Abstract
Purpose.
We hypothesize that growth factors, including epidermal growth factor (EGF) and bovine pituitary extract (BPE), induce proliferation, but not differentiation (e.g., lipid accumulation), of human meibomian gland epithelial cells. We also hypothesize that these actions involve a significant upregulation of genes linked to cell cycle processes, and a significant downregulation of genes associated with differentiation. Our objective was to test these hypotheses.
Methods.
Immortalized human meibomian gland and conjunctival epithelial cells were cultured for varying time periods in the presence or absence of EGF, BPE, EGF + BPE, or serum, followed by cell counting, neutral lipid staining, or RNA isolation for molecular biological procedures.
Results.
Our studies show that growth factors stimulate a significant, time-dependent proliferation of human meibomian gland epithelial cells. These effects are associated with a significant upregulation of genes linked to cell cycle, DNA replication, ribosomes, and translation, and a significant decrease in those related to cell differentiation, tissue development, lipid metabolic processes, and peroxisome proliferator-activated receptor signaling. Serum-induced differentiation, but not growth factor-related proliferation, elicits a pronounced lipid accumulation in human meibomian gland epithelial cells. This lipogenic response is unique, and is not duplicated by human conjunctival epithelial cells.
Conclusions.
Our results demonstrate that EGF and BPE stimulate human meibomian gland epithelial cells to proliferate. Further, our findings show that action is associated with an upregulation of cell cycle and translation ontologies, and a downregulation of genetic pathways linked to differentiation and lipid biosynthesis.
Keywords: meibomian gland, growth factors, gene expression
Our study demonstrates that growth factors stimulate human meibomian gland epithelial cells to proliferate, and that this action is associated with an upregulation of cell cycle and translation ontologies, and a downregulation of genetic pathways linked to differentiation and lipid biosynthesis.
Introduction
Meibomian gland secretion is critically dependent upon the active proliferation of glandular epithelial cells.1 Once generated, these sebaceous-like cells undergo a maturation process toward terminal differentiation, lipid production, and holocrine secretion. Such secretion involves the death and disintegration of fully mature, lipid-rich epithelial cells, their release into glandular ductules, and ultimately, delivery to the ocular surface.1,2 Given this continual loss of cells, stimulation of epithelial cell proliferation is extremely important and promotes not only meibocyte replenishment, but also the production of meibum.
The stimuli that induce proliferation of human meibomian gland epithelial cells are unknown. We hypothesize that epidermal growth factor (EGF), as well as pituitary hormones and growth factors (i.e., in the components of bovine pituitary extract [BPE]) will induce such proliferation, but not differentiation (e.g., lipid accumulation). We also hypothesize that these actions will involve a significant upregulation of genes linked to cell cycle processes, and a significant downregulation of genes associated with differentiation.
In support of our hypotheses, researchers have shown that EGF is a strong mitogen that stimulates the clonal growth of rabbit meibomian gland epithelial cells,3 enhances the mitotic activity of hamster sebocytes,4,5 and promotes the proliferation, but not differentiation, of human amnion epithelial cells by regulating cell cycle genes.6 Similarly, several pituitary hormones increase sebaceous gland epithelial cell growth,2 and BPE augments the proliferation of human mammary, thymic, ureteral, and conjunctival epithelial cells.7–10 Scientists also have reported that EGF and BPE increase the proliferation of normal and immortalized keratinocytes, whereas factors, such as serum, induce their terminal differentiation.11 In addition, hamster sebocytes exposed to EGF do not accumulate intracellular lipids.5
The purpose of our study was to test our hypotheses with human meibomian gland epithelial cells. Toward that end, we examined the effects of EGF, BPE, and both factors in combination, on the proliferation, lipid accumulation, and gene expression of these cells in vitro.
Methods
Meibomian Gland Epithelial Cell Culture Procedures
Immortalized human meibomian gland epithelial cells, recently generated in our laboratory,12 were cultured in 6-well plates (BD Falcon, San Jose, CA) or 10 cm culture dishes (Corning Incorporated, Corning, NY) at 37°C in 5% CO2 in one of the following media: MCDB 153 medium (MCDB; Sigma-Aldrich, St. Louis, MO), which was developed originally for the serum-free growth of specific cell types; serum-free keratinocyte basal medium (SFM; Invitrogen-Gibco, Grand Island, NY); SFM with 5 ng/mL EGF (Invitrogen-Gibco); SFM with 50 μg/mL BPE (Invitrogen-Gibco); SFM with EGF + BPE; or serum-containing medium, which consisted of an equal volume of Dulbecco's modified Eagle's medium (DMEM) and Ham's F12 (Invitrogen-Gibco), supplemented with 10% fetal bovine serum (Invitrogen-Gibco) and 10 ng/mL EGF. After varying time intervals, total cells were counted with a hemocytometer under a phase-contrast microscope (Nikon Eclipse TS100; Nikon, Avon, MA).
For statistical evaluation of cell counts, ANOVA and Bonferroni multiple comparisons tests were performed with Prism 5 (GraphPad Software, Inc., La Jolla, CA).
LipidTox Staining
Immortalized human meibomian gland and conjunctival epithelial cells (gift of Ilene K. Gipson, Boston, MA) were cultured on glass coverslips (Electron Microscopy Sciences, Hatfield, PA) placed in 6-well plates in SFM supplemented with 5 ng/mL EGF and either 50 μg/mL BPE (meibomian cells) or 25 μg/mL BPE (conjunctival cells). When cells reached approximately 75% confluence, medium was changed to SFM, SFM supplemented with 50 μg/mL BPE and 5 ng/mL EGF, or serum-containing medium. Media were replaced every two to three days for seven days, when cells were washed, then fixed in 4% paraformaldehyde for 30 minutes. Following additional washes, coverslips were exposed to LipidTOX Green neutral lipid stain (Invitrogen, Grand Island, NY) in a humid chamber for 30 minutes. Coverslips were mounted on slides with ProLong Gold antifade reagent with DAPI (Invitrogen) and allowed to dry overnight before imaging with a Nikon Eclipse E800 (Nikon Instruments, Melville, NY).
Microarray Procedures
Total RNA was extracted from cells, evaluated for integrity, and processed for the determination of mRNA levels at Asuragen (Austin, TX), as described previously.13 Our studies used Illumina HumanHT-12 v3 and v4 Expression BeadChips (Illumina, San Diego, CA). Data were obtained with Illumina BeadStudio software, and used background subtraction and cubic spline normalization. Normalized hybridization intensity values were adjusted by adding a constant, so that the lowest intensity value for any sample was equal to 16.14
Standardized data were analyzed without log transformation and statistical analyses were performed with Student's t-test (two-tailed, unpaired). These evaluations were conducted with GeneSifter software (Geospiza, Seattle, WA), which also generated gene ontology, KEGG pathway, and Z-score (zsc) reports organized according to the guidelines of the Gene Ontology Consortium (available in the public domain at http://www.geneontology.org/GO).15,16 Comparison of gene data between groups was facilitated by the use of the GeneSifter intersector program (Geospiza; available in the public domain at www.public.genesifter.net). All data from the Illumina BeadChips are accessible for download through the National Center for Biotechnology Information's Gene Expression Omnibus (available in the public domain at http://www.ncbi.nlm.nih.gov/geo) via series accession numbers GSE18099 and GSE 37,089.
