Abstract
DAL5 is a constitutively expressed allantoin system gene whose product is required for allantoate transport. Its simple pattern of expression prompted us to use this gene for identifying the element(s) that mediates transcriptional activation of allantoin system genes. Deletion analysis of the DAL5 5'-flanking sequences resulted in identification of two small regions required for DAL5 expression. Analysis of these two regions with synthetic oligonucleotides localized the sequences supporting transcriptional activation to two DNA fragments of 10 to 12 base pairs, each containing one copy of the pentanucleotide 5'-GATAA-3'. The 5'-flanking region of DAL5 contained eight copies of this sequence. Synthetic constructions containing single copies of these fragments were unable to support transcriptional activation, while those containing two or more copies supported high-level activation. The 5'-GATAA-3' sequence was also found beneath the footprint of a DNA-binding protein. These observations are consistent with the suggestion that DNA fragments containing the sequence 5'-GATAA-3' play an important role in DAL5 gene expression, probably representing a portion of the binding site for a transcriptional activation factor.
Full text
PDF

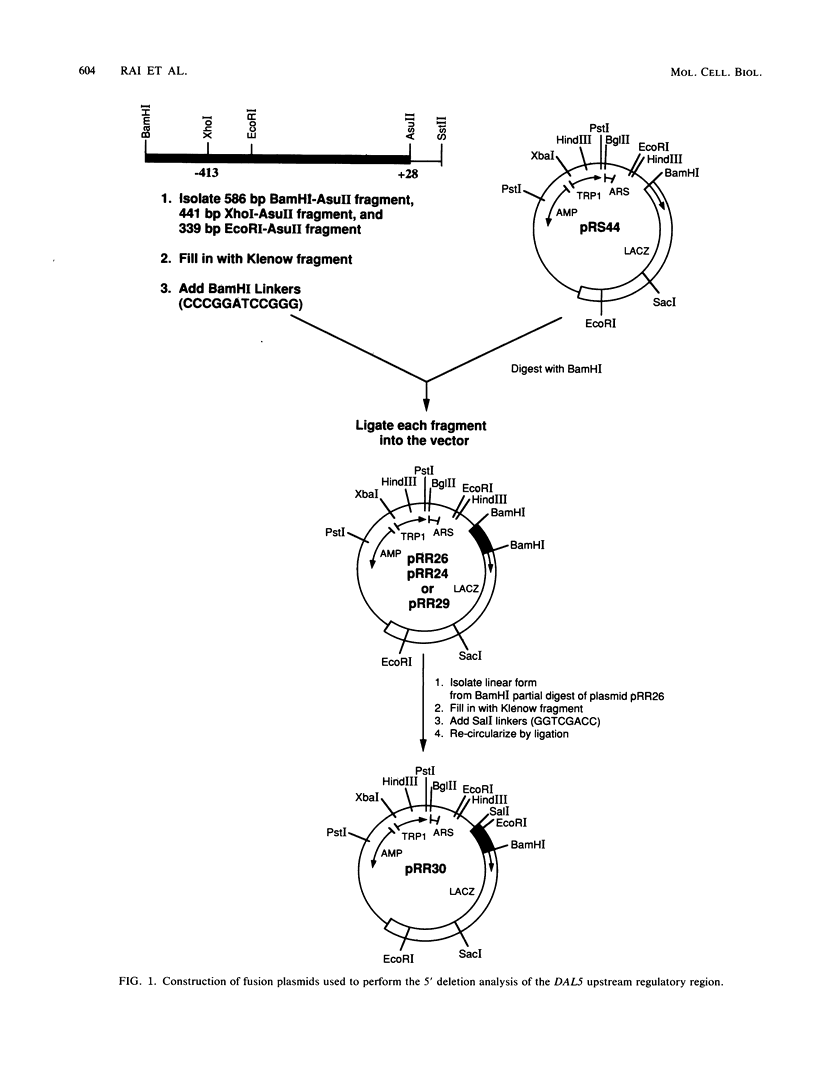




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blumberg H., Hartshorne T. A., Young E. T. Regulation of expression and activity of the yeast transcription factor ADR1. Mol Cell Biol. 1988 May;8(5):1868–1876. doi: 10.1128/mcb.8.5.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigan A. M., Pabich E. K., Donahue T. F. Mutational analysis of the HIS4 translational initiator region in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jul;8(7):2964–2975. doi: 10.1128/mcb.8.7.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. G., Lawther R. P. Induction of the allantoin degradative enzymes in Saccharomyces cerevisiae by the last intermediate of the pathway. Proc Natl Acad Sci U S A. 1973 Aug;70(8):2340–2344. doi: 10.1073/pnas.70.8.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue T. F., Daves R. S., Lucchini G., Fink G. R. A short nucleotide sequence required for regulation of HIS4 by the general control system of yeast. Cell. 1983 Jan;32(1):89–98. doi: 10.1016/0092-8674(83)90499-3. [DOI] [PubMed] [Google Scholar]
- Genbauffe F. S., Cooper T. G. Induction and repression of the urea amidolyase gene in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Nov;6(11):3954–3964. doi: 10.1128/mcb.6.11.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Yocum R. R., Gifford P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7410–7414. doi: 10.1073/pnas.79.23.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepburn A. G., Hindley J. Small-scale techniques for the analysis of recombinant plasmids. J Biochem Biophys Methods. 1979 Oct;1(5):299–308. doi: 10.1016/0165-022x(79)90004-6. [DOI] [PubMed] [Google Scholar]
- Hochschild A., Ptashne M. Cooperative binding of lambda repressors to sites separated by integral turns of the DNA helix. Cell. 1986 Mar 14;44(5):681–687. doi: 10.1016/0092-8674(86)90833-0. [DOI] [PubMed] [Google Scholar]
