Odds ratios (95% CI) | |
|---|---|---|---|---|---|---|
| Cases | Controls | |||||
| Australian |
rs10490924 (ARMS2) |
GG |
189 |
271 |
- |
Ref |
| GT |
282 |
138 |
<0.0001 |
2.93 (2.22 - 3.86) |
||
| TT |
145 |
16 |
<0.0001 |
12.99 (7.51 - 22.49) |
||
| Indel -EU427539 (ARMS2) |
Wt/Wt |
190 |
269 |
- |
Ref |
|
| Wt/Indel |
306 |
137 |
<0.0001 |
3.16 (2.41 - 4.16) |
||
| Indel/Indel |
119 |
15 |
<0.0001 |
11.23 (6.36 – 19.82) |
||
|
rs11200638 (HTRA1) |
GG |
194 |
271 |
- |
Ref |
|
| GA |
292 |
138 |
<0.0001 |
2.96 (2.25 – 3.89) |
||
| AA |
130 |
17 |
<0.0001 |
10.68 (6.24 – 18.29) |
||
| Indian |
rs10490924 (ARMS2) |
GG |
39 |
85 |
- |
Ref |
| GT |
84 |
94 |
0.004 |
1.95 (1.21 – 3.15) |
||
| TT |
99 |
26 |
<0.0001 |
8.30 (4.67 – 14.74) |
||
| Indel -EU427539 (ARMS2) |
Wt/Wt |
44 |
84 |
- |
Ref |
|
| Wt/Indel |
79 |
88 |
0.017 |
1.71 (1.16 – 2.75) |
||
| Indel/Indel |
104 |
34 |
<0.0001 |
5.84 (3.43 – 9.94) |
||
| rs11200638 (HTRA1) | GG |
44 |
61 |
- |
Ref |
|
| GA |
70 |
67 |
0.01 |
1.45 (0.87 – 2.42) |
||
| AA | 84 | 17 | <0.0001 | 6.85 (3.58 – 13.12) | ||
Homozygosity of the indel and the other variants were strongly associated with an increased risk of AMD in both the Indian and Australian cohorts. Combined homozygosities at the A69S and the rs11200638 along with the indel variant did not alter the risk of AMD significantly either in the Indian (OR=7.69, 95%CI, 4.07–14.51) or Australian cohorts (OR=10.61, 95%CI, 7.05–15.96).
The measure of linkage disequilibrium (LD) between the A69S, del443ins54 and rs11200638 variants were remarkably high across this 10q26 region with relatively higher values in the Australian (D’=0.99; r2=0.98) compared to the Indian (D’=0.87; r2=0.71) cohorts (Figure 1). Two major haplotypes (frequency >5%) were identified across the three variants with “T-Indel-A” being the risk haplotype and “G-WT (wild-type)-G” being protective in the Indian and Australian cohorts, respectively (Table 3). Different pairwise haplotype combinations with either the ‘T’ or ‘A’ allele at A69S in presence of the del443ins54 or its wild-type form along with the ‘G’ or ‘A’ allele of rs11200638, did not substantially alter the p values or ORs observed for either the risk or protective haplotypes as opposed to when all three variants were assessed together, reinforcing the observation of the high LD between variants in these two genes (Table 3).
Figure 1.
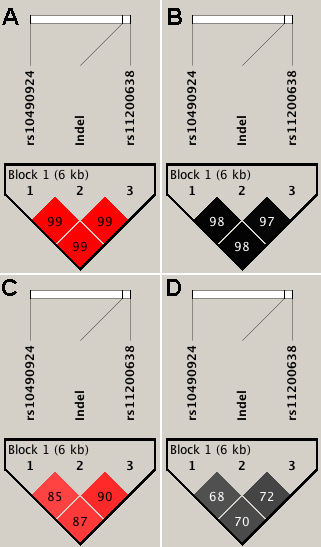
Linkage disequilibrium plots showing the three variants in the Australian and Indian cohorts. The D’ and r2 values between the SNPs are indicated inside the quadrants for the Australian (A and B) and the Indian (C and D), respectively.
