Abstract
Purpose
Polymorphisms in the interleukin 1 alpha (IL1A) and IL1B gene regions were previously associated with keratoconus in a Korean population. In the present study, we investigated whether the IL1A and IL1B polymorphisms are associated with keratoconus in a Japanese population.
Methods
A total of 169 Japanese patients with keratoconus and 390 Japanese healthy controls were recruited. We genotyped one IL1A single nucleotide polymorphism (SNP; rs2071376) and two IL1B SNPs (rs1143627 and rs16944) to compare the frequencies of alleles, genotypes, and haplotypes between cases and controls.
Results
Statistically significant association was observed for rs1143627 (−31 T>C) in the IL1B promoter region; the T allele of rs1143627 was associated with an increased risk of keratoconus (p=0.014, corrected p value [pc]=0.043, odds ratio=1.38). The C allele of rs16944 (−511 C>T) in the IL1B promoter region had a 1.33-fold increased risk of keratoconus, although this increase did not reach statistical significance (p=0.033, pc=0.098). The TT genotype of rs1143627 was weakly associated with an increased risk of keratoconus (p=0.033, pc=0.099, odds ratio=1.52). However, no significant differences were found in the allele and genotype frequencies between the cases and controls for rs2071376 in IL1A. Regarding haplotypic diversity, the haplotype created by the T allele of rs1143627 and C allele of rs16944 was associated with a 1.72-fold increased risk of keratoconus (p=4.0×10−5, pc=1.6×10−4).
Conclusions
Our results replicate associations reported recently in a Korean population. Thus, IL1B may play an important role in the development of keratoconus through genetic polymorphisms.
Introduction
Keratoconus (OMIM 148300) is a non-inflammatory corneal disorder characterized by progressive thinning of the corneal tissue, which can lead to severe visual impairment. Prevalence rates for keratoconus vary widely across geographic areas and studies, ranging from 0.0002% to 2.34% [1]. The exact etiology of keratoconus remains uncertain, but the disease is currently thought to be triggered by various genetic, as well as environmental, factors. Family and twin studies provide convincing evidence of the importance of genetic factors in the development of keratoconus. The prevalence of keratoconus in relatives of keratoconus cases is greater than that of the general population [2-4], and monozygotic twins show a high concordance rate for keratoconus [5-7].
To date, many genetic loci associated with keratoconus have been reported with linkage analysis: 1p36.23–36.21, 2p24, 3p14-q13, 4q, 5q14-q21, 5q32-q33, 8q13.1-q21.11, 9q, 12p, 13q32, 14q11.2, 14q24.3, 15q22.33-q24.2, 16q22.3-q23.1, 20p11.21, and 20q12 [8,9]. Of these loci, 20p11.21 harbors the visual system homeobox 1 (VSX1) gene. Several studies have identified mutations in VSX1 as a cause of keratoconus development in multiple ethnic populations [9-17], and VSX1 could be a strong candidate gene for keratoconus. However, a similar number of studies did not find VSX1 mutations to be specific to the disease [18-26]; therefore, VSX1 mutations may be responsible for a small fraction of keratoconus cases, suggesting that other genetic factors have more powerful effects on the development of keratoconus. In addition to VSX1, several other candidate genes have been reported, including superoxide dismutase 1 [16,27,28], lipoxygenase [29], transforming growth factor, beta 1 [30], secreted protein acidic and rich in cysteine [16], human leukocyte antigen [31-33], mitochondrial complex I genes [34], and collagen type IV, alpha 3 and collagen type IV, alpha four [35]. In recent genome-wide association studies, hepatocyte growth factor [36] and 2q21.3 including RAB3 GTPase activating protein subunit 1 (catalytic) [37] were reported to be associated with keratoconus. However, studies in other populations failed to replicate previously reported associations for some of these candidate genes [16,17,21,22,38], and for other candidates, replication studies have not been reported.
Interleukin (IL)-1 is a proinflammatory cytokine that induces the production of cytokines and chemokines and plays an important role in inflammatory processes. IL-1 is involved in various cellular activities, including cell proliferation, differentiation, and apoptosis. The IL-1 family consists of two proinflammatory cytokines (IL-1α and IL-1β) and the IL-1 receptor antagonist (IL-1Ra). These proteins are encoded by IL1A, IL1B, and IL1RN, respectively, which comprise a cluster spanning 360 kb on chromosome 2q14. Recently, Kim et al. [39] investigated the association of polymorphisms in IL1A, IL1B, and IL1RN with keratoconus in a Korean population and reported that IL1B promoter polymorphisms rs1143627 (−31 T>C) and rs16944 (−511 C>T) are significantly associated with an increased risk of keratoconus. The study also suggested that the heterozygous genotype of the IL1A intronic polymorphism rs2071376 (+376 C>A) is associated with a decreased risk of the disease. However, no replication study has been performed in other ethnic populations. The aim of the present study was to investigate whether genetic polymorphisms in the IL1A-IL1B region are associated with the risk of keratoconus in Japanese patients.