Real-Time PCR Procedures
The differential expression of selected genes was confirmed by using quantitative real-time PCR (qPCR) methods. The cDNAs were transcribed by using SuperScript III Reverse Transcriptase (Invitrogen) and random hexamer primers (Invitrogen). The qPCR reactions were performed in triplicate by using TaqMan Gene Assays (Applied Biosystems, Inc., Foster City, CA), TaqMan-specific primers and probes for small proline-rich protein 3 (Hs01878180_s1), keratin 10 (Hs00166289_m1), TIMP metallopeptidase inhibitor 1 (Hs00171558_m1), laminin alpha 3 (Hs00165042_m1), and GAPDH (4326317E). Differential gene expression was calculated by following the Comparative Ct method, as described in Applied Biosystems User Bulletin 2 (updated 2001).
Results
Effect of EGF and BPE on the Proliferation of Human Meibomian Gland Epithelial Cells
To determine whether EGF and/or BPE stimulate the proliferation of human meibomian gland epithelial cells, we conducted an extensive series of time course experiments.
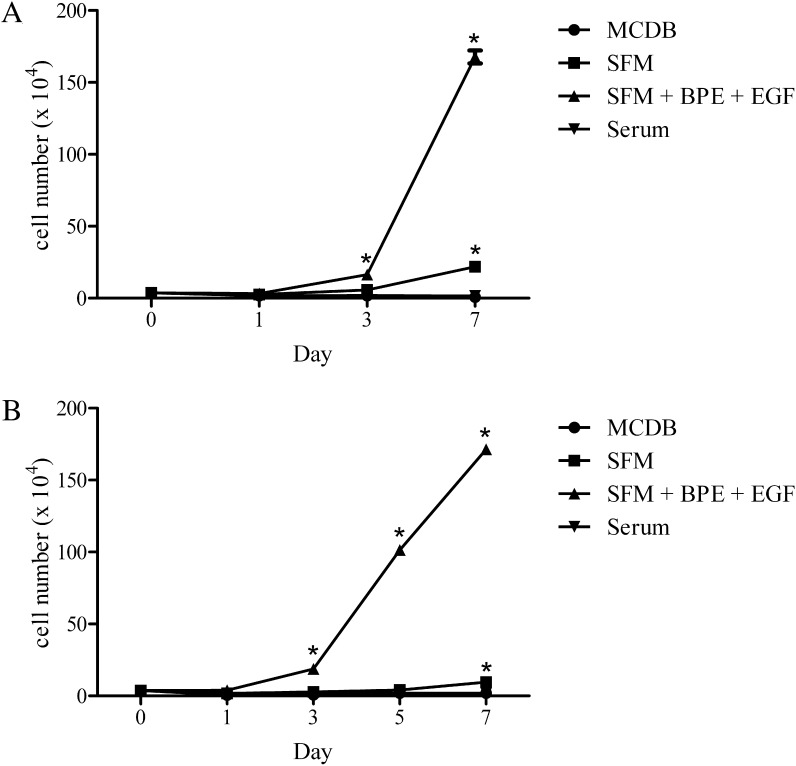
Our first set of studies was performed after culturing cells (3.76 × 104cells/well; 3 wells/condition) to 20% to 30% confluence, then for an additional 1, 3, and 7 days in one of 4 different media. These included MCDB, SFM, SFM + EGF + BPE, and serum-containing media. Our results demonstrate that SFM + EGF + BPE induced a marked rise in cell proliferation (Fig. 1A). The total number of cells increased approximately 50-fold from day 1, with the largest rise occurring between days 3 and 7. The cells appeared to reach confluence by day 7. In contrast, MCDB supported minimal to no cell proliferation; rather, the total number of cells decreased by approximately 80% over the 7-day period. SFM promoted a gradual increase in the cell proliferation rate. Compared to the cell count at day 1, the total number of cells rose 2-fold, then 8-fold by days 3 and 7. Serum-containing media induced little to no proliferation. The cell count remained the same between days 1 and 7. In addition, cellular morphology appeared flat and enlarged (data not shown).
Figure 1.
Effect of EGF + BPE, serum, and media on the proliferation of human meibomian gland epithelial cells. Cells (n = 3 wells/condition) at passages 44 (A) and 49 (B) were cultured as described in the text. Values equal the mean ± SE. *Significantly (P < 0.0001) greater than values of all other conditions on that day.
To confirm and extend these experiments, we cultured 20% to 30% confluent cells for 1, 3, 5, and 7 days in the 4 different media. As shown in Figure 1B, our findings again demonstrated that SFM + EGF + BPE stimulated an ever-increasing cell proliferation rate, and a 45-fold rise in the total cell count between days 1 and 7. MCDB did not support cell proliferation, and most cells detached from the well within 3 days of culture. SFM permitted slight, but steady, cell proliferation. The total number of cells, relative to the quantity at day 1, increased by 5.4-fold during the 7-day time course. Serum-containing media supported little or no proliferation, and the cell count remained constant from days 1 to 7.
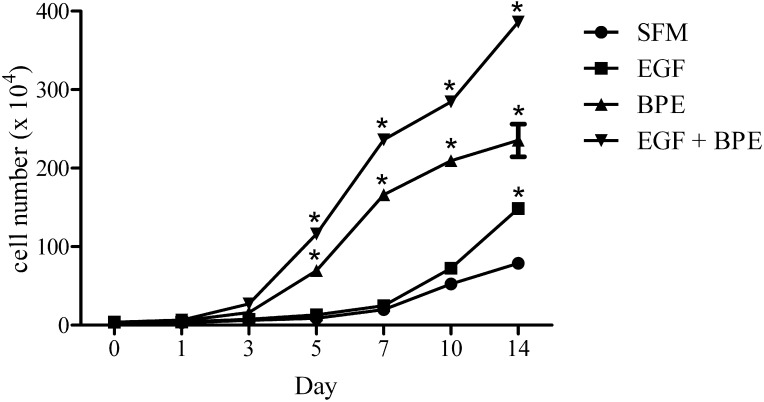
To examine the impact of individual supplements on cellular proliferation, we cultured cells in SFM in the presence or absence of EGF, BPE, or EGF + BPE. Cells were 20% to 30% confluent on day 0, and were maintained for 1, 3, 5, 7, 10, and 14 days in culture. Our results demonstrated that cell proliferation rates increased during the time course in all media conditions (Fig. 2). The relative rates were as follows: EGF + BPE > BPE > EGF > SFM. Within 5 days of culture, total cell counts had increased 2.9-, 3.3-, 12.8-, and 18.2-fold in SFM, SFM + EGF, SFM + BPE, and SFM + EGF + BPE media, respectively. By day 5 of culture, cells cultured in the SFM + EGF + BPE media appeared to be 90% to 95% confluent. After 14 days of culture, total cell counts had risen 25.0-, 36.8-, 43.6-, and 60.7-fold in SFM, SFM + EGF, SFM + BPE, and SFM + EGF + BPE media, respectively. These findings indicated that cell proliferation may continue after cells reach confluence. However, the rate of proliferation appeared to decline after confluence was achieved.
Figure 2.
Influence of EGF, BPE, and EGF + BPE on the proliferation of human meibomian gland epithelial cells. Cells at passage 50 were cultured as explained in the text. Values represent the mean ± SE. *Significantly (P < 0.0001) greater than SFM control.
