Abstract
Background
Eosinophilic airway inflammation is heterogeneous in asthmatic patients. We recently described a distinct subtype of asthma defined by the expression of genes inducible by TH2 cytokines in bronchial epithelium. This gene signature, which includes periostin, is present in approximately half of asthmatic patients and correlates with eosinophilic airway inflammation. However, identification of this subtype depends on invasive airway sampling, and hence noninvasive biomarkers of this phenotype are desirable.
Objective
We sought to identify systemic biomarkers of eosinophilic airway inflammation in asthmatic patients.
Methods
We measured fraction of exhaled nitric oxide (Feno), peripheral blood eosinophil, periostin, YKL-40, and IgE levels and compared these biomarkers with airway eosinophilia in asthmatic patients.
Results
We collected sputum, performed bronchoscopy, and matched peripheral blood samples from 67 asthmatic patients who remained symptomatic despite maximal inhaled corticosteroid treatment (mean FEV1, 60% of predicted value; mean Asthma Control Questionnaire [ACQ] score, 2.7). Serum periostin levels are significantly increased in asthmatic patients with evidence of eosinophilic airway inflammation relative to those with minimal eosinophilic airway inflammation. A logistic regression model, including sex, age, body mass index, IgE levels, blood eosinophil numbers, Feno levels, and serum periostin levels, in 59 patients with severe asthma showed that, of these indices, the serum periostin level was the single best predictor of airway eosinophilia (P = .007).
Conclusion
Periostin is a systemic biomarker of airway eosinophilia in asthmatic patients and has potential utility in patient selection for emerging asthma therapeutics targeting TH2 inflammation.
Keywords: Asthma, biomarker, sputum, bronchoscopy, eosinophil, TH2, IL-13, periostin, IgE, Feno
Although asthma is traditionally thought to result from aeroallergen-induced inflammation driven by TH2 processes1 and is commonly characterized by eosinophilic infiltration of the airways, there is increasing evidence that there are other subtypes of asthma driven by alternative pathogenic mechanisms.2 An emerging concept holds that the nature and intensity of granulocytic infiltration of the airways, in particular the presence or absence of increased numbers of eosinophils, defines pathophysiologically and clinically distinct subsets of the disease.3-10 Eosinophilic disease is a strong predictor of corticosteroid responsiveness in asthmatic patients,11-13 and novel treatments targeting eosinophilic TH2-driven inflammation might only be expected to show efficacy in asthmatic patients in whom airway eosinophilia is present. For example, recent clinical trials of mepolizumab and reslizumab, mAbs directed against the eosinophilopoietic cytokine IL-5, have demonstrated the importance of selecting asthmatic patients with eosinophilic airway inflammation to show significant benefit from treatment.14-16 Direct airway sampling through sputum induction or bronchoscopy is technically challenging and often impractical in clinical practice, and hence noninvasive systemic biomarkers of airway eosinophilia are desirable for the rational management of asthma with existing12 and emerging targeted molecular therapies.
IL-13 is a pleiotropic TH2 cytokine produced by activated T cells, natural killer T cells, basophils, eosinophils, mast cells, alternatively activated macrophages, and nuocytes, and it has been strongly implicated in the pathogenesis of asthma in preclinical models.17 Increased levels of IL-13 have been detected in the airways of human asthmatic patients; however, this increase is only observed in a subset of asthmatic patients.18-22 We have shown that 3 of the most differentially expressed bronchial epithelial genes between asthmatic patients and healthy control subjects are periostin (POSTN), chloride-channel accessory protein 1 (CLCA1), and SERPINB2, and expression of these genes is sensitive to inhaled corticosteroid (ICS) treatment. All 3 genes are induced in bronchial epithelial cells by recombinant IL-13 treatment in vitro.23 The presence or absence of coordinated high expression levels of these genes defines 2 distinct populations of asthmatic patients, and this 3-gene TH2 signature correlates with IL13 and IL5 expression in the bronchial mucosa, airway and peripheral eosinophilia, airway remodeling, and clinical responsiveness to ICSs but not with atopy.13
To investigate whether the TH2 signature encodes systemic biomarkers of airway eosinophilia in patients with severe asthma that is persistently symptomatic despite maximal ICS treatment, we developed a highly sensitive assay for periostin protein in peripheral blood and found that it is detectable at increased levels in the sera or plasma of a subset of asthmatic patients and is correlated with airway eosinophilic inflammation. Periostin has the potential to be used as a biomarker to select patients for asthma therapies, such as ICSs or biologic agents, targeting TH2 inflammation, as we have recently demonstrated in a phase II proof-of-concept trial of lebrikizumab, a humanized mAb against IL-13.24
RESULTS
Our objective was to determine whether the bronchial epithelial TH2 signature associated with airway eosinophilia that we have described in patients with mild-to-moderate asthma not taking ICSs13 could be translated to a noninvasive biomarker of residual airway eosinophilia despite ICS treatment in patients with uncontrolled asthma.
Serum periostin is a biomarker of persistent airway eosinophilia despite steroid treatment in patients with severe asthma
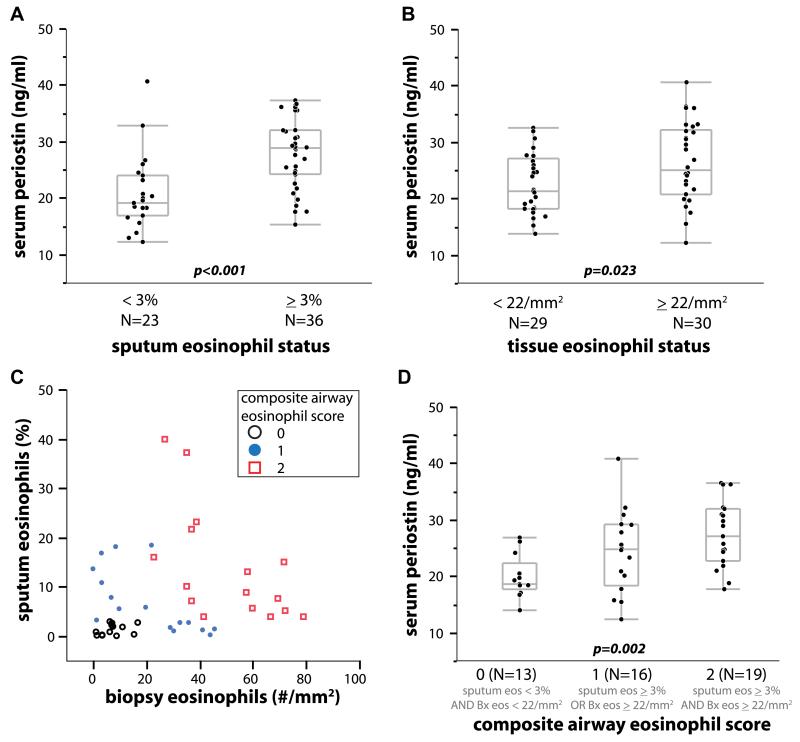
Although ICS treatment reduces airway periostin expression23 and airway eosinophilia25 in patients with mild-to-moderate ICS-responsive asthma, a subset of asthmatic patients has persistent eosinophilic airway inflammation and asthma symptoms despite high doses of corticosteroids,26 representing a significant unmet medical need. Thus we sought to establish whether systemic periostin levels might reflect persistent eosinophilic airway inflammation despite steroid treatment in patients with severe asthma. We conducted a multicenter 3-visit observational study (BOB-CAT) of patients with uncontrolled severe asthma (ACQ score, >1.50; FEV1, 40% to 80%) taking high doses of ICSs (>1000 μg/d fluticasone dipropionate or equivalent) with collection of induced sputum, endobronchial biopsies, and collection of peripheral blood. We obtained matched blood and complete airway data from 59 subjects (see the study overview in Fig 1). We used prespecified cutoff values from previous studies of 3% for sputum eosinophils12,15 and 22 eosinophils/mm2 total biopsy area.27,28 Serum periostin levels were stable within individual subjects across the 3 visits spanning up to 5 weeks (see Fig E2 in this article’s Online Repository at www.jacionline.org). Mean periostin levels were significantly higher in “eosinophil-high” compared with “eosinophil-low” subjects, as defined by sputum (P < .001, Wilcoxon rank sum test; Fig 2, A) or tissue (P = .023; Fig 2, B) eosinophil measurements.