Methods
We recruited 169 unrelated Japanese patients with keratoconus and 390 unrelated healthy Japanese controls at Yokohama City University, Ryogoku Eye Clinic, Tokyo Dental College Ichikawa General Hospital, and Yamaguchi University in Japan. The diagnosis of keratoconus was based on slit-lamp biomicroscopic findings in one or both eyes, including corneal stromal thinning, Fleischer ring, Vogt's striae, or Munson’s sign by cornea specialists at each institute. The diagnosis was also based on characteristic patterns on video keratography: inferior corneal steepening, inferocentral corneal thinning, or an asymmetric bowtie with skewed radial axis. The mean age of the patients was 33.8±9.6 years; 76.2% of patients were male, and 23.8% were female. Controls were healthy volunteers from a geographic region similar to that for the keratoconus patients, and the control population was comparable in age and sex to the patients (mean age, 33.4±9.4 years; 74.6% male and 25.4% female). According to the Online Mendelian Inheritance in Man database [40], several diseases have been associated with the IL1A and/or IL1B polymorphisms: periodontitis, Alzheimer disease, osteomyelitis, end stage renal disease, gastric cancer, inflammatory bowel disease, Parkinson disease, and diabetic nephropathy. These diseases were not included in the patient and control groups. This study was approved by the ethics committee of each participating institute and complied with the guidelines of the Declaration of Helsinki. All study details were explained to all patients and controls before consent was obtained for genetic screening.
Genomic DNA was extracted from peripheral blood samples using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany). Procedures were performed under standardized conditions to prevent variation in DNA quality. We evaluated three single nucleotide polymorphisms (SNPs) reportedly associated with keratoconus in a Korean population: rs2071376 in IL1A, and rs1143627 and rs16944 in IL1B. Genotyping of all SNPs was performed using the TaqMan 5′ exonuclease assay with validated TaqMan primer-probe sets supplied by Applied Biosystems (Foster City, CA). Polymerase chain reaction (PCR) was performed in a reaction mixture with a total volume of 10 μl containing 1X TaqMan Universal PCR Master Mix (Applied Biosystems), 24 nm of each primer-probe set, and 3 ng genomic DNA. The PCR conditions were as follows: 95 °C for 10 min, followed by 40 cycles of denaturation at 92 °C for 15 s and annealing/extension at 60 °C for 1 min. The probe fluorescence signal was detected using the StepOnePlus Real-Time PCR System (Applied Biosystems).
Allele and genotype frequencies were estimated with direct counting. Haplotype frequencies, Hardy–Weinberg equilibrium (HWE), and linkage disequilibrium (LD) were assessed using Haploview 4.1 software [41]. Differences in allele, genotype, and haplotype frequencies between cases and controls were assessed with χ2. To obtain a measure of significance corrected for multiple testing bias, we used the Bonferroni method. A corrected p (pc) value of <0.05 was considered statistically significant.
Results
Hardy–Weinberg equilibrium tests and haplotype block
The three SNPs were in HWE among the cases and the controls. Figure 1 shows the strength of LD for the three SNPs in all 559 participants. IL1B promoter SNPs rs1143627 and rs16944 were located in one haplotype block, and the magnitude of LD between the SNPs was extremely high (D’=0.83, r2=0.67). IL1A SNP rs2071376 was not linked with the IL1B SNPs.
Figure 1.