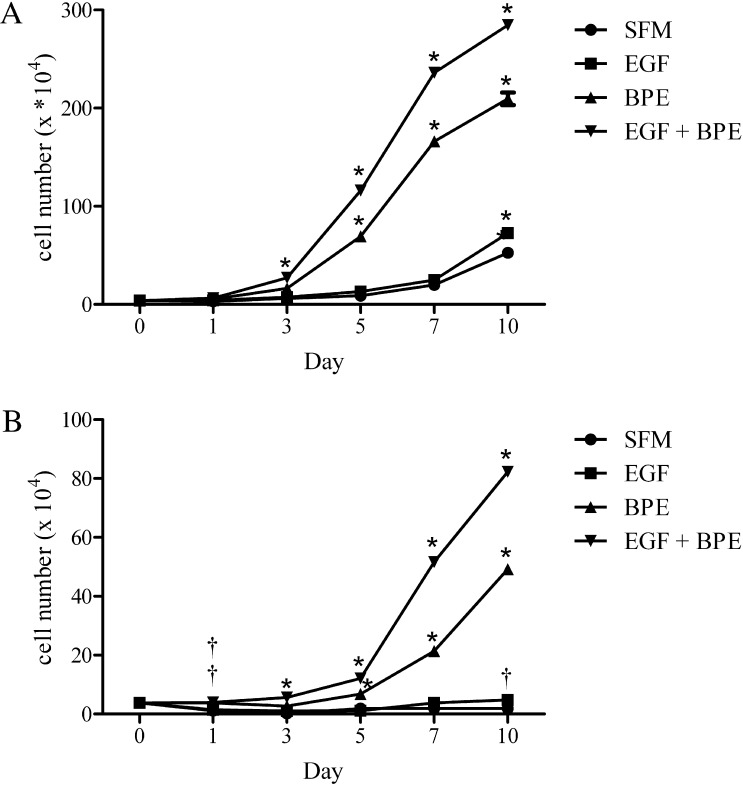
The rapidity and magnitude of the proliferative response to EGF and BPE were influenced by the cell passage number. As illustrated in Figure 3A, exposure of passage 50 human meibomian gland epithelial cells to EGF + BPE led to 1.7-, 4.3-, and 62.8-fold increases in cell number by 1, 3, and 7 days after treatment, respectively. By day 7, these cells were completely confluent and had begun to stratify. In contrast, earlier passage cells required more time to reach log phase growth. As shown in Figure 3B, the number of passage 16 cells increased 1.06-, 1.4-, and 13.7-fold within 1, 3, and 7 days after culture in EGF and BPE. At day 7, cells were approximately 90% confluent. Of particular note, these earlier passage cells did not proliferate in SFM, and most cells died and detached from the plates within 3 days of culture (Fig. 3A).
Figure 3.
Impact of EGF, BPE, and EGF + BPE on the proliferation of human meibomian gland epithelial cells. Cells (n = 3 wells/condition) at passages 50 (A) and 16 (B) were cultured as described in the text. Values equal the mean ± SE. Significantly (*P < 0.0001 and †P < 0.01) greater than SFM control. The double † in (B) indicates that BPE and EGF + BPE conditions were significantly different than the SFM control.
Influence of EGF and BPE on Gene Expression in Human Meibomian Gland Epithelial Cells
To examine whether growth factors upregulate genes linked to cell cycle processes, and downregulate genes associated with differentiation, we treated human meibomian gland epithelial cells with EGF and/or BPE for varying periods of time and then processed the cells for analyses with Illumina BeadChips and bioinformatic software.
In the first set of experiments, we cultured subconfluent, passage 16, human meibomian gland epithelial cells for 2 days in SFM (n = 3.76 × 104cells/well), followed by an additional 5 days in media containing EGF, BPE, or EGF + BPE (n = 3 wells/condition). Our findings demonstrate that growth factor-induced proliferation is associated with a significant (P < 0.05) change in the expression of thousands of genes (Table 1). As compared to the basal state, EGF, BPE, and their combination stimulated a significant increase in the expression of genes associated with the cell cycle, cell proliferation, DNA replication, ribosomes, and translation (Table 2). Conversely, these growth factors caused a significant decrease in ontologies related to cell differentiation, keratinization, tissue development, and fatty acid and lipid metabolic processes (Table 3).
Table 1. .
Effect of EGF and/or BPE on Gene Expression in Human Meibomian Gland Epithelial Cells
|
Group |
Genes ↑ |
Genes ↓ |
Total Genes |
| EGF | 1711 | 1709 | 3420 |
| BPE | 1941 | 1803 | 3744 |
| EGF + BPE | 1622 | 1670 | 3292 |
Data were evaluated without log transformation. The expression of listed genes was significantly (P < 0.05) up- (↑) or downregulated (↓) by growth factor exposure, compared to the basal SFM control group.
Table 2. .
Growth Factor Upregulation of Cell Cycle and Translational Process Ontologies
|
Ontology |
EGF Genes ↑ |
EGF Z-Score |
BPE Genes ↑ |
BPE Z-Score |
EGF + BPE Genes ↑ |
EGF + BPE Z-Score |
| Cell cycle | ||||||
| Cell cycle | 147 | 5.9 | 231 | 12.9 | 167 | 8.8 |
| Cell cycle arrest | 54 | 5.5 | 75 | 8.4 | 60 | 7.1 |
| Cell cycle checkpoint | 41 | 5.9 | 65 | 10.6 | 51 | 8.8 |
| Cell cycle phase | 95 | 5.1 | 169 | 13.4 | 123 | 9.6 |
| Cell cycle process | 112 | 5.3 | 190 | 13.0 | 138 | 9.2 |
| Cell division | 52 | 3.9 | 92 | 9.9 | 70 | 7.7 |
| Cell growth | 38 | 2.2 | 33 | 2.3 | ||
| Cell proliferation | 143 | 5.1 | 155 | 4.5 | 131 | 4.3 |
| Chromosome | 98 | 7.8 | 70 | 4.9 | ||
| Chromosome, centromeric region | 42 | 8.6 | 29 | 5.7 | ||
| Condensed chromosome | 19 | 2.4 | 46 | 9.8 | 30 | 6.0 |
| Condensed chromosome kinetochore | 11 | 2.2 | 27 | 8.3 | 18 | 5.4 |
| Condensed chromosome, centromeric region | 12 | 2.4 | 29 | 8.6 | 20 | 5.9 |
| DNA replication | 34 | 3.4 | 57 | 7.7 | 44 | 6.1 |
| G1/S transition checkpoint | 12 | 2.3 | 14 | 2.6 | 12 | 2.5 |
| G1/S transition of mitotic cell cycle | 31 | 4.8 | 43 | 7.1 | 35 | 6.2 |
| G2/M transition of mitotic cell cycle | 11 | 0.5 | 25 | 4.4 | 20 | 3.7 |
| Interphase | 51 | 4.7 | 80 | 9.1 | 63 | 7.5 |
| Interphase of mitotic cell cycle | 50 | 4.7 | 77 | 8.8 | 61 | 7.3 |
| Kinetochore | 29 | 7.9 | 20 | 5.3 | ||
| M phase | 56 | 3.3 | 112 | 11.4 | 79 | 7.8 |
| M phase of mitotic cell cycle | 43 | 3.3 | 93 | 11.8 | 65 | 8.1 |
| M/G1 transition of mitotic cell cycle | 14 | 3.1 | 26 | 7.2 | 19 | 5.4 |
| Mitosis | 42 | 3.3 | 90 | 11.6 | 64 | 8.2 |
| Mitotic cell cycle | 92 | 5.7 | 163 | 14.0 | 120 | 10.3 |
| Mitotic cell cycle checkpoint | 23 | 4.1 | 34 | 6.8 | 28 | 6.0 |
| Mitotic cell cycle G1/S transition checkpoint | 12 | 2.3 | 14 | 2.6 | 12 | 2.5 |
| Mitotic cell cycle G1/S transition DNA damage checkpoint | 11 | 2.4 | 13 | 2.7 | 10 | 2.1 |
| Mitotic metaphase/anaphase transition | 11 | 3.5 | 16 | 5.4 | 13 | 4.7 |
| Mitotic prometaphase | 32 | 9.0 | 23 | 6.7 | ||
| mRNA binding | 19 | 5.4 | 19 | 4.8 | 14 | 3.6 |
| Positive regulation of cell cycle | 16 | 3.6 | 12 | 2.7 | ||
| Positive regulation of cell cycle process | 12 | 1.2 | 20 | 3.4 | 15 | 2.5 |
| Positive regulation of cell proliferation | 59 | 3.4 | 63 | 2.9 | ||
| Regulation of cell cycle | 87 | 6.1 | 121 | 9.6 | 93 | 7.5 |
| Regulation of cell cycle arrest | 42 | 5.8 | 65 | 10.2 | 51 | 8.4 |
| Regulation of cell cycle process | 51 | 4.9 | 86 | 10.5 | 63 | 7.7 |
| Regulation of cell proliferation | 105 | 4.1 | 108 | 2.9 | 93 | 3.0 |
| Regulation of mitosis | 15 | 3.5 | 21 | 5.2 | 18 | 4.9 |