Table 3. Major haplotype frequencies at the three loci harboring the ARMS2 (A69S and del443ins54) and HTRA1 variants in the Indian and Australian cohorts.
| HAPLOTYPES (5’-3’) |
SOUTH INDIAN COHORT (n=433) |
AUSTRALIAN COHORT (n=1054) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| rs1094924 (ARMS2) | del443ins54 (ARMS2) | rs11200638 (HTRA1) | % Cases (N=227) | % Controls (N=206) | P values | OR (95%CI) | % Cases (N=624) | % Controls (N=430) | P values | OR (95%CI) |
|
T |
Indel |
A |
57.8 |
30.3 |
5.70x10-16 |
3.16 (2.34-4.19) |
44.1 |
20 |
6.33x10-30 |
3.15 (2.57-3.85) |
| G |
Wt* |
G |
33 |
55.6 |
2.35x10-11 |
0.39 (0.29-0.52) |
55.3 |
79.8 |
1.02x10-30 |
0.31 (0.25-0.38) |
| G |
- |
G |
35.8 |
59.5 |
4.49x10-12 |
0.38 (0.29-0.50) |
55.4 |
79.8 |
1.43x10-30 |
0.31 (0.26-0.38) |
|
T |
- |
A |
58.4 |
30.4 |
1.82x10-16 |
3.20 (2.42-4.25) |
44.1 |
20.2 |
1.55x10-29 |
3.11 (2.55-3.81) |
|
T |
Indel |
- |
59.1 |
31.8 |
1.32x10-15 |
3.09 (2.34-4.10) |
44.4 |
20 |
1.18x10-30 |
3.19 (2.61-3.91) |
| G |
Wt* |
- |
33.5 |
50.4 |
2.54x10-13 |
0.36 (0.27-0.47) |
55.4 |
79.8 |
1.43x10-30 |
0.31 (0.26-0.38) |
|
- |
Indel |
A |
50.8 |
31.8 |
2.41x10-15 |
3.06 (2.31-4.05) |
44.1 |
20 |
6.33x10-30 |
3.15 (2.57-3.86) |
| - | Wt* | G | 35.5 | 59.1 | 5.04x10-12 | 0.38 (0.29-0.50) | 55.3 | 79.8 | 1.02x10-30 | 0.31 (0.26-0.38) |
Combinations of the risk alleles at the three loci are bolded; *Wt = wildtype
Discussion
These data provide an independent replication of the association of the ARMS2 del443ins54 variant in two cohorts, and to the best of our knowledge for the first time among South Indians with AMD. The strong association of the del443ins54 along with the A69S and rs11200638 variants in the Indian and Australian cohorts were consistent with that observed in other populations [6-10]. Haplotype analysis with these three variants indicated that inclusion of del443ins54 in the haplotype neither increased nor decreased the risk of AMD in either cohort (Table 3).
The ARMS2 and HTRA1 genes are in high LD in European populations and thus dissecting out the role of one gene over the other has proved difficult. The advantage of undertaking a comparative analysis of genetic variants in populations of differing ethnicities expands the genetic diversity available and may provide the opportunity of identifying a more defined but associated region for further study. The current study highlighted similar degrees of associations across these three variants despite a relatively lower LD between the A69S, del443ins54 and rs11200638 variants in the Indian compared to the Australian cohort. However, it did confirm the presence of stratification differences between ethnicities with the allele frequency of the Indel of del443ins54 in South Indians being higher at 0.63 in cases compared to that in European populations (0.36–0.53) but lower than other Asian populations (0.73–0.88). The allele frequency in cases is similar to that previously shown by us in the assessment of the A69S (0.63) and the rs11200638 (0.60) variants of the ARMS2 and HTRA1 genes respectively, in the South Indian cohort. Evidence of population stratification has also been observed in AMD studies of the protective CFHR3–1 deletion, with the highest frequencies of the deleted allele being present in African populations (16%–20%) compared to Asians (<2%) [15,16].
The potential role of the ARMS2 and HTRA1 gene in AMD is still unclear but functional dissection of the effect of the rs11200638 promoter variant in the HTRA1 gene has revealed that this variant resides within a putative transcription binding site for the factors AP2α and SRF (serum response factor) [3,4]. Initial investigation of the influence of the homozygous risk genotype on HTRA1 expression levels revealed consistently higher levels of expression with the AA genotype compared to the GG genotype [3,4]. In contrast, other studies of the rs11200638 variant have revealed no functional effect on HTRA1 expression [2,6,17].
Analysis of the chromosome 10q26 risk haplotype inclusive of the ARMS2 del443ins54 indel found decreased ARMS2 expression and almost 3.0 fold increase in HTRA1 expression [18]. Interestingly, a subsequent study has shown that while the ARMS2 risk del443ins54 results in decrease in mRNA transcription levels of the ARMS2 gene, a non risk associated variant (rs2736911) also leads to significantly reduced ARMS2 transcript levels suggesting that ARMS2 protein deficiency alone is unlikely to be pathogenic in AMD [17]. A functional role for ARMS2 in mitochondrial homeostasis has also been suggested and the biology concerning mitochondrial dysfunction and the effects on age supports this notion [2,5]. However, subsequent immunofluorescence and immunoblot experiments localized ARMS2 in retinal epithelial ARPE-19 cells and COS7 transfected cells to the cytosol rather than the mitochondria suggesting that ARMS2 may not confer risk to AMD through the mitochondrial pathway [19]. Studies concerning the effects of AMD risk variants on HTRA1 expression are equivocal and further investigations on the functional role of these variants are required.