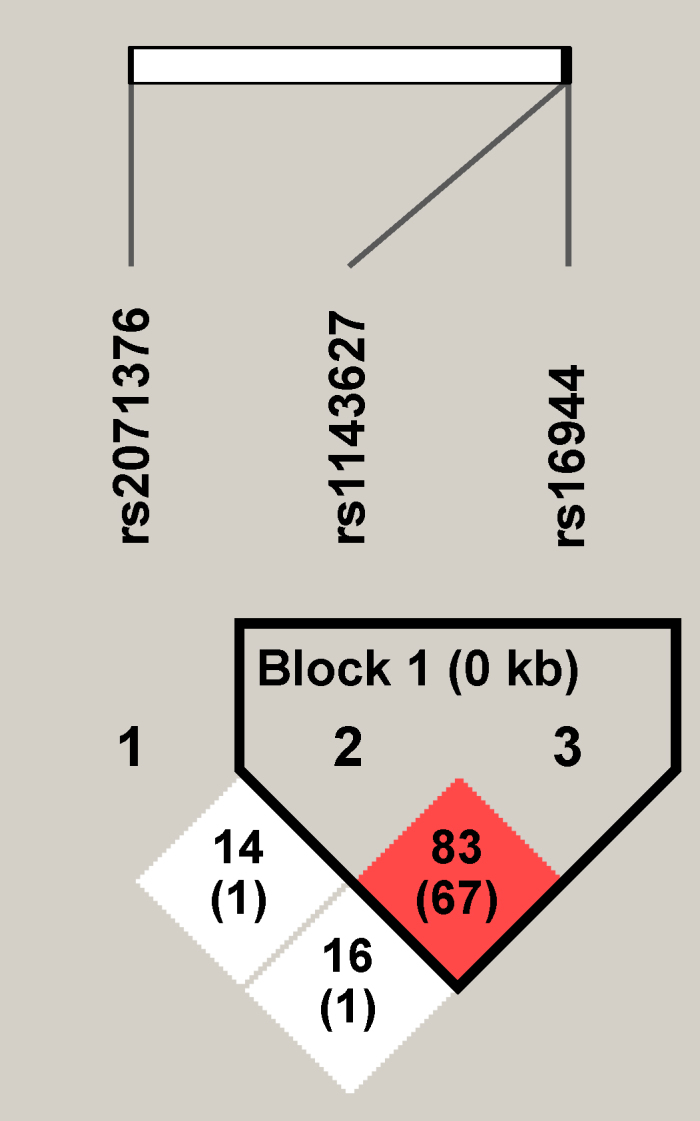
Linkage disequilibrium plot of three interleukin 1 alpha to interleukin 1 beta single nucleotide polymorphisms in 559 study participants. The D’ value and r2 value (in parentheses) corresponding to each single nucleotide polymorphism (SNP) pair are expressed as a percentage and shown within the respective square. Red represents a high-pairwise D' value.
Allele and genotype frequencies
Table 1 provides the genomic locations and allele frequencies for the three SNPs. In IL1B, the frequency of the T allele of rs1143627 was significantly increased in the cases compared to the controls (60.7% versus 52.7%, p=0.014, pc=0.043, odds ratio [OR]=1.38). The C allele frequency of rs16944 was also increased in the cases compared to the controls (60.7% versus 53.7%, p=0.033, OR=1.33), although this increase did not reach statistical significance after the Bonferroni correction was performed (pc=0.098). Conversely, the frequency of the A allele of rs2071376 in IL1A was slightly higher among the cases compared to the controls, but no significant association was observed (27.7% versus 24.9%, p=0.337, OR=1.15).
Table 1. Allele frequencies of three SNPs in IL1A and IL1B.
| SNPs | Chr. | Position (Build 37.1) | Gene | Gene location | Alleles (1>2) | Risk allele | Risk Allele Frequency, % |
P | Pc | OR (95%CI) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases (n=169) | Controls (n=390) | |||||||||||
|
rs2071376 |
(+376 C>A) |
2 |
113,535,395 |
IL1A |
Intron |
C>A |
A |
27.7 |
24.9 |
0.337 |
|
1.15 (0.86–1.54) |
|
rs1143627 |
(−31 T>C) |
2 |
113,594,387 |
IL1B |
Promoter |
T>C |
T |
60.7 |
52.7 |
0.014 |
0.043 |
1.38 (1.07–1.79) |
| rs16944 | (−511 C>T) | 2 | 113,594,867 | IL1B | Promoter | C>T | C | 60.7 | 53.7 | 0.033 | 0.098 | 1.33 (1.02–1.72) |
1, major allele; 2, minor allele; OR, odds ratio; CI, confidence interval. Allele frequency differences were tested using 2×2 contingency tables.
The genotype frequencies of the three SNPs are shown in Table 2. Individuals carrying two copies of the disease-risk allele of rs1143627 and rs16944 had 1.52- and 1.45-fold increased risks of keratoconus, respectively, and a marginal significant p value of 0.033 was observed for the TT genotype of rs1143627 (not significant after correction). For rs2071376 in IL1A, the AA genotype had a 1.76-fold increased risk of keratoconus, but it was not significant. In addition, the AC genotype of rs2071376 reported in a previous study [39] was not associated with the disease.