| Regulation of mitotic cell cycle | 34 | 3.1 | 54 | 6.6 | 40 | 4.8 |
| Regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 14 | 3.2 | 20 | 5.0 | 14 | 3.4 |
| RNA binding | 147 | 11.6 | 149 | 10.2 | 143 | 11.8 |
| S phase | 38 | 7.6 | 28 | 5.7 | ||
| S phase of mitotic cell cycle | 23 | 4.2 | 35 | 7.1 | 26 | 5.4 |
| Translation | ||||||
| Ribonucleoprotein complex | 116 | 13.4 | 126 | 12.9 | 120 | 13.6 |
| Ribosome | 69 | 15.2 | 52 | 9.3 | 56 | 11.9 |
| Regulation of translation | 25 | 3.7 | 26 | 4.2 | ||
| Structural constituent of ribosome | 67 | 16.4 | 47 | 9.3 | 55 | 13.3 |
| Translation | 114 | 14.7 | 86 | 8.1 | 103 | 13.2 |
| Translation factor activity, nucleic acid binding | 22 | 6.3 | 16 | 3.3 | 19 | 5.3 |
| Translation initiation factor activity | 15 | 5.8 | 9 | 2.2 | 13 | 5.0 |
| Translational elongation | 54 | 16.4 | 36 | 8.9 | 47 | 14.3 |
| Translational initiation | 19 | 6.0 | 12 | 2.4 | 16 | 4.9 |
| Translational termination | 50 | 16.2 | 33 | 8.7 | 42 | 13.5 |
Specific ontologies were selected after analyses of nontransformed data. A Z-score is a statistical rating of the relative expression of gene ontologies, and indicates how much a given ontology is over (positive)– or under (negative)–represented in a designated gene list. In other words, a Z-score is a normalized difference using the expected value and standard deviation of the number of genes.16 Positive Z-scores represent gene ontology terms with a higher number of genes meeting the criterion than is expected by chance, whereas negative Z-scores indicate gene ontology terms with a lower number of genes meeting the criterion than expected by chance. A Z-score near zero indicates that the number of genes meeting the criterion approximates the anticipated number.16 In this table, Z-scores with values > 2.0 are reported for selected ontologies with ≥14 genes. Genes ↑, number of genes upregulated in human meibomian gland epithelial cells after treatment with EGF, BPE, or EGF + BPE; Z-score, specific score for the upregulated genes in the growth factor-exposed cells.
Table 3. .
Growth Factor Downregulation of Cell Differentiation and Lipid Synthesis Ontologies
|
Ontology |
EGF Genes ↓ |
EGF Z-Score |
BPE Genes ↓ |
BPE Z-Score |
EGF + BPE Genes ↓ |
EGF + BPE Z-Score |
| Cell differentiation | ||||||
| Cell differentiation | 205 | 2.46 | 194 | 2.58 | ||
| Cornified envelope | 16 | 11.83 | 16 | 11.53 | 16 | 12.01 |
| Epidermal cell differentiation | 27 | 7.41 | 27 | 7.83 | ||
| Epidermis development | 50 | 8.29 | 47 | 7.16 | 44 | 6.91 |
| Epithelial cell differentiation | 41 | 6.3 | 40 | 5.7 | 39 | 5.92 |
| Epithelium development | 55 | 4.32 | 55 | 3.94 | 53 | 4.09 |
| Keratinization | 13 | 5.68 | 12 | 4.89 | 13 | 5.76 |
| Keratinocyte differentiation | 26 | 8.48 | 25 | 7.76 | 25 | 8.18 |
| Tissue development | 110 | 4.75 | 108 | 3.95 | 107 | 4.58 |
| Lipid synthesis | ||||||
| Cellular lipid catabolic process | 20 | 3.51 | 18 | 2.61 | 17 | 2.58 |
| Cellular lipid metabolic process | 84 | 4.41 | 88 | 4.51 | 84 | 4.6 |
| Cholesterol biosynthetic process | 12 | 4.68 | 12 | 4.95 | ||
| Cholesterol metabolic process | 15 | 2.27 | 17 | 2.77 | 15 | 2.35 |
| Fatty acid β-oxidation | 14 | 5.56 | 12 | 4.28 | 11 | 4.01 |
| Fatty acid biosynthetic process | 20 | 3.32 | ||||
| Fatty acid catabolic process | 14 | 4.46 | 12 | 3.31 | 11 | 3.09 |
| Fatty acid metabolic process | 38 | 4.05 | 37 | 3.5 | 31 | 2.53 |
| Fatty acid oxidation | 18 | 4.94 | 16 | 3.89 | 13 | 2.92 |
| Glycerolipid biosynthetic process | 19 | 3.17 | 18 | 2.61 | 18 | 2.92 |
| Glycerophospholipid biosynthetic process | 14 | 3.26 | 12 | 2.25 | 12 | 2.49 |
| Lipid biosynthetic process | 55 | 3.79 | 56 | 3.59 | 53 | 3.58 |
| Lipid catabolic process | 27 | 2.36 | ||||
| Lipid metabolic process | 121 | 5.13 | 124 | 4.91 | 116 | 4.74 |
| Lipid modification | 21 | 3.89 | 21 | 3.65 | 18 | 2.96 |
| Lipid oxidation | 18 | 4.94 | 16 | 3.89 | 13 | 2.92 |
| Phospholipid biosynthetic process | 19 | 3.21 | 17 | 2.61 | ||
| Phospholipid metabolic process | 25 | 2.28 | ||||
Specific ontologies were selected after evaluation of nontransformed data. Z-scores with values > 2.0 are reported for selected ontologies with ≥14 genes. Genes ↓, number of genes downregulated in human meibomian gland epithelial cells after treatment with EGF, BPE, or EGF + BPE; Z-score, specific score for the downregulated genes in the growth factor-exposed cells.
These modulatory effects were duplicated in a second series of experiments, which involved the treatment of passage 24 human meibomian gland epithelial cells (n = 3 wells/condition) under proliferating (i.e., EGF + BPE) or differentiating (i.e., serum) conditions for 4 days. The combined growth factors significantly (P < 0.05) increased ontologies associated with cell cycle processes (zsc = 17.1), cell proliferation (zsc = 3.6), DNA replication (zsc = 11.2), ribosome biogenesis (zsc = 8.6), and translation (zsc = 6.9), and reduced those linked to epithelial cell differentiation (zsc = 5.8), tissue development (zsc = 3.0), lipid metabolic processes (zsc = 3.1), and lipid modification (zsc = 4.5).
The influence of growth factors on human meibomian gland epithelial cells also was shown by the analysis of KEGG pathways. As demonstrated in Table 4, EGF, BPE, and EGF + BPE promoted the activity of cell cycle, DNA replication, and ribosome pathways, and decreased processes related to fatty acid metabolism and peroxisome proliferator–activated receptor (PPAR) signaling (e.g., PPARδ and retinoid X receptor α [RXRα]). Of interest, the growth factors also suppressed pathways associated with acid secretion and lysosomes.