In conclusion, we provide convincing evidence for the association of the del443ins54 variant with AMD, despite differences in allele, genotype and haplotype frequencies and LD across the 10q26 region reflecting population stratification differences in two different ethnicities. AMD, a complex multi-factorial disease is associated with multiple genomic regions with varying magnitudes of effect and the relevance of genetic associations differ between populations. Further, elucidation of the genetic basis of this disease through the analysis of individuals from different ethnic groups has the potential to provide useful insights into the genetic diversity of risk and protective variants within a gene as well as their contributions to disease. Also, meaningful genetic dissection of the ARMS2 and HTRA1 gene in this region will require much larger patient cohorts than have currently been assessed, or through the identification of other ethnic populations which show relatively lower levels of LD over this 10q26 region.
Acknowledgments
The authors thank all the Australian and Indian patients and the normal volunteers for their participation and Drs. Nazimul Hussain, Taraprasad Das, Anjli Hussain and Avinash Pathangay for their help in collecting some of the earlier patients’ data. RHG and PNB acknowledge an NHMRC Practitioner Award and NHMRC Fellowship, respectively. SK and Stuart Cantsilieris acknowledge a Senior Research Fellowship of the Council of Scientific and Industrial Research (CSIR), Government of India, and an Australian Postgraduate Award, respectively. CERA receives Operational Infrastructure Support from the Victorian Government, Australia. Grant support: This work was supported by the Australia-India Strategic Research Fund (AISRF) jointly funded through the Department of Biotechnology (DBT), Government of India (SC and IK) and the Department of Innovation, Industry, Science and Research (DIISR), Government of Australia (PNB and RHG), the National Health and Medical Research Council (NHMRC) Centre for Clinical Research Excellence #529923 - Translational Clinical Research in Major Eye Diseases.
References
- 1.Swaroop A, Branham KEH, Chen W, Abecasis G. Genetic susceptibility to age-related macular degeneration: a paradigm for dissecting complex disease traits. Hum Mol Genet. 2007;16 Spec No. 2:R174–82. doi: 10.1093/hmg/ddm212. [DOI] [PubMed] [Google Scholar]
- 2.Kanda A, Chen W, Othman M, Branham KE, Brooks M, Khanna R, He S, Lyons R, Abecasis GR, Swaroop A. A variant of mitochondrial protein LOC387715/ARMS2, not HTRA1, is strongly associated with age-related macular degeneration. Proc Natl Acad Sci USA. 2007;104:16227–32. doi: 10.1073/pnas.0703933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Z, Camp NJ, Sun H, Tong Z, Gibbs D, Cameron DJ, Chen H, Zhao Y, Pearson E, Li X, Chien J, Dewan A, Harmon J, Bernstein PS, Shridhar V, Zabriskie NA, Hoh J, Howes K, Zhang K. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science. 2006;314:992–3. doi: 10.1126/science.1133811. [DOI] [PubMed] [Google Scholar]
- 4.Dewan A, Liu M, Hartman S, Zhang SS, Liu DT, Zhao C, Tam PO, Chan WM, Lam DS, Snyder M, Barnstable C, Pang CP, Hoh J. HTRA1 Promoter Polymorphism in Wet Age-Related Macular Degeneration. Science. 2006;314:989–92. doi: 10.1126/science.1133807. [DOI] [PubMed] [Google Scholar]
- 5.Fritsche LG, Loenhardt T, Janssen A, Fisher SA, Rivera A, Keilhauer CN, Weber BH. Age-related macular degeneration is associated with an unstable ARMS2 (LOC387715) mRNA. Nat Genet. 2008;40:892–6. doi: 10.1038/ng.170. [DOI] [PubMed] [Google Scholar]
- 6.Wang G, Spencer KL, Scott WK, Whitehead P, Court BL, Ayala-Haedo J, Mayo P, Schwartz SG, Kovach JL, Gallins P, Polk M, Agarwal A, Postel EA, Haines JL, Pericak-Vance MA. Analysis of the indel at the ARMS2 3′UTR in age-related macular degeneration. Hum Genet. 2010;127:595–602. doi: 10.1007/s00439-010-0805-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hadley D, Orlin A, Brown G, Brucker AJ, Ho AC, Regillo CD, Donoso LA, Tian L, Kaderli B, Stambolian D. Analysis of six genetic risk factors highly associated with AMD in the region surrounding ARMS2 and HTRA1 on chromosome 10, region q26. Invest Ophthalmol Vis Sci. 2010;51:2191–6. doi: 10.1167/iovs.09-3798. [DOI] [PubMed] [Google Scholar]