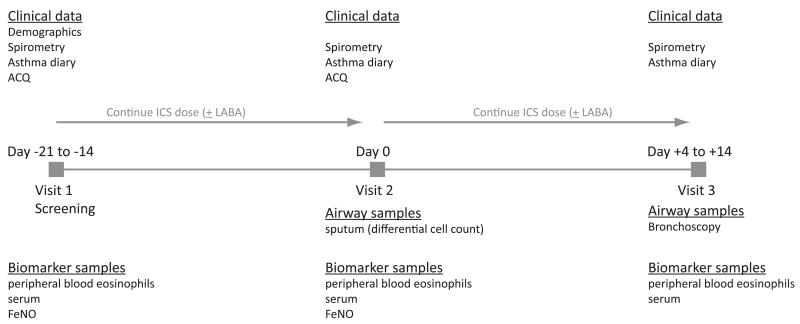
FIG 1.
BOBCAT study schema.
FIG 2.
Peripheral blood periostin levels differentiate patients with severe uncontrolled asthma taking high-dose ICSs according to airway eosinophilic inflammation. A, Dichotomized by sputum eosinophilia. B, Dichotomized by tissue eosinophilia. C, Sputum versus biopsy specimen eosinophilia. D, Composite airway eosinophil score demonstrating a strong positive progression of increasing serum periostin levels with increasing scores (P = .002, logistic regression). Bx, Biopsy.
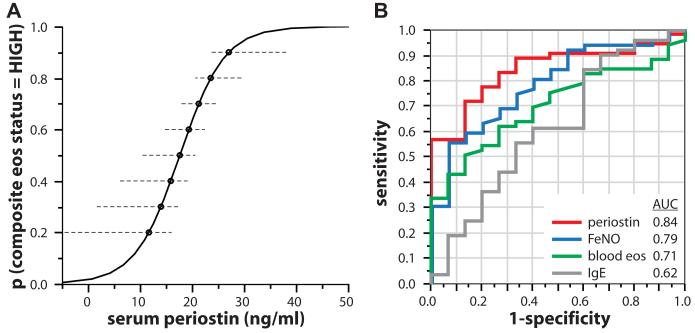
Although there was some overlap between asthmatic patients with increased tissue or sputum eosinophilia, a clear subset of asthmatic patients had values of less than the threshold for both measurements, whereas many asthmatic patients had increased numbers of either tissue or sputum eosinophils but not both (Fig 2, C), which is consistent with prior reports comparing sputum and tissue eosinophilia.29,30 Serum periostin levels were sensitive but not specific for either airway eosinophilia metric taken individually (Fig 2, A and B). However, subjects stratified by a composite score of 0 for neither, 1 for either, or 2 for both sputum and tissue eosinophilia exhibited a highly significant progression of increasing serum periostin levels with increasing eosinophil scores (P < .002, logistic regression; Fig 2, D). Subjects in BOBCAT with serum periostin levels of greater than 25 ng/mL had an 85% probability of a composite airway eosinophil score of 1 or 2 (Fig 3, A). Using 25 ng/mL serum periostin as an arbitrary cutoff, eosinophil-low and eosinophil-high subjects in BOBCAT are effectively differentiated, with a positive predictive value of 93% (see Table E3 in this article’s Online Repository at www.jacionline.org). Taken together, these data show that serum periostin is a systemic biomarker of persistent airway eosinophilia despite steroid treatment in patients with severe asthma.
FIG 3.
Sensitivity of biomarkers for eosinophilic airway inflammation. A, Probability of composite eosinophil status = “high” as a function of serum periostin. Dashed lines denote 95% CIs. B, Receiver operating characteristic curve analysis of the sensitivity and specificity of serum periostin, Feno, and serum IgE levels and blood eosinophil numbers for composite airway eosinophil status. AUC, Area under the curve.
Comparison of serum periostin levels with peripheral blood eosinophil numbers and fraction of exhaled nitric oxide, serum IgE, and serum YKL-40 levels as asthma biomarkers
In recent years, other noninvasive biomarkers of asthma severity and airway inflammation have been described. Four markers of particular interest are fraction of exhaled nitric oxide (Feno), an exhaled gas produced by the action of the enzymeinducible nitric oxide synthase in inflamed bronchial mucosa31; peripheral blood eosinophils; serum IgE; and YKL-40, a chitinase-like protein detectable in peripheral blood.32 We measured levels of these biomarkers in asthmatic patients and compared them with airway eosinophilia and other biomarkers.
Neither periostin, Feno, IgE levels, nor blood eosinophil numbers were significantly correlated with ACQ, FEV1, age, sex, or body mass index in BOBCAT. Feno levels, like serum periostin levels, were generally consistent across multiple visits, although Feno levels varied more than periostin levels (see Fig E2 in this article’s Online Repository at www.jacionline.org). However, blood eosinophil numbers were considerably more variable than periostin levels (r2 = 0.18 for blood eosinophil numbers between visits 1 and 3 [data not shown] compared with r2 = 0.65 for serum periostin levels between visits 1 and 3). Stratifying for sputum and biopsy eosinophil status, as in Fig 2, Feno levels were significantly higher in eosinophil-high asthmatic patients compared with eosinophil-low asthmatic patients. However, although both Feno and periostin levels were highly specific, exhibiting consistently low values for eosinophil-low subjects, Feno measurement detected fewer subjects with tissue eosinophilia and exhibited greater overlap between eosinophil-low and eosinophil-high subjects according to each metric used (see Fig E3 in this article’s Online Repository at www.jacionline.org). We fit a logistic regression model incorporating age, sex, body mass index, blood eosinophil numbers, serum IgE, Feno, and serum periostin levels (Table II) and found that the periostin level was the most significant single predictor of composite airway eosinophil status (P = .007). By using a cutoff value of 35 ppb, as previously described,33 Feno levels differentiate eosinophil-low and eosinophil-high asthmatic patients with comparable specificity to but lower sensitivity than a periostin cutoff of 25 ng/mL (see Table E3). Peripheral blood eosinophil numbers trended higher in asthmatic patients with high composite airway eosinophil status but did not reach statistical significance (data not shown). Periostin and Feno levels and blood eosinophil numbers were generally weakly continuously intercorrelated with each other and with airway eosinophil numbers (see Table E4 in this article’s Online Repository at www.jacionline.org). To assess the relative performance of each marker across its range of values in BOB-CAT, we performed receiver operating characteristic analysis of periostin, Feno, and serum IgE levels and blood eosinophil numbers versus composite airway eosinophil status and found that periostin performed favorably to Feno measurement (area under the curve, 0.84 and 0.79, respectively), whereas blood eosinophil numbers and serum IgE levels performed substantially less well (Fig 3, B).
TABLE II.
Logistic regression model of biomarkers versus eosinophil status in BOBCAT (n = 59)
| Estimate | SE | z Score | P value | |
|---|---|---|---|---|
| Age | −0.0396 | 0.039 | −1.015 | .31 |
| Sex (male) | −0.2031 | 0.889 | −0.229 | .82 |
| Body mass index | −0.1004 | 0.066 | −1.527 | .13 |
| Blood eosinophils | 1.7482 | 3.621 | 0.483 | .63 |
| Serum IgE | −0.0002 | 0.001 | −0.100 | .92 |
| Feno | 0.0476 | 0.038 | 1.238 | .22 |
| Serum periostin | 0.2491 | 0.092 | 2.719 | .007 |
We applied a logistic regression model to evaluate demographic and biomarker variables as predictors of composite airway eosinophil status (eosinophil low = sputum eosinophils <3% and biopsy eosinophils <22/mm2; eosinophil high = sputum eosinophils >3% or biopsy eosinophils >22/mm2).