Table 4. .
Growth Factor Influence on KEGG Pathways in Human Meibomian Gland Epithelial Cells
|
Ontology |
EGF Genes |
EGF Z-Score |
BPE Genes |
BPE Z-Score |
EGF + BPE Genes |
EGF + BPE Z-Score |
| Upregulation | ||||||
| Cell cycle | 27 | 4.5 | 38 | 7.1 | 32 | 6.4 |
| DNA replication | 11 | 4.3 | 17 | 7.3 | 14 | 6.4 |
| Ribosome | 53 | 15.8 | 35 | 8.7 | 43 | 12.9 |
| Downregulation | ||||||
| Collecting duct acid secretion | 6 | 2.4 | 9 | 4.5 | 8 | 3.9 |
| Fatty acid metabolism | 11 | 3.8 | 10 | 3.4 | 10 | 3.5 |
| Lysosome | 20 | 2.7 | 23 | 3.8 | 18 | 2.4 |
| PPAR signaling pathway | 14 | 3.1 | 14 | 3.2 | 12 | 2.5 |
Pathways were selected after analyses of nontransformed data. Analogous results were found in the experiments comparing cellular responses to EGF + BPE, compared to serum. In those studies, KEGG pathways for cell cycle (zsc = 8.3), DNA replication (zsc = 8.7), and ribosome (zsc = 3.7) all were upregulated, and those for collecting duct acid secretion (zsc = 3.4), fatty acid metabolism (zsc = 2.4), and lysosome (zsc = 5.8) all were downregulated.
Examples of cell cycle and translation genes upregulated by EGF + BPE treatment of human meibomian gland epithelial cells are listed in Table 5. An illustration of keratin- and lipid-related genes downregulated by combined growth factor exposure is shown in Table 6.
Table 5. .
EGF Plus BPE Upregulation of Selected Cell Cycle- and Translation-Related Genes in Human Meibomian Gland Epithelial Cells
|
Accession # |
Gene |
Ratio |
P
Value |
| Cell cycle | |||
| NM_015714 | G0/G1switch 2 | 16.11 | 0.0206 |
| NM_001255 | Cell division cycle 20 homolog | 11.94 | 0.0000 |
| NM_001067 | Topoisomerase (DNA) IIα | 10.96 | 0.0028 |
| NM_181803 | Ubiquitin-conjugating enzyme E2C | 10.18 | 0.0007 |
| NM_004701 | Cyclin B2 | 8.38 | 0.0052 |
| NM_005915 | Minichromosome maintenance complex component 6 | 7.37 | 0.0000 |
| NM_182776 | Minichromosome maintenance complex component 7 | 6.54 | 0.0001 |
| NM_030928 | Chromatin licensing and DNA replication factor 1 | 6.39 | 0.0026 |
| NM_002916 | Replication factor C (activator 1) 4 | 5.01 | 0.0012 |
| NM_002388 | Minichromosome maintenance complex component 3 | 4.96 | 0.0004 |
| NM_001237 | Cyclin A2 | 3.56 | 0.0130 |
| NM_004354 | Cyclin G2 | 3.49 | 0.0027 |
| NM_002689 | Polymerase (DNA directed), α2 (70kD subunit) | 2.73 | 0.0052 |
| NM_001760 | Cyclin D3 | 2.43 | 0.0011 |
| NM_053056 | Cyclin D1 | 2.34 | 0.0000 |
| NM_199246 | Cyclin G1 | 2.3 | 0.0156 |
| NM_057735 | Cyclin E2 | 2.07 | 0.0449 |
| NM_182851 | Cyclin B1 interacting protein 1 | 1.99 | 0.0011 |
| NM_001759 | Cyclin D2 | 1.43 | 0.0023 |
| NM_001259 | Cyclin-dependent kinase 6 | 1.34 | 0.0420 |
| NM_000075 | Cyclin-dependent kinase 4 | 1.31 | 0.0401 |
| Translation | |||
| NM_138957 | Mitogen-activated protein kinase 1 | 3.61 | 0.0199 |
| NM_003246 | Thrombospondin 1 | 3.28 | 0.0006 |
| NM_002759 | Eukaryotic translation initiation factor 2-alpha kinase 2 | 2.65 | 0.0154 |
| NM_004280 | Eukaryotic translation elongation factor 1 epsilon 1 | 2.16 | 0.0266 |
| NM_001402 | Eukaryotic translation elongation factor 1 alpha 1 | 1.81 | 0.0002 |
| NM_001568 | Eukaryotic translation initiation factor 3, subunit E | 1.81 | 0.0006 |
Relative ratios were calculated by comparing the degree of gene expression in meibomian gland epithelial cells exposed to EGF and BPE for 5 days, compared to that of the basal state.
Table 6. .
EGF Plus BPE Downregulation of Selected Keratin- and Lipid-Related Genes in Human Meibomian Gland Epithelial Cells
|
Accession # |
Gene |
Ratio |
P
Value |
| Keratin | |||
| NM_173086 | Keratin 6C | 251.79 | 0.0000 |
| NM_207392 | Keratinocyte differentiation-associated protein | 175.04 | 0.0001 |
| NM_005987 | Small proline-rich protein 1A | 166.23 | 0.0000 |
| NM_001024209 | Small proline-rich protein 2E | 63.06 | 0.0001 |
| NM_003125 | Small proline-rich protein 1B | 62.37 | 0.0000 |
| NM_005988 | Small proline-rich protein 2A | 56.75 | 0.0000 |
| NM_005557 | Keratin 16 | 52.72 | 0.0000 |
| NM_001014291 | Small proline-rich protein 2G | 51.78 | 0.0000 |
| NM_182507 | Keratin 80 | 51.01 | 0.0002 |
| NM_002964 | S100 calcium binding protein A8 | 10.86 | 0.0001 |
| NM_002965 | S100 calcium binding protein A9 | 8.72 | 0.0000 |
| Lipid | |||
| NM_004354 | Cyclin G2 | 3.49 | 0.0027 |
| NM_001444 | Fatty acid binding protein 5 | 37.75 | 0.0026 |
| NM_018404 | ArfGAP with dual PH domains 2 | 25.36 | 0.0001 |
| NM_001031615 | Aldehyde dehydrogenase 3 family, member B2 | 21.55 | 0.0001 |
| NM_004364 | CCAAT/enhancer binding protein (C/EBP), α | 20.52 | 0.0001 |
| NM_004605 | Sulfotransferase family, cytosolic, 2B, member 1 | 17.83 | 0.0001 |
| NM_001955 | Endothelin 1 | 10.72 | 0.0002 |
| NM_025153 | ATPase, class V, type 10B | 10.02 | 0.0000 |
| NM_152443 | Retinol dehydrogenase 12 (all-trans/9-cis/11-cis) | 9.35 | 0.0007 |
| NM_002899 | Retinol binding protein 1, cellular | 9.26 | 0.0000 |
Relative ratios were calculated by comparing the degree of gene expression in meibomian gland epithelial cells treated with EGF and BPE for 5 days, compared to that of the basal state.
To confirm in part the Illumina BeadChip results, selected genes were analyzed by qPCR. This experimental approach verified the regulatory effects of EGF, BPE, and EGF + BPE on laminin, α3; keratin 10; small proline-rich protein 3; and TIMP metallopeptidase inhibitor 1 (Table 7).
Table 7. .