- Johnson A. D., Herskowitz I. A repressor (MAT alpha 2 Product) and its operator control expression of a set of cell type specific genes in yeast. Cell. 1985 Aug;42(1):237–247. doi: 10.1016/s0092-8674(85)80119-7. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mueller P. P., Hinnebusch A. G. Multiple upstream AUG codons mediate translational control of GCN4. Cell. 1986 Apr 25;45(2):201–207. doi: 10.1016/0092-8674(86)90384-3. [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Stillman D., Kipling D. Both positive and negative regulators of HO transcription are required for mother-cell-specific mating-type switching in yeast. Cell. 1987 Feb 27;48(4):579–587. doi: 10.1016/0092-8674(87)90236-4. [DOI] [PubMed] [Google Scholar]
- Pfeifer K., Arcangioli B., Guarente L. Yeast HAP1 activator competes with the factor RC2 for binding to the upstream activation site UAS1 of the CYC1 gene. Cell. 1987 Apr 10;49(1):9–18. doi: 10.1016/0092-8674(87)90750-1. [DOI] [PubMed] [Google Scholar]
- Rai R., Genbauffe F. S., Cooper T. G. Structure and transcription of the allantoate permease gene (DAL5) from Saccharomyces cerevisiae. J Bacteriol. 1988 Jan;170(1):266–271. doi: 10.1128/jb.170.1.266-271.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R., Genbauffe F., Lea H. Z., Cooper T. G. Transcriptional regulation of the DAL5 gene in Saccharomyces cerevisiae. J Bacteriol. 1987 Aug;169(8):3521–3524. doi: 10.1128/jb.169.8.3521-3524.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rine J., Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987 May;116(1):9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarokin L., Carlson M. Short repeated elements in the upstream regulatory region of the SUC2 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jul;6(7):2324–2333. doi: 10.1128/mcb.6.7.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumrada R. A., Cooper T. G. Ubiquitous upstream repression sequences control activation of the inducible arginase gene in yeast. Proc Natl Acad Sci U S A. 1987 Jun;84(12):3997–4001. doi: 10.1073/pnas.84.12.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele D. J., Hamer D. H. Tandemly duplicated upstream control sequences mediate copper-induced transcription of the Saccharomyces cerevisiae copper-metallothionein gene. Mol Cell Biol. 1986 Apr;6(4):1158–1163. doi: 10.1128/mcb.6.4.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West R. W., Jr, Yocum R. R., Ptashne M. Saccharomyces cerevisiae GAL1-GAL10 divergent promoter region: location and function of the upstream activating sequence UASG. Mol Cell Biol. 1984 Nov;4(11):2467–2478. doi: 10.1128/mcb.4.11.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitomer R. S., Sellers J. W., McCarter D. W., Hastings G. A., Wick P., Lowry C. V. Elements involved in oxygen regulation of the Saccharomyces cerevisiae CYC7 gene. Mol Cell Biol. 1987 Jun;7(6):2212–2220. doi: 10.1128/mcb.7.6.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]