YKL-40 levels showed no significant correlations with periostin levels or with any measures of airway or peripheral eosinophilia (see Table E4). Consistent with these findings of exhaled and blood biomarker levels, we found that bronchial epithelial gene expression levels of POSTN and NOS2 (the gene that encodes inducible nitric oxide synthase) in the dataset described in our previous studies13,23 were significantly correlated with each other and with bronchial mucosal expression levels of IL13 and IL5, whereas expression of CHI3L1 (the gene that encodes YKL-40) was not correlated with POSTN, IL13, or IL5 expression levels (see Table E5 in this article’s Online Repository at www.jacionline.org). Taken together, these data suggest that peripheral blood periostin measurement is a more reliable indicator of airway TH2/eosinophilic inflammation than Feno levels and that periostin and Feno levels are substantially better indicators of airway TH2/eosinophilic inflammation than blood eosinophil numbers, serum IgE levels, or YKL-40 levels in asthmatic patients.
DISCUSSION
Although asthma is traditionally regarded as an allergic disease mediated by TH2-driven inflammation,1 there is emerging evidence of pathophysiologic heterogeneity.3-10 We have recently shown that, in patients with mild-to-moderate asthma not undergoing steroid treatment, only about half the subjects have evidence of TH2 inflammation in their airways. The TH2-high subset is distinguished by increased markers of allergy, eosinophilic airway inflammation, bronchial fibrosis, and sensitivity to ICSs.13 Because antagonists of the TH2 cytokines IL-4, IL-5, and IL-13 are under active development as asthma therapeutics,34-37 it will become important to identify the asthmatic patients most likely to benefit from these targeted therapies. Although bronchoscopy, induced sputum sampling, and measurement of exhaled gases enable the direct characterization of inflammatory pathways in the airways, these modalities can be time consuming, expensive, and invasive and/or are not widely available in primary care settings. Furthermore, assay procedures are not standardized across the relatively few centers equipped to analyze airway samples, which makes implementation in multicenter clinical trials challenging. Thus it will be beneficial to develop noninvasive biomarkers of TH2-driven eosinophilic airway inflammation widely available on accessible assay platforms to select patients with evidence of TH2-driven eosinophilic inflammation in their airways for targeted therapies. To address this need, we have used gene expression profiling in asthmatic airway samples to enable the discovery and characterization of clinically useful peripheral biomarkers of TH2-driven eosinophilic airway inflammation.
Periostin is a secreted matricellular protein associated with fibrosis, and its expression is upregulated by recombinant IL-4 and IL-13 in cultured bronchial epithelial cells23,38 and bronchial fibroblasts.39 It is expressed at increased levels in vivo in a murine model40 and a rhesus model41 of asthma, as well as in bronchial epithelial cells23 and the subepithelial bronchial layer39 of human asthmatic patients. In human asthmatic patients periostin expression levels correlate with reticular basement membrane thickness, an indicator of subepithelial fibrosis.42 Unlike chloride-channel accessory protein 1, which is associated with mucus secretion and localized to the apical surface of bronchial epithelial and goblet cells,43 periostin is secreted into the extracellular matrix from the basolateral surface of bronchial epithelial cells stimulated with IL-1342 and thus might be more likely to be detectable systemically. Periostin is also overexpressed in nasal polyps associated with aspirin-sensitive asthma44,45 and in the esophageal epithelia of patients with eosinophilic esophagitis in an IL-13–dependent manner46 and thus might play a role in the tissue infiltration of eosinophils in TH2-driven disease processes.47 Future studies should examine blood periostin levels in diseases characterized by TH2 inflammation, tissue eosinophilia, and periostin expression, such as nasal polyposis, allergic rhinitis, atopic dermatitis, and eosinophilic esophagitis. In addition, extrapulmonary TH2/eosinophilic diseases are often comorbid with asthma, and thus the dominant tissue source of systemic periostin in patients with multiple TH2/eosinophilic diseases cannot be determined from this study and should be addressed in future studies. Some studies have described “neutrophilic” and “mixed granulocytic” phenotypes of severe asthma on the basis of increased sputum neutrophil numbers with or without sputum eosinophilia.9 In BOBCAT we observed that serum periostin levels were correlated with airway eosinophilia, regardless of sputum or tissue neutrophil counts, and we did not observe a consistent relationship between serum periostin levels and sputum or tissue neutrophilia (data not shown). Increased periostin expression has also been observed in patients with several types of epithelially derived cancers,48-52 and increased levels of soluble periostin have been reported in the sera of some patients with cancer.48,49,53-55 Whether the local and systemic expression of periostin in patients with asthma or other conditions is primarily due to the direct or indirect actions of IL-13 is as yet unclear and will best be addressed by assessments comparing periostin expression before and after therapeutic blockade of IL-13.
Feno levels are associated with airway inflammation and predict ICS responsiveness in patients with asthma of varying severity.11,33 However, Feno levels do not reliably reflect airway eosinophilia in patients with severe steroid-dependent asthma, and there are discrepancies between sputum and mucosal eosinophil quantification with respect to Feno levels.29,56,57 Titrating ICS treatment to suppress sputum eosinophil numbers reduces the rate of severe asthma exacerbations,12 but titrating ICS treatment to Feno levels does not.58 Serum YKL-40 has been described as a marker of asthmatic airway inflammation, but its levels were not correlated with measures of TH2 inflammation, such as IgE levels or eosinophil numbers.32 Accordingly, in the present study we found that bronchial epithelial gene expression levels of POSTN and NOS2, but not CHI3L1, were correlated with bronchial IL-13 and IL-5 expression (see Table E5). Although we observed relatively strong positive correlations between bronchial or systemic eosinophilia and serum periostin levels, the correlations between eosinophilia and Feno levels were weaker, and we did not observe a correlation between serum YKL-40 levels and eosinophilic inflammation in the asthmatic patients studied. Sputum and blood eosinophil counts and Feno levels are subject to significant temporal variability, depending on allergen exposure, exacerbations, and steroid treatment.56,57,59,60 We observed relatively little intrasubject variability in serum periostin levels in 3 measurements over the course of up to 5 weeks in BOBCAT (see Fig E2). The coefficient of variation (CV) across the first 2 visits was 2.2% (95% CI, 1.2% to 5.7%) for periostin levels and 8.2% (95% CI, 6.0% to 22.5%) for Feno levels. In our recent phase II study of lebrikizumab in 218 patients with moderate-to-severe asthma, we recorded 2 baseline measurements 1 week apart for serum periostin and Feno levels and found that the CV for periostin was 5% versus 19.8% for Feno levels and 21.3% for blood eosinophil numbers.24 Taken together, comparative analyses of periostin and Feno levels and blood eosinophil numbers comprising a logistic regression model (Table II), a receiver operating characteristic analysis (Fig 3, B), fixed cutoffs (see Table E3), continuous correlation analysis (see Table E4), and longitudinal stability (see Fig E2 in this article’s Online Repository at www.jacionline.org) collectively and consistently show that although each marker reflects airway eosinophilia, to some degree, periostin predicts the airway phenotype with greater fidelity than Feno levels or blood eosinophil numbers. Future studies should be directed at comparatively assessing longitudinal intrasubject variability in serum periostin levels, airway eosinophilia, Feno levels, and other candidate biomarkers of TH2 inflammation over extended periods of time.