Confirmation of Selected BeadChip Gene Expression Results
|
Gene |
EGF | EGF | BPE | BPE | EGF + BPE | EGF + BPE |
|
Array |
qPCR |
Array |
qPCR |
Array |
qPCR |
|
| Upregulation | ||||||
| LAMA3 | 11.2 | 8.2 | 9.6 | 5.7 | 10.3 | 4.9 |
| TIMP1 | 11.7 | 13.6 | 4.8 | 4.9 | 8.4 | 6.9 |
| Downregulation | ||||||
| KRT10 | 10.6 | 10.8 | 10.2 | 13.7 | 12.4 | 16.3 |
| SPRR3 | 14.0 | 10.5 | 1.6 | 3.0 | 22.9 | 11.4 |
The expression of selected genes that were shown to be up- or downregulated by growth factors in human meibomian gland epithelial cells by using Illumina BeadChips (“Array”), were reexamined with qPCR procedures. LAMA3, Laminin, α3; TIMP1, TIMP metallopeptidase inhibitor 1; KRT10, keratin 10; SPRR3, small proline-rich protein 3.
The influence of EGF and BPE on human meibomian gland epithelial cells was not completely the same. In fact, BPE significantly (P < 0.05) increased the expression of 1088 genes compared to EGF. In turn, EGF significantly (P < 0.05) upregulated the activity of 918 genes relative to BPE. Major differences occurred between these growth factors in the extent of their impact on ontologies and KEGG pathways. For example, BPE induced far greater effects on cell cycle processes (zsc = 14.1) and DNA replication (zsc = 8.5), whereas EGF exerted a stronger influence on ribosomes (zsc = 11.3) and translation (zsc = 11.9).
Impact of EGF + BPE on Lipid Accumulation in Human Meibomian Gland Epithelial Cells
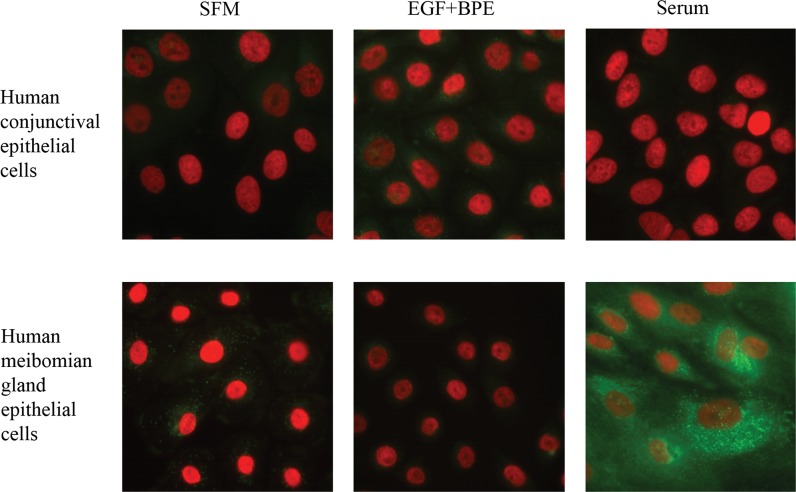
To test our hypothesis that growth factors do not promote differentiated cell functions, such as lipid accumulation, we induced human meibomian gland epithelial cells to proliferate with EGF + BPE, or to differentiate with serum. In addition, to demonstrate the specificity of the lipid-related response of meibomian (i.e., sebaceous-like) cells, we exposed human conjunctival epithelial cells to the same conditions. Following 7 days of treatment, cells were fixed and stained with LipidTox (green) and DAPI (red).
As shown in Figure 4, EGF + BPE exposure had no apparent effect on cellular lipid levels compared to those of vehicle-treated cells. In contrast, serum treatment elicited a pronounced lipid accumulation in human meibomian gland epithelial cells. This lipogenic response is unique, and is not duplicated in human conjunctival epithelial cells (Fig. 4).
Figure 4.
Effect of EGF + BPE, and serum on lipid accumulation in human meibomian gland and conjunctival epithelial cells. Cells were cultured in SFM or in media supplemented with EGF + BPE or serum. Cells were fixed and stained with LipidTox Green neutral lipid stain and DAPI (red). Magnification: ×400.
Discussion
Our studies showed that EGF and BPE, alone or in combination, stimulated a significant, time-dependent proliferation of human meibomian gland epithelial cells. These effects were associated with a significant upregulation of genes linked to cell cycle, DNA replication, ribosomes, and translation, and a significant decrease in those related to cell differentiation, tissue development, lipid metabolic processes, and PPAR signaling. Serum-induced differentiation, but not growth factor-related proliferation, elicits a pronounced lipid accumulation in human meibomian gland epithelial cells. This lipogenic response is unique, and is not duplicated by human conjunctival epithelial cells. Overall, our results supported our hypothesis that growth factors stimulate human meibomian gland epithelial cells to proliferate through processes involving gene regulation.
Our finding that EGF and BPE increased the proliferation, but not the differentiation, of human meibomian gland epithelial cells is consistent with the responses of other cell types to these growth factors.2–11 EGF + BPE induced the highest rate of proliferation, followed by BPE alone, then EGF alone. In contrast, MCDB and serum-containing media supported minimal to no proliferation. SFM permitted slight growth in late-, but not early-passage cells. Of particular interest, the rapidity and magnitude of the proliferative response to EGF and BPE was influenced by the cell passage number. Late-passage (i.e., 50) human meibomian gland epithelial cells proliferated more quickly than early-passage (i.e., 16) cells, despite being exposed to the same culture conditions. This difference may reflect a positive selection of faster-growing cells,17,18 as well as the development of an altered karyotype,12 with increasing passage number. We found previously that these human meibomian gland epithelial cells express slight changes in karyotype after passage 34.12 For this reason, we performed our microarray analyses on early-passage cells, which feature a normal karyotype.12
The growth factor stimulation of human meibomian gland epithelial cell proliferation was associated with a significant upregulation of genes linked to DNA replication, cell cycle, ribosomes, and translation. Stimulated genes related to DNA replication included DNA directed polymerase, minichromosome maintenance complex components, replication factor C, and replication proteins. EGF and BPE also increased the expression of genes for cyclin B2 and D1-3, cyclin-dependent kinase (CDK) 4 and 6, and E2F transcription factor, and downregulated those genes encoding CDK inhibitors. Collectively, these genes are the major molecular components that drive cell cycle progression. Cyclin is a regulatory subunit that modulates the activation state of the catalytic subunit CDK in the cyclin-CDK complex.19 Cyclin B is involved in the control of G2/M transition and mitosis.19 Cyclin Ds are proto-oncogenic components of the retinoblastoma pathway, and are able to form complexes with Cdk2, Cdk4, Cdk5, and Cdk6.20 Indeed, the accumulation of cyclin D1-Cdk4/6 complexes is of great importance for cell cycle progression.21 These complexes promote release of E2F transcription factors, which allow the transcription of genes required for the transition from G1 through the S phase.22–24 The growth factor upregulation of ribosome- and translation-associated genes was to be expected, given that ribosome biogenesis consumes approximately 80% of the energy of a proliferating cell,25,26 and that ribosomal protein mRNAs monopolize the translational capacity of cells.26
We discovered that EGF and BPE cause a significant decrease in the expression of genes related to cell differentiation and keratinization, such as small proline-rich proteins (SPRR), keratins, the S100 calcium binding proteins A8 (S100A8) and A9 (S100A9), and keratinocyte differentiation-associated protein. The SPRR are precursors of the cornified cell envelope of terminally differentiating, stratified squamous epithelia and influence the rigidity of this envelope.27–31 The SPRR, whether alone27 or combined with S100A8 and A9,32 are known to promote keratinization in the conjunctiva33,34 and elsewhere in the body.35,36 Of particular interest, the expression of SPRR, S100A8, and S100A9 genes is increased significantly in meibomian glands of patients with meibomian gland dysfunction (MGD).13 It is possible application of these growth factors might be able to suppress the keratinization process, which is a primary cause of MGD.2,37
We also found that EGF and BPE significantly reduce the expression of genes associated with lipid biosynthesis and PPAR signaling. This growth factor-induced gene effect could account for the profound lack of lipid accumulation in proliferating meibomian (this study)12 and sebaceous38 gland epithelial cells. As concerns PPARs, these ligand-activated nuclear factors form heterodimers with retinoid X receptors (RXR) to regulate transcription.39–42 PPARs have been identified in mitochondria, peroxisomes, and microsomes of sebocytes, and are important modulators of multiple lipid metabolic genes.43 There are three subclasses of PPARs: α, β/δ, and γ, each with different tissue localizations and transcriptional activities.39,44,45 PPARδ is a potent metabolic regulator in various tissues, such as fat, skeletal muscle, and the heart. PPARδ enhances fatty acid catabolism and energy uncoupling in adipose tissue, and suppresses macrophage-derived inflammation.46,47 PPARδ also regulates the late stages of sebocyte differentiation and stimulates sebaceous lipid synthesis.48 In our study, we demonstrated that EGF and BPE significantly downregulate the genes encoding PPARδ and RXRα in human meibomian gland epithelial cells. These growth factor actions possibly may contribute to the suppression of lipid accumulation in these cells.