- 8.Yang Z, Tong Z, Chen Y, Zeng J, Lu F, Sun X, Zhao C, Wang K, Davey L, Chen H, London N, Muramatsu D, Salasar F, Carmona R, Kasuga D, Wang X, Bedell M, Dixie M, Zhao P, Yang R, Gibbs D, Liu X, Li Y, Li C, Li Y, Campochiaro B, Constantine R, Zack DJ, Campochiaro P, Fu Y, Li DY, Katsanis N, Zhang K. Genetic and functional dissection of HTRA1 and LOC387715 in age-related macular degeneration. PLoS Genet. 2010;6:e1000836. doi: 10.1371/journal.pgen.1000836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ricci F, Zampatti S, D’Abbruzzi F, Missiroli F, Martone C, Lepre T, Pietrangeli I, Sinibaldi C, Peconi C, Novelli G, Giardina E. Typing of ARMS2 and CFH in age-related macular degeneration: case-control study and assessment of frequency in the Italian population. Arch Ophthalmol. 2009;127:1368–72. doi: 10.1001/archophthalmol.2009.237. [DOI] [PubMed] [Google Scholar]
- 10.Gotoh N, Nakanishi H, Hayashi H, Yamada R, Otani A, Tsujikawa A, Yamashiro K, Tamura H, Saito M, Saito K, Lida T, Matsuda F, Yoshimura N. ARMS2 (LOC387715) variants in Japanese patients with exudative age-related macular degeneration and polypoidal choroidal vasculopathy. Am J Ophthalmol. 2009;147:1037–41. doi: 10.1016/j.ajo.2008.12.036. [DOI] [PubMed] [Google Scholar]
- 11.Kaur I, Katta S, Hussain A, Hussain N, Mathai A, Narayanan R, Hussain A, Reddy RK, Majji AB, Das T, Chakrabarti S. Variants in the 10q26 gene cluster (LOC387715 and HTRA1) exhibit enhanced risk of age-related macular degeneration along with CFH in Indian patients. Invest Ophthalmol Vis Sci. 2008;49:1771–6. doi: 10.1167/iovs.07-0560. [DOI] [PubMed] [Google Scholar]
- 12.Richardson AJ, Islam FA, Aung KZ, Guymer RH, Baird PN. An intergenic region between the tagSNP rs3793917 and rs11200638 in the HTRA1 gene indicates association with age-related macular degeneration. Invest Ophthalmol Vis Sci. 2010;51:4932–6. doi: 10.1167/iovs.09-5114. [DOI] [PubMed] [Google Scholar]
- 13.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genomeassociation and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 15.Hageman GS, Hancox LS, Taiber AJ, Gehrs KM, Anderson DH, Johnson LV, Radeke MJ, Kavanagh D, Richards A, Atkinson J, Meri S, Bergeron J, Zernant J, Merriam J, Gold B, Allikmets R, Dean M, AMD Clinical Study Group Extended haplotypes in the complement factor H (CFH) and CFH-related (CFHR) family of genes protect against age-related macular degeneration: Characterization, ethnic distribution and evolutionary implications. Ann Med. 2006;38:592–604. [PMC free article] [PubMed] [Google Scholar]
- 16.Sudmant PH, Kitzman JO, Antonacci F, Alkan C, Malig M, Tsalenko M, Sampas N, Bruhn L, Shendure J. 1000 Genomes Project, Eichler EE. Diversity of Human Copy Number Variation and Multicopy Genes. Science. 2010;330:641–6. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedrich U, Myers CA, Fritsche LG, Milenkovich A, Wolf A, Corbo JC, Weber BH. Risk- and non-risk-associated variants at the 10q26 AMD locus influence ARMS2 mRNA expression but exclude pathogenic effects due to protein deficiency. Hum Mol Genet. 2011;20:1387–99. doi: 10.1093/hmg/ddr020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang G, Scott WK, Haines JL, Pericak-Vance MA. Genotype at Polymorphism rs11200638 and HTRA1 Expression Level. Arch Ophthalmol. 2010;128:1491–3. doi: 10.1001/archophthalmol.2010.256. [DOI] [PubMed] [Google Scholar]
- 19.Wang G, Spencer KL, Court BL, Olson LM, Scott WK, Haines JL, Pericak-Vance MA. Localization of Age-Related Macular Degeneration-Associated ARMS2 in Cytosol, Not Mitochondria. Invest Ophthalmol Vis Sci. 2009;50:3084–90. doi: 10.1167/iovs.08-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]