The standard of care for bronchial asthma that is not well controlled with symptomatic therapy (eg, β-agonists) is ICS. In patients with mild-to-moderate asthma with increased levels of IL-13 in the airway20 and patients with eosinophilic esophagitis with increased expression levels of IL-13 in esophageal tissue,46 ICS treatment substantially reduces the level of IL-13 and IL-13–induced genes in the affected tissues. In the airway epithelia of asthmatic patients after 1 week of ICS treatment and in cultured bronchial epithelial cells, we have shown that corticosteroid treatment substantially reduces IL-13μinduced expression levels of TH2 signature genes.13,23 This downregulation could be the result of ICS-mediated reduction in IL-13 levels, ICS-mediated reduction of target gene expression, or a combination of the two. In patients with severe asthma refractory to ICS treatment, a similar fraction of subjects (approximately 40%) was found to have detectable sputum IL-13 levels to those seen in patients with mild ICS-naive asthma,20 which is comparable to the proportion of subjects with the bronchial epithelial TH2 signature we have described.13 Analogous observations have been reported for persistence in steroid-refractory asthmatic patients of IL-4– and IL-5–expressing cells in bronchoalveolar lavage fluid61 and eosinophilic inflammation in bronchial biopsy specimens and sputum (Fig 2). These observations suggest that although TH2-driven eosinophilic inflammation is suppressed by ICS treatment in patients with moderate asthma, it reappears in a subset of patients with severe asthma incompletely controlled by steroid treatment. An additional complication is brought on by incomplete adherence to prescribed ICS therapy, which might underlie poor control in some patients with severe asthma. Hence future studies should be directed at assessing blood periostin levels in the context of ICS treatment status, ICS dose, intrinsic steroid sensitivity, and adherence to ICS therapy in patients with controlled and uncontrolled asthma.
Currently, there are numerous biological therapeutics in clinical development targeting IL-13 and related factors driving TH2 inflammation in asthmatic patients.34-37 It is critical that these treatments are targeted to patients with relevant molecular pathology because otherwise important treatment effects might be underestimated; studies of anti–IL-5 therapy highlight this point.15,16,62 Accordingly, we have shown in 2 phase II studies of lebrikizumab, a humanized mAb against IL-13, that asthmatic patients with high pretreatment serum periostin levels experienced substantially greater treatment benefit from IL-13 blockade than patients with low pretreatment periostin levels (Scheerens et al, unpublished data).24 Our findings suggest that approximately half of patients with steroid-naive mild-to-moderate asthma might exhibit activity of the TH2 pathway in their airways, and a similar fraction of patients with severe steroid-refractory asthma exhibits the activity of this pathway. Therefore biomarkers that identify asthmatic patients likely to have TH2-driven inflammation in their airways might aid in the identification and selection of the patients most likely to respond to these experimental targeted therapies.
METHODS
Patients’ samples and microarray data
Microarray data and bronchial epithelial RNA from 42 nonsmoking patients with mild-to-moderate asthma were obtained from a previous study (data from this study are shown in Table E5).E1,E2
BOBCAT was a multicenter study conducted in the United States, Canada, and the United Kingdom to collect matched airway and blood samples in 67 patients with moderate-to-severe asthma. Inclusion criteria required a diagnosis of moderate-to-severe asthma (confirmed by an FEV1 between 40% and 80% of predicted value, as well as evidence within the past 5 years of >12% reversibility of airway obstruction with a short-acting bronchodilator or methacholine sensitivity [PC20] <8 mg/mL) that was uncontrolled (as defined by at least 2 exacerbations in the prior year or a score of >1.50 on the ACQE3 while receiving a stable dose regimen [>6 weeks] of a high-dose ICS [>1000 μg fluticasone or equivalent per day]) with or without a long-acting β-agonist. Permitted concomitant medications also included leukotriene receptor antagonists and oral corticosteroids. The BOBCAT study scheme is depicted in Fig 1.
Feno measurement, bronchoscopy, bronchoalveolar lavage, induced sputum, and immunohistochemistry for eosinophil counts were performed as previously described.E2,E4-E6 All research protocols were approved by the relevant institutional review boards, and informed consent was obtained from all subjects before enrollment. Patients’ demographics and lung function and biomarker data are summarized in Table I.
TABLE I.
Demographic and clinical data: range of values for biomarker assessments in the BOBCAT study
| No. of subjects | 67 |
| Age (y) | 46 (12) |
| Sex (M/F) | 32/35 |
| FEV1 (% predicted) | 60 (11) |
| ACQ score | 2.7 (0.8) |
| Daily ICS dose (μg FPI) equivalent | >1000* |
| BAL eosinophils (% nonsquamous cells) | 0.7 (0.1-2.4), n = 59 |
| Sputum eosinophils (% nonsquamous cells) | 5.2 (1.8-16.4), n = 59 |
| Biopsy eosinophils/mm2 | 23.0 (7.0-44.0), n = 59 |
| Blood eosinophils (× 109/L) | 0.24 (0.13-0.38) |
| Serum IgE (IU/mL) | 160 (40-373) |
| Feno (ppb) | 25 (17-46), n = 66 |
| Periostin (ng/mL), serum or plasma | 24.5 (19.6-30.6) |
| YKL-40 (ng/mL), serum or plasma | 67 (42-111), n = 65 |
Values are presented as means (SD) or medians (interquartile ranges). In cases of missing data, the number of subjects for whom data are available is indicated next to the value. Blood eosinophil numbers and periostin levels represent the mean from visits 1, 2, and 3. Feno represents the mean from visits 1 and 2.
BAL, Bronchoalveolar lavage; F, female; FPI, fluticasone dipropionate; M, male.
Of these, 7 patients were also receiving systemic corticosteroids, receiving between 5 and 40 mg of prednisolone equivalent per day.
Periostin assay development
Generation of murine anti-periostin mAbs
BALB/c mice (Charles River, Hollister, Calif) were immunized with recombinant human periostin protein (rhuPOSTN; residues N22-Q836; R&D Systems, Minneapolis, Minn) mixed with MPL+TDM adjuvant through footpad injection, intraperitoneal injection, or both in an additional protocol. Mice received 6 doses, followed by a prefusion boost in PBS alone through the footpad and intraperitoneal routes 3 days before fusion. Popliteal lymph nodes were harvested from these mice, the sera of which demonstrated strong binding titers to the immunization protein by using ELISA, and lymphocytes were fused with X63-Ag8.653 murine myeloma cells (American Type Culture Collection, Rockville, Md) through electrofusion (Cytopulse, Glen Burnie, Md) and plated on 96-well plates. Supernatants were screened 11 days after fusion for binding to immunization protein by using ELISA. Hybridomas demonstrating strong periostin-specific binding by means of ELISA were expanded and subcloned by limiting dilution. Final clones demonstrating the highest ELISA binding after the second round of subcloning were expanded for large-scale production in bioreactors (Integra Biosciences, Chur, Switzerland). Supernatants were then purified by means of Protein A affinity chromatography, as previously described.E7
Generation of rabbit polyclonal and monoclonal anti-periostin antibodies
rhuPOSTN-immunized NZW rabbits were bled serially to obtain antisera for the purification of periostin-specific polyclonal antibodies. The spleen from 1 of these rabbits was also subsequently used in a rabbit monoclonal production protocol carried out by Epitomics (Burlingame, Calif), as previously described.E8,E9
Recombinant periostin splice variants
Human periostin is expressed as at least 4 alternatively spliced transcripts,E10 and each splice variant is expressed in bronchial epithelia from asthmatic patients (data not shown). Periostin isoforms 1 to 4 were cloned from cDNA derived from “TH2-high” bronchial epithelial brushings from asthmatic patients described by Woodruff et al,E2 and recombinant proteins were generated from supernatants of transfected HEK293 cells as tools for use in the characterization of novel anti-periostin mAbs by their epitope binding patterns (see Fig E1, A and B).
mAb evaluation
Eight hybridoma lines were identified for individual subcloning. After 2 rounds of selection, 7 murine clone lines (designated A through G) and 4 rabbit clone lines (H though K) were isolated.
mAb affinity measurements
Simple BiaCore (GE Healthcare, Fairfield, Conn) experiments were carried out to characterize the relative affinities of the murine mAbs for full-length periostin. Antibodies E and F showed the highest apparent affinities, demonstrating both fast on rates and relatively slow off rates compared with the other antibodies tested. Monoclonal antibodies A, B, and C behaved similarly in this experiment, exhibiting lower relative affinity compared with mAbs E and F, mostly because of faster off rates. Monoclonal antibody D demonstrated poor affinity because of a very fast off-rate. Rough affinities were estimated at less than 1 nmol/L for E and F; 1 to 10 nmol/L for A, B, and C; and approximately 25 nmol/L for D.