The influence of EGF and BPE on human meibomian gland epithelial cells was not identical. There were significant differences between these factors in terms of their impact on specific genes, ontologies, and KEGG pathways. For example, BPE induced far greater effects on genes related to cell cycle processes, compared to EGF. This BPE influence may reflect the fact that pituitary extracts should contain adrenocorticotrophic hormone, fibroblast growth factor, growth hormone, insulin-like growth factor 1, thyroid-stimulating hormone, α-melanocyte stimulating hormone, and transforming growth factor-α,49,50 as well as EGF, all of which have been shown to stimulate the proliferation of sebocytes.37 However, this stimulatory effect of BPE may be countered somewhat by β-endorphin, which also is present in the pituitary and reportedly decreases sebocyte proliferation.51
Our results demonstrated that serum-induced differentiation, but not growth factor-elicited proliferation, promotes a striking accumulation of lipids within human meibomian gland epithelial cells. This response, which was not found in human conjunctival epithelial cells, also may occur after a decrease in the proliferation of meibomian gland epithelial cells12 and sebocytes.38 These findings are consistent with earlier observations that neutral lipids accumulate in sebaceous gland epithelial cells during differentiation.38,52
Overall, our investigation demonstrated that EGF and BPE exert a significant effect on the proliferation and gene expression of human meibomian gland epithelial cells.
Acknowledgments
The authors thank Stephen M. Richards (Boston, MA) for his technical assistance, and Ilene K. Gipson and Sandra Michaud (Boston, MA) for the human conjunctival epithelial cells.
Supported by grants from NIH (EY05612) and Alcon Research, Ltd.
Disclosure: S. Liu, None; W.R. Kam, None; J. Ding, None; M.P. Hatton, None; D.A. Sullivan, Alcon Research, Ltd. (F)
References
- 1. Thody AJ, Shuster S. Control and function of sebaceous glands. Physiol Rev. 1989; 69: 383–416 [DOI] [PubMed] [Google Scholar]
- 2. Knop E, Knop N, Millar T, Obata H, Sullivan DA. The International Workshop on Meibomian Gland Dysfunction: report of the Subcommittee on Anatomy, Physiology, and Pathophysiology of the Meibomian Gland. Invest Ophthalmol Vis Sci. 2011; 52: 1938–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Maskin SL, Tseng SC. Clonal growth and differentiation of rabbit meibomian gland epithelium in serum-free culture: differential modulation by EGF and FGF. Invest Ophthalmol Vis Sci. 1992; 33: 205–217 [PubMed] [Google Scholar]
- 4. Matias JR, Orentreich N. Stimulation of hamster sebaceous glands by epidermal growth factor. J Invest Dermatol. 1983; 80: 516–519 [DOI] [PubMed] [Google Scholar]
- 5. Akimoto N, Sato T, Sakiguchi T, Kitamura K, Kohno Y, Ito A. Cell proliferation and lipid formation in hamster sebaceous gland cells. Dermatology. 2002; 204: 118–123 [DOI] [PubMed] [Google Scholar]
- 6. Fatimah SS, Tan GC, Chua KH, Tan AE, Hayati AR. Effects of epidermal growth factor on the proliferation and cell cycle regulation of cultured human amnion epithelial cells. J Biosci Bioeng. 2012; 114: 220–227 [DOI] [PubMed] [Google Scholar]
- 7. Hammond SL, Ham RG, Stampfer MR. Serum-free growth of human mammary epithelial cells: rapid clonal growth in defined medium and extended serial passage with pituitary extract. Proc Natl Acad Sci U S A. 1984; 81: 5435–5439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Galy A, Jolivet M, Jolivet-Reynaud C, Hadden J. Fibroblast growth factor (FGF) and an FGF-like molecule in pituitary extracts stimulate thymic epithelial cell proliferation. Thymus. 1990; 15: 199–211 [PubMed] [Google Scholar]
- 9. Wille JJ, Park J, Elgavish A. Effects of growth factors, hormones, bacterial lipopolysaccharides, and lipotechoic acids on the clonal growth of normal ureteral epithelial cells in serum-free culture. J Cell Physiol. 1992; 150: 52–58 [DOI] [PubMed] [Google Scholar]
- 10. Ang LP, Tan DT, Seah CJ, Beuerman RW. The use of human serum in supporting the in vitro and in vivo proliferation of human conjunctival epithelial cells. Br J Ophthalmol. 2005; 89: 748–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Creek KE, Geslani G, Batova A, Pirisi L. Progressive loss of sensitivity to growth control by retinoic acid and transforming growth factor-beta at late stages of human papillomavirus type 16-initiated transformation of human keratinocytes. Adv Exp Med Biol. 1995; 375: 117–135 [DOI] [PubMed] [Google Scholar]
- 12. Liu S, Khandelwal P, Hatton M, Sullivan DA. Culture, immortalization and characterization of human meibomian gland epithelial cells. Invest Ophthalmol Vis Sci. 2010; 51: 3993–4005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu S, Richards SM, Lo K, Hatton M, Fay AM, Sullivan DA. Changes in gene expression in meibomian gland dysfunction. Invest Ophthalmol Vis Sci. 2011; 52: 2727–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shi L, Reid LH, Jones WD, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nature Biotechnol. 2006; 24: 1151–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genetics. 2000; 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003; 4: R7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lu S, Gough AW, Bobrowski WF, Stewart BH. Transport properties are not altered across Caco-2 cells with heightened TEER despite underlying physiological and ultrastructural changes. J Pharm Sci. 1996; 85: 270–273 [DOI] [PubMed] [Google Scholar]
- 18. Hughes P, Marshall D, Reid Y, Parkes H, Gelber C. The costs of using unauthenticated, over-passaged cell lines: how much more data do we need? BioTechniques. 2007; 43: 575, 577–578, 581–582 passim [DOI] [PubMed] [Google Scholar]
- 19. Buolamwini JK. Cell cycle molecular targets in novel anticancer drug discovery. Curr Pharm Des. 2000; 6: 379–392 [DOI] [PubMed] [Google Scholar]
- 20. Yu Q, Wu J. Involvement of cyclins in mammalian spermatogenesis. Mol Cell Biochem. 2008; 315: 17–24 [DOI] [PubMed] [Google Scholar]
- 21. Gladden AB, Diehl JA. Location, location, location: the role of cyclin D1 nuclear localization in cancer. J Cell Biochem. 2005; 96: 906–913 [DOI] [PubMed] [Google Scholar]