Table 2. Genotype frequencies of three SNPs in IL1A and IL1B.
| SNPs | Alleles (1>2) | Genotypes | Frequency, % |
P | Pc | OR (95%CI) | ||
|---|---|---|---|---|---|---|---|---|
| Cases (n=169) | Controls (n=390) | |||||||
|
rs2071376 |
(+376 C>A) |
C>A |
AA |
8.3 |
4.9 |
0.117 |
|
1.76 (0.86–3.60) |
| |
|
|
AC |
38.7 |
40.1 |
0.763 |
|
0.94 (0.65–1.37) |
| |
|
|
CC |
53.0 |
55.0 |
0.654 |
|
0.92 (0.64–1.32) |
|
rs1143627 |
(−31 T>C) |
T>C |
CC |
15.4 |
22.2 |
0.064 |
|
0.64 (0.39–1.03) |
| |
|
|
CT |
47.9 |
50.1 |
0.633 |
|
0.92 (0.64–1.31) |
| |
|
|
TT |
36.7 |
27.6 |
0.033 |
0.099 |
1.52 (1.03–2.23) |
|
rs16944 |
(−511 C>T) |
T>C |
CC |
36.7 |
28.6 |
0.058 |
|
1.45 (0.99–2.12) |
| |
|
|
CT |
47.9 |
50.3 |
0.613 |
|
0.91 (0.63–1.31) |
| TT | 15.4 | 21.1 | 0.115 | 0.68 (0.42–1.10) | ||||
1, major allele; 2, minor allele; OR, odds ratio; CI, confidence interval. Individual genotype frequency differences were tested using 2×2 contingency tables for each genotype by combining the remaining genotypes into one category.
Haplotype analysis
A haplotype consisting of rs1143627 and rs16944 was observed (Table 3). The frequency of haplotype TC created by the risk alleles was significantly increased in cases compared to controls (60.4% versus 47.1%, p=4.0×10−5, pc=1.6×10−4, OR=1.72). In contrast, the frequencies of haplotype CC and TT were decreased in the cases compared to the controls with OR=0.03 (CC: 0.2% versus 6.7%, p=2.9× 10−6, pc=1.2×10−5; TT: 0.2% versus 5.7%, p=2.1× 10−5, pc=8.4×10−5).
Table 3. Haplotype frequencies of rs1143627 and rs16944 in IL1B.
| Haplotypes | Frequency, % |
P | Pc | OR (95%CI) | |
|---|---|---|---|---|---|
| Cases (n=169) | Controls (n=390) | ||||
|
rs1143627-rs16944 |
|
|
|
||
| TC |
60.4 |
47.1 |
4.0×10−5 |
1.6×10−4 |
1.72 (1.33–2.23) |
| CT |
39.1 |
40.6 |
0.643 |
|
0.94 (0.72–1.22) |
| CC |
0.2 |
6.7 |
2.9×10−6 |
1.2×10−5 |
0.03 (0.003–0.31) |
| TT | 0.2 | 5.7 | 2.1×10−5 | 8.4×10−5 | 0.03 (0.003–0.37) |
1, major allele; 2, minor allele; OR, odds ratio; CI, confidence interval. Individual haplotype frequency differences were tested using 2×2 contingency tables for each haplotype by combining the remaining haplotypes into one category.
Discussion
The aim of the present study was to assess whether polymorphisms in the IL1A-IL1B region affect the development of keratoconus in a Japanese population. Therefore, we genotyped three polymorphisms in the region. Here, we report a significant association between IL1B promoter polymorphism rs1143627 and keratoconus in Japanese patients, suggesting that the IL1B promoter polymorphisms contribute to the risk of keratoconus in Japanese and Korean populations.
The SNPs rs1143627 is at the −31 position in the promoter region of IL1B. As promoter sequences are potential sources of polymorphisms affecting gene expression, rs1143627 may play critical roles in IL1B gene expression, which may contribute to the risk of keratoconus. This hypothesis is supported by previous studies showing that IL1B promoter polymorphisms, especially the T allele of rs1143627 in the TATA box, can enhance the expression of IL1B [42-45]. The IL1-B protein has been detected in human corneal epithelial, stromal fibroblast, and endothelial cells [46], and expression of this protein has reportedly been enhanced in keratoconus corneas compared to normal corneas [47]. In keratoconus, keratocyte apoptosis has been suggested to contribute to the corneal thinning process [48-50], which suggests that the enhanced IL1B expression caused by the promoter polymorphism, rs1143627, can induce the overexpression of IL1-B protein, resulting in the increased corneal apoptotic activity observed in patients with keratoconus. None of the previous linkage studies of keratoconus has ever detected 2q14 locus harboring IL1B [8]. Linkage studies have less power than case-control association studies to detect small or modest genetic effects [51]. IL1B polymorphisms have conferred modest risk for keratoconus (e.g., OR=1.3–1.5) in the present and previous studies [39], and therefore, the previous linkage studies may not have been able to find 2q14.