Periostin-depleted normal human serum
Affinity-purified rabbit polyclonal anti-periostin was coupled to CarboLink resin (Pierce, Rockford, Ill) by using a standard kit protocol. This anti-periostin resin was then used to deplete endogenous periostin from a normal human serum pool. A periostin-depleted normal human serum pool serum was used as a negative control in all ELISA assay development to demonstrate absolute anti-periostin antibody specificity.
Prototype murine/rabbit mAb ELISAs
Capture ELISAs were constructed by using each murine mAb as a capture reagent paired with rabbit mAb K as the detection antibody, which can detect all 4 isoforms of human periostin protein (Fig E1, C). An anti-rabbit IgG–horseradish peroxidase conjugate was used to detect bound mAb K. Serum samples were diluted 1:200 before analysis. rhuPOSTN (full-length) was used to generate standard curves. Murine mAbs E and G paired best with the rabbit detection mAb K, allowing the most sensitive detection of the periostin standard (Fig E1, D).
Although rabbit mAbs clearly demonstrated utility as effective and specific ELISA reagents, the parent hybridoma lines produced minimal antibody in cell culture. Historically, rabbit hybridomas have also proved to be less robust than their murine counterparts and significantly more susceptible to both line mortality and loss of specific antibody production. For these reasons, murine capture and detection mAbs were preferred for the final format of the total periostin ELISA.
Construction of a murine/murine mAb ELISA
Each murine anti-periostin mAb (with the exception of mAb F because of insufficient material) was biotinylated by using a standard method. Preparations of unlabeled murine mAbs were used to compete with these biotinylated mAbs for binding to coated full-length periostin in a direct ELISA. Epitope groups were loosely defined by inhibitory patterns.
Thirty-six unique murine mAb pairs were evaluated as capture and detection antibodies in a standard indirect sandwich ELISA. Amdex streptavidin–horseradish peroxidase was used to identify bound biotinylated antibodies. Varying quantities of rhuPOSTN (0-300 pg/mL in assay diluent) were analyzed, and signal/noise ratios at 33 pg/mL rhuPOSTN were computed for each assay format. Multiple mAb pairs allowed the specific and sensitive detection of periostin, as shown in Table E1. Using mAb G as the capture (coat) antibody and biotinylated mAb E as the detection antibody, we observed a 7.3-fold increase over background absorbance at 33 pg/mL rhuPOSTN.
Multiple assay formats were also evaluated for their ability to recognize all periostin isoforms, a factor necessary for the accurate quantitation of total analyte levels in human serum. Periostin isoforms 1, 2, 3, and 4 were each transfected into HEK293 cells and generated as recombinant proteins in cell-culture supernatants. These supernatants were analyzed in parallel by using 4 different murine mAb assay configurations. Each assay format detected all periostin isoforms. Cell-culture supernatant from vector-transfected K293 cells was used as a negative control (Fig E1, E). Importantly, this experiment also confirmed the reactivity and specificity of the murine mAbs to novel preparations of human periostin isoforms that were unique from the original commercial periostin preparation used as immunogen. On the basis of its optimal sensitivity and ability to detect all periostin isoforms, the mAb G (coat) and biotinylated mAb E pair was selected for final assay development.
Assay qualification
The murine/murine total periostin ELISA was run on 11 plates over 3 days to evaluate assay accuracy and precision, control performance, limit of detection, and linearity of dilution. Minimum dilution for samples and controls was 1:200 in assay diluent (PBS, 0.5% BSA, 0.05% Tween 20, and 0.05% ProClin 300 [Sigma-Aldrich, St Louis, Mo], pH 7.4). Standard curves were generated by using rhuPOSTN (full-length) 2-fold serially diluted in assay diluent from 600 to 9.4 pg/mL. A 4-parameter logistic curve fit was applied to the OD data, and the interassay relative error was calculated.
Matrix controls were constructed by using a normal human serum pool. Unspiked pooled sera were used as the normal control, whereas the high control was generated by spiking 100 ng/mL rhuPOSTN to the serum pool. Normal and high controls were diluted 1:200 in assay diluent and measured in single wells at final dilutions of 1:200, 1:400, 1:800, and 1:1600. All dilutions were quantified within assay limits of quantitation. Means, SDs, and percentage CVs for the 4 dilutions were computed for each control on every plate, and these mean concentrations were then evaluated across all plates (n = 11) to determine interplate precision (Table E2). This control table was then used to define the normal and high control pass/fail criteria, setting allowable variance to ± 20% of the mean concentration for each control.
Because no individual human serum could be identified with extremely low endogenous periostin levels, a unique low control was constructed in assay diluent at 2.8 ng/mL rhuPOSTN. This control was measured in duplicate at the minimum (1:200) dilution only. For the low control, the pass/fail criterion was ± 20% of the nominal analyte concentration: allowable variance (of the duplicate mean) was therefore from 2.24 to 3.36 ng/mL.
The limit of detection for the assay was confirmed at the level of the lowest standard (9.4 pg/mL, in well), but the assay sensitivity limit was conservatively set equal to the serum periostin concentration of the low control (2.8 ng/mL). This control was also used to define the lower limit of quantitation at 14 pg/mL (in well), whereas the upper limit of quantitation was set at 594 pg/mL, which is equivalent to the in-well concentration of the high control.
rhuPOSTN was spiked into a normal human serum pool to a final concentration of 23.5 μg/mL to assess sample dilutional linearity. This ultrahigh periostin sample was minimally diluted and then further serially diluted past 1:107. Accurate (within ± 20%) recovery of this sample was observed for all dilutions within the assay’s quantitative range (1:40,000 to 1:2,560,000).
Comparison of sample matrices
Blood was obtained by means of venipuncture from 10 healthy subjects using 4 typical clinical specimen tubes for each donor: (1) serum, (2) plasma anticoagulated with EDTA, (3) plasma anticoagulated with sodium citrate, and (4) plasma anticoagulated with heparin. Serum or plasma was then harvested from the collection tube, placed in aliquots, and frozen at −70°C. Thawed aliquots were subsequently evaluated in the total periostin ELISA. Total measured periostin levels were comparable within each subject within assay limits independently of matrix and anticoagulant (data not shown).
Supplementary Material
Acknowledgments
We thank Jiabing Ding, Meredith Hazen, and Jo-Anne Hongo for assistance developing the periostin assay; Jon Hidayat and the BOBCAT protocol execution team for running the BOBCAT study; and Flavius Martin, John Monroe, Merdad Parsey, and John Matthews for advice and comments on the manuscript.
Subjects enrolled by the BOBCAT Study Group included the following investigators and sites: Steven Kelsen, Temple University Hospital; Michel Laviolette, Hopital Laval; Ammar Hatab, West Coast Clinical Trials; Mario Castro, Washington University School of Medicine; Eugene Bleecker, Wake Forest University; Neil Thomson, Gartnaval General Hospital; Richard Leigh, University of Calgary; Ron Olivenstein, McGill University; James Good, National Jewish Medical and Research Center; Gail Gauvreau, McMaster University; Nizar Jarjour, University of Wisconsin-Madison; Adel Mansur, Birmingham Heartlands; Pedro Avila, Northwestern University School of Medicine; Joel Kline, University of Iowa Hospitals and Clinics; Irvin Mayers, University of Alberta Hospital; Kyle Hogarth, University of Chicago; and Liam Heaney, Queen’s University Belfast.
Supported by Genentech; by grants to J.V.F., P.G.W., P.B., and Q.H. from Genentech; National Institutes of Health (NIH) grants (HL56385, HL080414, and HL66564 to J.V.F. and HL097591 and HL095372 to P.G.W.); and the Sandler Asthma Basic Research Center (to J.V.F.).