- 22. Sherr CJ. Mammalian G1 cyclins. Cell. 1993; 73: 1059–1065 [DOI] [PubMed] [Google Scholar]
- 23. Dirks PB, Rutka JT. Current concepts in neuro-oncology: the cell cycle--a review. Neurosurgery. 1997; 40: 1000–1013, discussion 1013–1005 [DOI] [PubMed] [Google Scholar]
- 24. Besson A, Yong VW. Mitogenic signaling and the relationship to cell cycle regulation in astrocytomas. J Neuro-Oncol. 2001; 51: 245–264 [DOI] [PubMed] [Google Scholar]
- 25. Schmidt EV. The role of c-myc in cellular growth control. Oncogene. 1999; 18: 2988–2996 [DOI] [PubMed] [Google Scholar]
- 26. Thomas G. An encore for ribosome biogenesis in the control of cell proliferation. Nat Cell Biol. 2000; 2: E71–E72 [DOI] [PubMed] [Google Scholar]
- 27. Hohl D, de Viragh PA, Amiguet-Barras F, Gibbs S, Backendorf C, Huber M. The small proline-rich proteins constitute a multigene family of differentially regulated cornified cell envelope precursor proteins. J Invest Dermatol. 1995; 104: 902–909 [DOI] [PubMed] [Google Scholar]
- 28. Tesfaigzi J, Carlson DM. Expression, regulation, and function of the SPR family of proteins. A review. Cell Biochem Biophys. 1999; 30: 243–265 [DOI] [PubMed] [Google Scholar]
- 29. Fischer DF, Sark MW, Lehtola MM, Gibbs S, van de Putte P, Backendorf C. Structure and evolution of the human SPRR3 gene: implications for function and regulation. Genomics. 1999; 55: 88–99 [DOI] [PubMed] [Google Scholar]
- 30. Song HJ, Poy G, Darwiche N, et al. Mouse Sprr2 genes: a clustered family of genes showing differential expression in epithelial tissues. Genomics. 1999; 55: 28–42 [DOI] [PubMed] [Google Scholar]
- 31. Candi E, Schmidt R, Melino G. The cornified envelope: a model of cell death in the skin. Nature Rev. 2005; 6: 328–340 [DOI] [PubMed] [Google Scholar]
- 32. Mischke D, Korge BP, Marenholz I, Volz A, Ziegler A. Genes encoding structural proteins of epidermal cornification and S100 calcium-binding proteins form a gene complex ("epidermal differentiation complex”) on human chromosome 1q21. J Invest Dermatol. 1996; 106: 989–999.2 [DOI] [PubMed] [Google Scholar]
- 33. Kawasaki S, Kawamoto S, Yokoi N, et al. Up-regulated gene expression in the conjunctival epithelium of patients with Sjögren's syndrome. Exp Eye Res. 2003; 77: 17–26 [DOI] [PubMed] [Google Scholar]
- 34. Li S, Gallup M, Chen YT, McNamara NA. Molecular mechanism of proinflammatory cytokine-mediated squamous metaplasia in human corneal epithelial cells. Invest Ophthalmol Vis Sci. 2010; 51: 2466–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Iizuka H, Takahashi H, Honma M, Ishida-Yamamoto A. Unique keratinization process in psoriasis: late differentiation markers are abolished because of the premature cell death. J Dermatol. 2004; 31: 271–276 [DOI] [PubMed] [Google Scholar]
- 36. Ishida-Yamamoto A, Iizuka H, Manabe M, et al. Altered distribution of keratinization markers in epidermolytic hyperkeratosis. Arch Dermatol Res. 1995; 287: 705–711 [DOI] [PubMed] [Google Scholar]
- 37. Jester JV, Nicolaides N, Kiss-Palvolgyi I, Smith RE. Meibomian gland dysfunction. II. The role of keratinization in a rabbit model of MGD. Invest Ophthalmol Vis Sci. 1989; 30: 936–945 [PubMed] [Google Scholar]
- 38. Ito A, Sakiguchi T, Kitamura K, Akamatsu H, Horio T. Establishment of a tissue culture system for hamster sebaceous gland cells. Dermatology. 1998; 197: 238–244 [DOI] [PubMed] [Google Scholar]
- 39. Berger J, Moller DE. The mechanisms of action of PPARs. Ann Rev Med. 2002; 53: 409–435 [DOI] [PubMed] [Google Scholar]
- 40. Fajas L, Debril MB, Uwerx J. Peroxisome proliferator-activated receptor-gamma: from adipogenesis to carcinogenesis. J Mol Endocrinol. 2001; 27: 1–9 [DOI] [PubMed] [Google Scholar]
- 41. Qi C, Zhu Y, Reddy JK. Peroxisome proliferator-activated receptors, coactivators, and downstream targets. Cell Biochem Biophys. 2000; 32: 187–204 [DOI] [PubMed] [Google Scholar]
- 42. Ferre P. The biology of peroxisome proliferator-activated receptors: relationship with lipid metabolism and insulin sensitivity. Diabetes. 2004; 53 (suppl 1): S43–S50 [DOI] [PubMed] [Google Scholar]
- 43. Zouboulis CC, Schagen S, Alestas T. The sebocyte culture: a model to study the pathophysiology of the sebaceous gland in sebostasis, seborrhoea and acne. Arch Dermatol Res. 2008; 300: 397–413 [DOI] [PubMed] [Google Scholar]
- 44. Desvergne B, Michalik L, Wahli W. Be fit or be sick: peroxisome proliferator-activated receptors are down the road. Mol Endocrinol. 2004; 18: 1321–1332 [DOI] [PubMed] [Google Scholar]
- 45. van Raalte DH, Li M, Pritchard PH, Wasan KM. Peroxisome proliferator-activated receptor (PPAR)-alpha: a pharmacological target with a promising future. Pharm Res. 2004; 21: 1531–1538 [DOI] [PubMed] [Google Scholar]
- 46. Wang YX, Lee CH, Tiep S, et al. Peroxisome-proliferator-activated receptor delta activates fat metabolism to prevent obesity. Cell. 2003; 113: 159–170 [DOI] [PubMed] [Google Scholar]
- 47. Barish GD, Narkar VA, Evans RM. PPAR delta: a dagger in the heart of the metabolic syndrome. J Clin Invest. 2006; 116: 590–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Di-Poi N, Michalik L, Desvergne B, Wahli W. Functions of peroxisome proliferator-activated receptors (PPAR) in skin homeostasis. Lipids. 2004; 39: 1093–1099 [DOI] [PubMed] [Google Scholar]
- 49. Thorner MO, Vance ML, Laws ER Jr, Horvath E, Kovacs K. The anterior pituitary. In: Wilson JD, DW Foster, Kronenberg HM, Larsen PR. eds Williams Textbook of Endocrinology, 9th ed. Philadelphia: WB Saunders; 1998: 249–340 [Google Scholar]
- 50. Halper J, Parnell PG, Carter BJ, Ren P, Scheithauer BW. Presence of growth factors in human pituitary. Lab Invest. 1992; 66: 639–645 [PubMed] [Google Scholar]
- 51. Zouboulis CC, Bohm M. Neuroendocrine regulation of sebocytes -- a pathogenetic link between stress and acne. Exp Dermatol. 2004; 13 (suppl 4): 31–35 [DOI] [PubMed] [Google Scholar]
- 52. Xia LQ, Zouboulis C, Detmar M, Mayer-da-Silva A, Stadler R, Orfanos CE. Isolation of human sebaceous glands and cultivation of sebaceous gland-derived cells as an in vitro model. J Invest Dermatol. 1989; 93: 315–321 [PubMed] [Google Scholar]