The previous study showed that only the heterozygous genotype of IL1A SNP rs2071376 was associated with keratoconus in the Korean population, whereas the allele frequencies were not significantly different between the patients and the controls [39]. In the present study, there was no association between keratoconus and any allele or genotype of rs2071376 in the Japanese population. Since rs2071376 was not linked with the IL1B SNPs, rs1143627 and rs16944, in the Japanese and Korean populations [39], the possibility that IL1A SNP rs2071376 is primarily associated with the development of keratoconus is low.
Keratoconus is genetically heterogeneous, and detecting susceptibility genes could provide useful information regarding the etiology of this poorly understood disease. In the present study, we found
that the IL1B promoter polymorphism, rs1143627, is associated with keratoconus in the Japanese population. This finding is in line with a previous study in a Korean population, suggesting that the IL1B promoter polymorphism is an important risk factor for susceptibility to keratoconus. Further genetic and functional studies are needed to clarify the contribution of the IL1B promoter region to the development of keratoconus.
Acknowledgments
This study was supported by a grant from the Johnson & Johnson KK Vision Care Company. We sincerely thank all of the participants for their participation in this study and all of the medical staff involved in sample collection and diagnosis.
References
- 1.Gordon-Shaag A, Millodot M, Shneor E. The Epidemiology and Etiology of Keratoconus. Int J Keratoco. Ectatic Corneal Dis. 2012;1:7–15. [Google Scholar]
- 2.Ihalainen A. Clinical and epidemiological features of keratoconus genetic and external factors in the pathogenesis of the disease. Acta Ophthalmol Suppl. 1986;178:1–64. [PubMed] [Google Scholar]
- 3.Rabinowitz YS. Keratoconus. Surv Ophthalmol. 1998;42:297–319. doi: 10.1016/s0039-6257(97)00119-7. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Rabinowitz YS, Rotter JI, Yang H. Genetic epidemiological study of keratoconus: evidence for major gene determination. Am J Med Genet. 2000;93:403–9. [PubMed] [Google Scholar]
- 5.Parker J, Ko WW, Pavlopoulos G, Wolfe PJ, Rabinowitz YS, Feldman ST. Videokeratography of keratoconus in monozygotic twins. J Refract Surg. 1996;12:180–3. doi: 10.3928/1081-597X-19960101-31. [DOI] [PubMed] [Google Scholar]
- 6.Bechara SJ, Waring GO, 3rd, Insler MS. Keratoconus in two pairs of identical twins. Cornea. 1996;15:90–3. [PubMed] [Google Scholar]
- 7.Tuft SJ, Hassan H, George S, Frazer DG, Willoughby CE, Liskova P. Keratoconus in 18 pairs of twins. Acta Ophthalmol (Copenh) 2012;90:e482–6. doi: 10.1111/j.1755-3768.2012.02448.x. [DOI] [PubMed] [Google Scholar]
- 8.Nielsen K, Hjortdal J, Pihlmann M, Corydon TJ. Update on the keratoconus genetics. Acta Ophthalmol (Copenh) 2013;91:106–13. doi: 10.1111/j.1755-3768.2012.02400.x. [DOI] [PubMed] [Google Scholar]
- 9.Héon E, Greenberg A, Kopp KK, Rootman D, Vincent AL, Billingsley G, Priston M, Dorval KM, Chow RL, McInnes RR, Heathcote G, Westall C, Sutphin JE, Semina E, Bremner R, Stone EM. VSX1: a gene for posterior polymorphous dystrophy and keratoconus. Hum Mol Genet. 2002;11:1029–36. doi: 10.1093/hmg/11.9.1029. [DOI] [PubMed] [Google Scholar]