Abbreviations used
- ACQ
Asthma Control Questionnaire
- BOBCAT
Bronchoscopic Exploratory Research Study of Biomarkers in Corticosteroid-refractory Asthma
- CV
Coefficient of variation
- Feno
Fraction of exhaled nitric oxide
- ICS
Inhaled corticosteroid
- POSTN
Periostin gene
- rhuPOSTN
Recombinant human periostin protein
Footnotes
Clinical implications: Periostin levels in peripheral blood identify asthmatic patients with increased airway eosinophil numbers and therefore might be a clinically useful biomarker to select patients for TH2-targeted asthma therapies.
Disclosure of potential conflict of interest: G. Jia is employed by and has patents with Genentech. D. F. Choy is employed by, has received patents from, and has stock options in Genentech. S. Mosesova is employed by and has stock options in Genentech/Roche. L. C. Wu is employed by Genentech and holds equity in the Roche group. R. Carter has received research support from Genentech and has received travel expenses from Chiesi. S. Audusseau has received travel support for an investigator meeting. Q. Hamid has received research support from Meakins Christie Labs. P. Bradding has received research support from Genentech. J. V. Fahy has received grants from the National Heart, Lung, and Blood Institute and Boehringer Ingelheim; has received travel reimbursement from GlaxoSmithKline, Merck, Amgen, and the National Heart, Lung, and Blood Institute; is a member of the scientific Advisory Board for Cytokinetics; has consultant arrangements with Gilead, GlaxoSmithKline, Amgen, Portola Pharmaceuticals, Five Prime Therapeutics, and Merck; and is named inventor on a patent application for periostin as a biomarker in asthma. P. G. Woodruff has received research support from Genentech, is coinventor on a patent application related to periostin, is on the advisory board for Boehringer Ingelheim, and has received consultancy fees from MedImmune. J. M. Harris is employed by and has stock options in Genentech. J. R. Arron is employed by Genentech, has received payment for lectures from the American Asthma Association, has patents with Genentech, and has stock in Roche Holdings. The rest of the authors declare that they have no relevant conflicts of interest.
REFERENCES
- 1.Galli SJ, Tsai M, Piliponsky AM. The development of allergic inflammation. Nature. 2008;454:445–54. doi: 10.1038/nature07204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haldar P, Pavord ID. Noneosinophilic asthma: a distinct clinical and pathologic phenotype. J Allergy Clin Immunol. 2007;119:1043–54. doi: 10.1016/j.jaci.2007.02.042. [DOI] [PubMed] [Google Scholar]
- 3.Wenzel SE, Szefler SJ, Leung DY, Sloan SI, Rex MD, Martin RJ. Bronchoscopic evaluation of severe asthma. Persistent inflammation associated with high dose glucocorticoids. Am J Respir Crit Care Med. 1997;156:737–43. doi: 10.1164/ajrccm.156.3.9610046. [DOI] [PubMed] [Google Scholar]
- 4.Anderson GP. Endotyping asthma: new insights into key pathogenic mechanisms in a complex, heterogeneous disease. Lancet. 2008;372:1107–19. doi: 10.1016/S0140-6736(08)61452-X. [DOI] [PubMed] [Google Scholar]
- 5.Fahy JV. Eosinophilic and neutrophilic inflammation in asthma: insights from clinical studies. Proc Am Thorac Soc. 2009;6:256–9. doi: 10.1513/pats.200808-087RM. [DOI] [PubMed] [Google Scholar]
- 6.Fahy JV. Identifying clinical phenotypes of asthma: steps in the right direction. Am J Respir Crit Care Med. 2010;181:296–7. doi: 10.1164/rccm.200911-1702ED. [DOI] [PubMed] [Google Scholar]
- 7.Moore WC, Meyers DA, Wenzel SE, Teague WG, Li H, Li X, et al. Identification of asthma phenotypes using cluster analysis in the Severe Asthma Research Program. Am J Respir Crit Care Med. 2010;181:315–23. doi: 10.1164/rccm.200906-0896OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siddiqui S, Brightling CE. Airways disease: phenotyping heterogeneity using measures of airway inflammation. Allergy Asthma Clin Immunol. 2007;3:60–9. doi: 10.1186/1710-1492-3-2-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simpson JL, Scott R, Boyle MJ, Gibson PG. Inflammatory subtypes in asthma: assessment and identification using induced sputum. Respirology. 2006;11:54–61. doi: 10.1111/j.1440-1843.2006.00784.x. [DOI] [PubMed] [Google Scholar]
- 10.Wardlaw AJ, Silverman M, Siva R, Pavord ID, Green R. Multi-dimensional phenotyping: towards a new taxonomy for airway disease. Clin Exp Allergy. 2005;35:1254–62. doi: 10.1111/j.1365-2222.2005.02344.x. [DOI] [PubMed] [Google Scholar]
- 11.Cowan DC, Cowan JO, Palmay R, Williamson A, Taylor DR. Effects of steroid therapy on inflammatory cell subtypes in asthma. Thorax. 2010;65:384–90. doi: 10.1136/thx.2009.126722. [DOI] [PubMed] [Google Scholar]
- 12.Green RH, Brightling CE, McKenna S, Hargadon B, Parker D, Bradding P, et al. Asthma exacerbations and sputum eosinophil counts: a randomised controlled trial. Lancet. 2002;360:1715–21. doi: 10.1016/S0140-6736(02)11679-5. [DOI] [PubMed] [Google Scholar]
- 13.Woodruff PG, Modrek B, Choy DF, Jia G, Abbas AR, Ellwanger A, et al. T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am J Respir Crit Care Med. 2009;180:388–95. doi: 10.1164/rccm.200903-0392OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castro M, Mathur S, Hargreave F, Boulet LP, Xie F, Young J, et al. Reslizumab for poorly controlled, eosinophilic asthma: a randomized, placebo-controlled study. Am J Respir Crit Care Med. 2011;184:1125–32. doi: 10.1164/rccm.201103-0396OC. [DOI] [PubMed] [Google Scholar]
- 15.Haldar P, Brightling CE, Hargadon B, Gupta S, Monteiro W, Sousa A, et al. Mepolizumab and exacerbations of refractory eosinophilic asthma. N Engl J Med. 2009;360:973–84. doi: 10.1056/NEJMoa0808991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nair P, Pizzichini MM, Kjarsgaard M, Inman MD, Efthimiadis A, Pizzichini E, et al. Mepolizumab for prednisone-dependent asthma with sputum eosinophilia. N Engl J Med. 2009;360:985–93. doi: 10.1056/NEJMoa0805435. [DOI] [PubMed] [Google Scholar]
- 17.Hershey GK. IL-13 receptors and signaling pathways: an evolving web. J Allergy Clin Immunol. 2003;111:677–91. doi: 10.1067/mai.2003.1333. [DOI] [PubMed] [Google Scholar]
- 18.Berry MA, Parker D, Neale N, Woodman L, Morgan A, Monk P, et al. Sputum and bronchial submucosal IL-13 expression in asthma and eosinophilic bronchitis. J Allergy Clin Immunol. 2004;114:1106–9. doi: 10.1016/j.jaci.2004.08.032. [DOI] [PubMed] [Google Scholar]
- 19.Humbert M, Durham SR, Kimmitt P, Powell N, Assoufi B, Pfister R, et al. Elevated expression of messenger ribonucleic acid encoding IL-13 in the bronchial mucosa of atopic and nonatopic subjects with asthma. J Allergy Clin Immunol. 1997;99:657–65. doi: 10.1016/s0091-6749(97)70028-9. [DOI] [PubMed] [Google Scholar]
- 20.Saha SK, Berry MA, Parker D, Siddiqui S, Morgan A, May R, et al. Increased sputum and bronchial biopsy IL-13 expression in severe asthma. J Allergy Clin Immunol. 2008;121:685–91. doi: 10.1016/j.jaci.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shikotra A, Choy DF, Ohri CM, Doran E, Butler C, Hargadon B, et al. Increased expression of immunoreactive thymic stromal lymphopoietin in patients with severe asthma. J Allergy Clin Immunol. 2012;129:104–11. e9. doi: 10.1016/j.jaci.2011.08.031. [DOI] [PubMed] [Google Scholar]
- 22.Truyen E, Coteur L, Dilissen E, Overbergh L, Dupont LJ, Ceuppens JL, et al. Evaluation of airway inflammation by quantitative Th1/Th2 cytokine mRNA measurement in sputum of asthma patients. Thorax. 2006;61:202–8. doi: 10.1136/thx.2005.052399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woodruff PG, Boushey HA, Dolganov GM, Barker CS, Yang YH, Donnelly S, et al. Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc Natl Acad Sci U S A. 2007;104:15858–63. doi: 10.1073/pnas.0707413104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corren J, Lemanske RF, Hanania NA, Korenblat PE, Parsey MV, Arron JR, et al. Lebrikizumab treatment in adults with asthma. N Engl J Med. 2011;365:1088–98. doi: 10.1056/NEJMoa1106469. [DOI] [PubMed] [Google Scholar]
- 25.Boushey HA, Sorkness CA, King TS, Sullivan SD, Fahy JV, Lazarus SC, et al. Daily versus as-needed corticosteroids for mild persistent asthma. N Engl J Med. 2005;352:1519–28. doi: 10.1056/NEJMoa042552. [DOI] [PubMed] [Google Scholar]
- 26.Chanez P, Wenzel SE, Anderson GP, Anto JM, Bel EH, Boulet LP, et al. Severe asthma in adults: what are the important questions? J Allergy Clin Immunol. 2007;119:1337–48. doi: 10.1016/j.jaci.2006.11.702. [DOI] [PubMed] [Google Scholar]
- 27.Miranda C, Busacker A, Balzar S, Trudeau J, Wenzel SE. Distinguishing severe asthma phenotypes: role of age at onset and eosinophilic inflammation. J Allergy Clin Immunol. 2004;113:101–8. doi: 10.1016/j.jaci.2003.10.041. [DOI] [PubMed] [Google Scholar]
- 28.Silkoff PE, Lent AM, Busacker AA, Katial RK, Balzar S, Strand M, et al. Exhaled nitric oxide identifies the persistent eosinophilic phenotype in severe refractory asthma. J Allergy Clin Immunol. 2005;116:1249–55. doi: 10.1016/j.jaci.2005.09.029. [DOI] [PubMed] [Google Scholar]
- 29.Lemiere C, Ernst P, Olivenstein R, Yamauchi Y, Govindaraju K, Ludwig MS, et al. Airway inflammation assessed by invasive and noninvasive means in severe asthma: eosinophilic and noneosinophilic phenotypes. J Allergy Clin Immunol. 2006;118:1033–9. doi: 10.1016/j.jaci.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 30.Macedo P, Hew M, Torrego A, Jouneau S, Oates T, Durham A, et al. Inflammatory biomarkers in airways of patients with severe asthma compared with non-severe asthma. Clin Exp Allergy. 2009;39:1668–76. doi: 10.1111/j.1365-2222.2009.03319.x. [DOI] [PubMed] [Google Scholar]
- 31.Pavord ID, Shaw D. The use of exhaled nitric oxide in the management of asthma. J Asthma. 2008;45:523–31. doi: 10.1080/02770900801978557. [DOI] [PubMed] [Google Scholar]
- 32.Chupp GL, Lee CG, Jarjour N, Shim YM, Holm CT, He S, et al. A chitinase-like protein in the lung and circulation of patients with severe asthma. N Engl J Med. 2007;357:2016–27. doi: 10.1056/NEJMoa073600. [DOI] [PubMed] [Google Scholar]
- 33.Dweik RA, Sorkness RL, Wenzel S, Hammel J, Curran-Everett D, Comhair SA, et al. Use of exhaled nitric oxide measurement to identify a reactive, at-risk phenotype among patients with asthma. Am J Respir Crit Care Med. 2010;181:1033–41. doi: 10.1164/rccm.200905-0695OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barnes PJ. The cytokine network in asthma and chronic obstructive pulmonary disease. J Clin Invest. 2008;118:3546–56. doi: 10.1172/JCI36130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holgate ST, Polosa R. Treatment strategies for allergy and asthma. Nat Rev Immunol. 2008;8:218–30. doi: 10.1038/nri2262. [DOI] [PubMed] [Google Scholar]
- 36.Oh CK, Geba GP, Molfino N. Investigational therapeutics targeting the IL-4/IL-13/STAT-6 pathway for the treatment of asthma. Eur Respir Rev. 2010;19:46–54. doi: 10.1183/09059180.00007609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walsh GM. Emerging drugs for asthma. Expert Opin Emerg Drugs. 2008;13:643–53. doi: 10.1517/14728210802591378. [DOI] [PubMed] [Google Scholar]
- 38.Yuyama N, Davies DE, Akaiwa M, Matsui K, Hamasaki Y, Suminami Y, et al. Analysis of novel disease-related genes in bronchial asthma. Cytokine. 2002;19:287–96. doi: 10.1006/cyto.2002.1972. [DOI] [PubMed] [Google Scholar]
- 39.Takayama G, Arima K, Kanaji T, Toda S, Tanaka H, Shoji S, et al. Periostin: a novel component of subepithelial fibrosis of bronchial asthma downstream of IL-4 and IL-13 signals. J Allergy Clin Immunol. 2006;118:98–104. doi: 10.1016/j.jaci.2006.02.046. [DOI] [PubMed] [Google Scholar]
- 40.Hayashi N, Yoshimoto T, Izuhara K, Matsui K, Tanaka T, Nakanishi K. T helper 1 cells stimulated with ovalbumin and IL-18 induce airway hyperresponsiveness and lung fibrosis by IFN-gamma and IL-13 production. Proc Natl Acad Sci U S A. 2007;104:14765–70. doi: 10.1073/pnas.0706378104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abbas AR, Jackman JK, Bullens SL, Davis SM, Choy DF, Fedorowicz G, et al. Lung gene expression in a rhesus allergic asthma model correlates with physiologic parameters of disease and exhibits common and distinct pathways with human asthma and a mouse asthma model. Am J Pathol. 2011;179:1667–80. doi: 10.1016/j.ajpath.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sidhu SS, Yuan S, Innes AL, Kerr S, Woodruff PG, Hou L, et al. Roles of epithelial cell-derived periostin in TGF-beta activation, collagen production, and collagen gel elasticity in asthma. Proc Natl Acad Sci U S A. 2010;107:14170–5. doi: 10.1073/pnas.1009426107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Winpenny JP, Marsey LL, Sexton DW. The CLCA gene family: putative therapeutic target for respiratory diseases. Inflamm Allergy Drug Targets. 2009;8:146–60. doi: 10.2174/187152809788462590. [DOI] [PubMed] [Google Scholar]
- 44.Plager DA, Kahl JC, Asmann YW, Nilson AE, Pallanch JF, Friedman O, et al. Gene transcription changes in asthmatic chronic rhinosinusitis with nasal polyps and comparison to those in atopic dermatitis. PLoS One. 2010;5:e11450. doi: 10.1371/journal.pone.0011450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stankovic KM, Goldsztein H, Reh DD, Platt MP, Metson R. Gene expression profiling of nasal polyps associated with chronic sinusitis and aspirin-sensitive asthma. Laryngoscope. 2008;118:881–9. doi: 10.1097/MLG.0b013e31816b4b6f. [DOI] [PubMed] [Google Scholar]
- 46.Blanchard C, Mingler MK, Vicario M, Abonia JP, Wu YY, Lu TX, et al. IL-13 involvement in eosinophilic esophagitis: transcriptome analysis and reversibility with glucocorticoids. J Allergy Clin Immunol. 2007;120:1292–300. doi: 10.1016/j.jaci.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 47.Blanchard C, Mingler MK, McBride M, Putnam PE, Collins MH, Chang G, et al. Periostin facilitates eosinophil tissue infiltration in allergic lung and esophageal responses. Mucosal Immunol. 2008;1:289–96. doi: 10.1038/mi.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baril P, Gangeswaran R, Mahon PC, Caulee K, Kocher HM, Harada T, et al. Periostin promotes invasiveness and resistance of pancreatic cancer cells to hypoxiainduced cell death: role of the beta4 integrin and the PI3k pathway. Oncogene. 2007;26:2082–94. doi: 10.1038/sj.onc.1210009. [DOI] [PubMed] [Google Scholar]