- 10.Bisceglia L, Ciaschetti M, De Bonis P, Campo PA, Pizzicoli C, Scala C, Grifa M, Ciavarella P, DelleNoci N, Vaira F, Macaluso C, Zelante L. VSX1 mutational analysis in a series of Italian patients affected by keratoconus: detection of a novel mutation. Invest Ophthalmol Vis Sci. 2005;46:39–45. doi: 10.1167/iovs.04-0533. [DOI] [PubMed] [Google Scholar]
- 11.Eran P, Almogit A, David Z, Wolf HR, Hana G, Yaniv B, Elon P, Isaac A. The D144E substitution in the VSX1 gene: a non-pathogenic variant or a disease causing mutation? Ophthalmic Genet. 2008;29:53–9. doi: 10.1080/13816810802008242. [DOI] [PubMed] [Google Scholar]
- 12.Mok JW, Baek SJ, Joo CK. VSX1 gene variants are associated with keratoconus in unrelated Korean patients. J Hum Genet. 2008;53:842–9. doi: 10.1007/s10038-008-0319-6. [DOI] [PubMed] [Google Scholar]
- 13.Paliwal P, Singh A, Tandon R, Titiyal JS, Sharma A. A novel VSX1 mutation identified in an individual with keratoconus in India. Mol Vis. 2009;15:2475–9. [PMC free article] [PubMed] [Google Scholar]
- 14.Dash DP, George S, O'Prey D, Burns D, Nabili S, Donnelly U, Hughes AE, Silvestri G, Jackson J, Frazer D, Héon E, Willoughby CE. Mutational screening of VSX1 in keratoconus patients from the European population. Eye (Lond) 2010;24:1085–92. doi: 10.1038/eye.2009.217. [DOI] [PubMed] [Google Scholar]
- 15.Paliwal P, Tandon R, Dube D, Kaur P, Sharma A. Familial segregation of a VSX1 mutation adds a new dimension to its role in the causation of keratoconus. Mol Vis. 2011;17:481–5. [PMC free article] [PubMed] [Google Scholar]
- 16.De Bonis P, Laborante A, Pizzicoli C, Stallone R, Barbano R, Longo C, Mazzilli E, Zelante L, Bisceglia L. Mutational screening of VSX1, SPARC, SOD1, LOX, and TIMP3 in keratoconus. Mol Vis. 2011;17:2482–94. [PMC free article] [PubMed] [Google Scholar]
- 17.Saee-Rad S, Hashemi H, Miraftab M, Noori-Daloii MR, Chaleshtori MH, Raoofian R, Jafari F, Greene W, Fakhraie G, Rezvan F, Heidari M. Mutation analysis of VSX1 and SOD1 in Iranian patients with keratoconus. Mol Vis. 2011;17:3128–36. [PMC free article] [PubMed] [Google Scholar]
- 18.Aldave AJ, Yellore VS, Salem AK, Yoo GL, Rayner SA, Yang H, Tang GY, Piconell Y, Rabinowitz YS. No VSX1 gene mutations associated with keratoconus. Invest Ophthalmol Vis Sci. 2006;47:2820–2. doi: 10.1167/iovs.05-1530. [DOI] [PubMed] [Google Scholar]
- 19.Liskova P, Ebenezer ND, Hysi PG, Gwilliam R, El-Ashry MF, Moodaley LC, Hau S, Twa M, Tuft SJ, Bhatacharya SS. Molecular analysis of the VSX1 gene in familial keratoconus. Mol Vis. 2007;13:1887–91. [PMC free article] [PubMed] [Google Scholar]
- 20.Tang YG, Picornell Y, Su X, Li X, Yang H, Rabinowitz YS. Three VSX1 gene mutations, L159M, R166W, and H244R, are not associated with keratoconus. Cornea. 2008;27:189–92. doi: 10.1097/ICO.0b013e31815a50e7. [DOI] [PubMed] [Google Scholar]
- 21.Gajecka M, Radhakrishna U, Winters D, Nath SK, Rydzanicz M, Ratnamala U, Ewing K, Molinari A, Pitarque JA, Lee K, Leal SM, Bejjani BA. Localization of a gene for keratoconus to a 5.6-Mb interval on 13q32. Invest Ophthalmol Vis Sci. 2009;50:1531–9. doi: 10.1167/iovs.08-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stabuc-Silih M, Strazisar M, Hawlina M, Glavac D. Absence of pathogenic mutations in VSX1 and SOD1 genes in patients with keratoconus. Cornea. 2010;29:172–6. doi: 10.1097/ICO.0b013e3181aebf7a. [DOI] [PubMed] [Google Scholar]
- 23.Stabuc-Silih M, Strazisar M, Ravnik-Glavac M, Hawlina M, Glavac D. Genetics and clinical characteristics of keratoconus. Acta Dermatovenerol Alp Panonica Adriat. 2010;19:3–10. [PubMed] [Google Scholar]
- 24.Tanwar M, Kumar M, Nayak B, Pathak D, Sharma N, Titiyal JS, Dada R. VSX1 gene analysis in keratoconus. Mol Vis. 2010;16:2395–401. [PMC free article] [PubMed] [Google Scholar]
- 25.Abu-Amero KK, Kalantan H, Al-Muammar AM. Analysis of the VSX1 gene in keratoconus patients from Saudi Arabia. Mol Vis. 2011;17:667–72. [PMC free article] [PubMed] [Google Scholar]
- 26.Jeoung JW, Kim MK, Park SS, Kim SY, Ko HS, Wee WR, Lee JH. VSX1 gene and keratoconus: genetic analysis in Korean patients. Cornea. 2012;31:746–50. doi: 10.1097/ICO.0b013e3181e16dd0. [DOI] [PubMed] [Google Scholar]
- 27.Udar N, Atilano SR, Brown DJ, Holguin B, Small K, Nesburn AB, Kenney MC. SOD1: a candidate gene for keratoconus. Invest Ophthalmol Vis Sci. 2006;47:3345–51. doi: 10.1167/iovs.05-1500. [DOI] [PubMed] [Google Scholar]
- 28.Udar N, Atilano SR, Brown DJ, Holguin B, Small K, Nesburn AB, Kenney MC. SOD1: a candidate gene for keratoconus. Invest Ophthalmol Vis Sci. 2006;47:3345–51. doi: 10.1167/iovs.05-1500. [DOI] [PubMed] [Google Scholar]
- 29.Bykhovskaya Y, Li X, Epifantseva I, Haritunians T, Siscovick D, Aldave A, Szczotka-Flynn L, Iyengar SK, Taylor KD, Rotter JI, Rabinowitz YS. Variation in the lysyl oxidase (LOX) gene is associated with keratoconus in family-based and case-control studies. Invest Ophthalmol Vis Sci. 2012;53:4152–7. doi: 10.1167/iovs.11-9268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guan T, Liu C, Ma Z, Ding S. The point mutation and polymorphism in keratoconus candidate gene TGFBI in Chinese population. Gene. 2012;503:137–9. doi: 10.1016/j.gene.2012.04.061. [DOI] [PubMed] [Google Scholar]
- 31.Klouda PT, Syrbopoulos EK, Entwistle CC, Goffin RB, Easty DL, Bradley BA. HLA and keratoconus. Tissue Antigens. 1983;21:397–9. doi: 10.1111/j.1399-0039.1983.tb00190.x. [DOI] [PubMed] [Google Scholar]
- 32.Gorskova EN, Sevost'ianov EN. Associations of HLA class I haplotype antigens with various patterns of keratoconus. Vestn Oftalmol. 1997;113:31–3. [PubMed] [Google Scholar]
- 33.Adachi W, Mitsuishi Y, Terai K, Nakayama C, Hyakutake Y, Yokoyama J, Mochida C, Kinoshita S. The association of HLA with young-onset keratoconus in Japan. Am J Ophthalmol. 2002;133:557–9. doi: 10.1016/s0002-9394(01)01368-x. [DOI] [PubMed] [Google Scholar]
- 34.Pathak D, Nayak B, Singh M, Sharma N, Tandon R, Sinha R, Titiyal JS, Dada R. Mitochondrial complex 1 gene analysis in keratoconus. Mol Vis. 2011;17:1514–25. [PMC free article] [PubMed] [Google Scholar]
- 35.Stabuc-Silih M, Ravnik-Glavac M, Glavac D, Hawlina M, Strazisar M. Polymorphisms in COL4A3 and COL4A4 genes associated with keratoconus. Mol Vis. 2009;15:2848–60. [PMC free article] [PubMed] [Google Scholar]
- 36.Burdon KP, Macgregor S, Bykhovskaya Y, Javadiyan S, Li X, Laurie KJ, Muszynska D, Lindsay R, Lechner J, Haritunians T, Henders AK, Dash D, Siscovick D, Anand S, Aldave A, Coster DJ, Szczotka-Flynn L, Mills RA, Iyengar SK, Taylor KD, Phillips T, Montgomery GW, Rotter JI, Hewitt AW, Sharma S, Rabinowitz YS, Willoughby C, Craig JE. Association of polymorphisms in the hepatocyte growth factor gene promoter with keratoconus. Invest Ophthalmol Vis Sci. 2011;52:8514–9. doi: 10.1167/iovs.11-8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li X, Bykhovskaya Y, Haritunians T, Siscovick D, Aldave A, Szczotka-Flynn L, Iyengar SK, Rotter JI, Taylor KD, Rabinowitz YS. A genome-wide association study identifies a potential novel gene locus for keratoconus, one of the commonest causes for corneal transplantation in developed countries. Hum Mol Genet. 2012;21:421–9. doi: 10.1093/hmg/ddr460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Udar N, Kenney MC, Chalukya M, Anderson T, Morales L, Brown D, Nesburn A, Small K. Keratoconus–no association with the transforming growth factor beta-induced gene in a cohort of American patients. Cornea. 2004;23:13–7. doi: 10.1097/00003226-200401000-00003. [DOI] [PubMed] [Google Scholar]