- 49.Ben QW, Zhao Z, Ge SF, Zhou J, Yuan F, Yuan YZ. Circulating levels of periostin may help identify patients with more aggressive colorectal cancer. Int J Oncol. 2009;34:821–8. doi: 10.3892/ijo_00000208. [DOI] [PubMed] [Google Scholar]
- 50.Kudo Y, Ogawa I, Kitajima S, Kitagawa M, Kawai H, Gaffney PM, et al. Periostin promotes invasion and anchorage-independent growth in the metastatic process of head and neck cancer. Cancer Res. 2006;66:6928–35. doi: 10.1158/0008-5472.CAN-05-4540. [DOI] [PubMed] [Google Scholar]
- 51.Puglisi F, Puppin C, Pegolo E, Andreetta C, Pascoletti G, D’Aurizio F, et al. Expression of periostin in human breast cancer. J Clin Pathol. 2008;61:494–8. doi: 10.1136/jcp.2007.052506. [DOI] [PubMed] [Google Scholar]
- 52.Siriwardena BS, Kudo Y, Ogawa I, Kitagawa M, Kitajima S, Hatano H, et al. Periostin is frequently overexpressed and enhances invasion and angiogenesis in oral cancer. Br J Cancer. 2006;95:1396–403. doi: 10.1038/sj.bjc.6603431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sasaki H, Dai M, Auclair D, Fukai I, Kiriyama M, Yamakawa Y, et al. Serum level of the periostin, a homologue of an insect cell adhesion molecule, as a prognostic marker in nonsmall cell lung carcinomas. Cancer. 2001;92:843–8. doi: 10.1002/1097-0142(20010815)92:4<843::aid-cncr1391>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 54.Sasaki H, Lo KM, Chen LB, Auclair D, Nakashima Y, Moriyama S, et al. Expression of Periostin, homologous with an insect cell adhesion molecule, as a prognostic marker in non-small cell lung cancers. Jpn J Cancer Res. 2001;92:869–73. doi: 10.1111/j.1349-7006.2001.tb01174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sasaki H, Yu CY, Dai M, Tam C, Loda M, Auclair D, et al. Elevated serum periostin levels in patients with bone metastases from breast but not lung cancer. Breast Cancer Res Treat. 2003;77:245–52. doi: 10.1023/a:1021899904332. [DOI] [PubMed] [Google Scholar]
- 56.Dweik RA, Boggs PB, Erzurum SC, Irvin CG, Leigh MW, Lundberg JO, et al. An official ATS clinical practice guideline: interpretation of exhaled nitric oxide levels (FENO) for clinical applications. Am J Respir Crit Care Med. 2011;184:602–15. doi: 10.1164/rccm.9120-11ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nair P, Kjarsgaard M, Armstrong S, Efthimiadis A, O’Byrne PM, Hargreave FE. Nitric oxide in exhaled breath is poorly correlated to sputum eosinophils in patients with prednisone-dependent asthma. J Allergy Clin Immunol. 2010;126:404–6. doi: 10.1016/j.jaci.2010.05.032. [DOI] [PubMed] [Google Scholar]
- 58.Shaw DE, Berry MA, Thomas M, Green RH, Brightling CE, Wardlaw AJ, et al. The use of exhaled nitric oxide to guide asthma management: a randomized controlled trial. Am J Respir Crit Care Med. 2007;176:231–7. doi: 10.1164/rccm.200610-1427OC. [DOI] [PubMed] [Google Scholar]
- 59.D’Silva L, Cook RJ, Allen CJ, Hargreave FE, Parameswaran K. Changing pattern of sputum cell counts during successive exacerbations of airway disease. Respir Med. 2007;101:2217–20. doi: 10.1016/j.rmed.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 60.Pavord ID, Jeffery PK, Qiu Y, Zhu J, Parker D, Carlsheimer A, et al. Airway inflammation in patients with asthma with high-fixed or low-fixed plus as-needed budesonide/formoterol. J Allergy Clin Immunol. 2009;123:1083–9. e1–7. doi: 10.1016/j.jaci.2009.02.034. [DOI] [PubMed] [Google Scholar]
- 61.Leung DY, Martin RJ, Szefler SJ, Sher ER, Ying S, Kay AB, et al. Dysregulation of interleukin 4, interleukin 5, and interferon gamma gene expression in steroidresistant asthma. J Exp Med. 1995;181:33–40. doi: 10.1084/jem.181.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Flood-Page P, Swenson C, Faiferman I, Matthews J, Williams M, Brannick L, et al. A study to evaluate safety and efficacy of mepolizumab in patients with moderate persistent asthma. Am J Respir Crit Care Med. 2007;176:1062–71. doi: 10.1164/rccm.200701-085OC. [DOI] [PubMed] [Google Scholar]
- E1.Woodruff PG, Boushey HA, Dolganov GM, Barker CS, Yang YH, Donnelly S, et al. Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc Natl Acad Sci U S A. 2007;104:15858–63. doi: 10.1073/pnas.0707413104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E2.Woodruff PG, Modrek B, Choy DF, Jia G, Abbas AR, Ellwanger A, et al. T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am J Respir Crit Care Med. 2009;180:388–95. doi: 10.1164/rccm.200903-0392OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E3.Juniper EF, Bousquet J, Abetz L, Bateman ED. Identifying “well-controlled” and “not well-controlled” asthma using the Asthma Control Questionnaire. Respir Med. 2006;100:616–21. doi: 10.1016/j.rmed.2005.08.012. [DOI] [PubMed] [Google Scholar]
- E4.Boushey HA, Sorkness CA, King TS, Sullivan SD, Fahy JV, Lazarus SC, et al. Daily versus as-needed corticosteroids for mild persistent asthma. N Engl J Med. 2005;352:1519–28. doi: 10.1056/NEJMoa042552. [DOI] [PubMed] [Google Scholar]
- E5.Brightling CE, Symon FA, Birring SS, Bradding P, Wardlaw AJ, Pavord ID. Comparison of airway immunopathology of eosinophilic bronchitis and asthma. Thorax. 2003;58:528–32. doi: 10.1136/thorax.58.6.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E6.Lemiere C, Ernst P, Olivenstein R, Yamauchi Y, Govindaraju K, Ludwig MS, et al. Airway inflammation assessed by invasive and noninvasive means in severe asthma: eosinophilic and noneosinophilic phenotypes. J Allergy Clin Immunol. 2006;118:1033–9. doi: 10.1016/j.jaci.2006.08.003. [DOI] [PubMed] [Google Scholar]
- E7.Hongo JA, Tsai SP, Moffat B, Schroeder KA, Jung C, Chuntharapai A, et al. Characterization of novel neutralizing monoclonal antibodies specific to human neurturin. Hybridoma. 2000;19:303–15. doi: 10.1089/027245700429855. [DOI] [PubMed] [Google Scholar]
- E8.Huang Z, Zhu W, Szekeres G, Xia H. Development of new rabbit monoclonal antibody to estrogen receptor: immunohistochemical assessment on formalin-fixed, paraffin-embedded tissue sections. Appl Immunohistochem Mol Morphol. 2005;13:91–5. doi: 10.1097/00129039-200503000-00015. [DOI] [PubMed] [Google Scholar]
- E9.Spieker-Polet H, Sethupathi P, Yam PC, Knight KL. Rabbit monoclonal anti-bodies: generating a fusion partner to produce rabbit-rabbit hybridomas. Proc Natl Acad Sci U S A. 1995;92:9348–52. doi: 10.1073/pnas.92.20.9348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E10.Hoersch S, Andrade-Navarro MA. Periostin shows increased evolutionary plasticity in its alternatively spliced region. BMC Evol Biol. 2010;10:30. doi: 10.1186/1471-2148-10-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.