- 39.Kim SH, Mok JW, Kim HS, Joo CK. Association of −31T>C and −511 C>T polymorphisms in the interleukin 1 beta (IL1B) promoter in Korean keratoconus patients. Mol Vis. 2008;14:2109–16. [PMC free article] [PubMed] [Google Scholar]
- 40.Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick's Online Mendelian Inheritance in Man (OMIM). Nucleic Acids Res. 2009;37(Database issue):D793–6. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 42.Hall SK, Perregaux DG, Gabel CA, Woodworth T, Durham LK, Huizinga TW, Breedveld FC, Seymour AB. Correlation of polymorphic variation in the promoter region of the interleukin-1 beta gene with secretion of interleukin-1 beta protein. Arthritis Rheum. 2004;50:1976–83. doi: 10.1002/art.20310. [DOI] [PubMed] [Google Scholar]
- 43.Chang YW, Jang JY, Kim NH, Lee JW, Lee HJ, Jung WW, Dong SH, Kim HJ, Kim BH, Lee JI, Chang R. Interleukin-1B (IL-1B) polymorphisms and gastric mucosal levels of IL-1beta cytokine in Korean patients with gastric cancer. Int J Cancer. 2005;114:465–71. doi: 10.1002/ijc.20724. [DOI] [PubMed] [Google Scholar]
- 44.Chen H, Wilkins LM, Aziz N, Cannings C, Wyllie DH, Bingle C, Rogus J, Beck JD, Offenbacher S, Cork MJ, Rafie-Kolpin M, Hsieh CM, Kornman KS, Duff GW. Single nucleotide polymorphisms in the human interleukin-1B gene affect transcription according to haplotype context. Hum Mol Genet. 2006;15:519–29. doi: 10.1093/hmg/ddi469. [DOI] [PubMed] [Google Scholar]
- 45.Landvik NE, Hart K, Skaug V, Stangeland LB, Haugen A, Zienolddiny S. A specific interleukin-1B haplotype correlates with high levels of IL1B mRNA in the lung and increased risk of non-small cell lung cancer. Carcinogenesis. 2009;30:1186–92. doi: 10.1093/carcin/bgp122. [DOI] [PubMed] [Google Scholar]
- 46.Weng J, Mohan RR, Li Q, Wilson SE. IL-1 upregulates keratinocyte growth factor and hepatocyte growth factor mRNA and protein production by cultured stromal fibroblast cells: interleukin-1 beta expression in the cornea. Cornea. 1997;16:465–71. [PubMed] [Google Scholar]
- 47.Zhou L, Yue BY, Twining SS, Sugar J, Feder RS. Expression of wound healing and stress-related proteins in keratoconus corneas. Curr Eye Res. 1996;15:1124–31. doi: 10.3109/02713689608995144. [DOI] [PubMed] [Google Scholar]
- 48.Wilson SE, Kim WJ. Keratocyte apoptosis: implications on corneal wound healing, tissue organization, and disease. Invest Ophthalmol Vis Sci. 1998;39:220–6. [PubMed] [Google Scholar]
- 49.Kim WJ, Rabinowitz YS, Meisler DM, Wilson SE. Keratocyte apoptosis associated with keratoconus. Exp Eye Res. 1999;69:475–81. doi: 10.1006/exer.1999.0719. [DOI] [PubMed] [Google Scholar]
- 50.Kaldawy RM, Wagner J, Ching S, Seigel GM. Evidence of apoptotic cell death in keratoconus. Cornea. 2002;21:206–9. doi: 10.1097/00003226-200203000-00017. [DOI] [PubMed] [Google Scholar]
- 51.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–7. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]


