Abstract
Polycyclic aromatic hydrocarbons (PAHs) are present in the environment as complex mixtures with components that have diverse carcinogenic potencies and mostly unknown interactive effects. Non-additive PAH interactions have been observed in regulation of cytochrome P450 (CYP) gene expression in the CYP1 family. To better understand and predict biological effects of complex mixtures, such as environmental PAHs, an 11 gene input-1 gene output fuzzy neural network (FNN) was developed for predicting PAH-mediated perturbations of dermal Cyp1b1 transcription in mice. Input values were generalized using fuzzy logic into low, medium, and high fuzzy subsets, and sorted using k-means clustering to create Mamdani logic functions for predicting Cyp1b1 mRNA expression. Model testing was performed with data from microarray analysis of skin samples from FVB/N mice treated with toluene (vehicle control), dibenzo[def,p]chrysene (DBC), benzo[a]pyrene (BaP), or 1 of 3 combinations of diesel particulate extract (DPE), coal tar extract (CTE) and cigarette smoke condensate (CSC) using leave one out cross-validation. Predictions were within 1 log2 fold change unit of microarray data, with the exception of the DBC treatment group, where the unexpected down-regulation of Cyp1b1 expression was predicted but did not reach statistical significance on the microarrays. Adding CTE to DPE was predicted to increase Cyp1b1 expression, whereas adding CSC to CTE and DPE was predicted to have no effect, in agreement with microarray results. The aryl hydrocarbon receptor repressor (Ahrr) was determined to be the most significant input variable for model predictions using back-propagation and normalization of FNN weights.
Keywords: PAHs, modeling, mixtures, Cyp1b1, skin Ahrr
Introduction
Polycyclic aromatic hydrocarbons (PAHs) are a class of persistent chemicals prevalent in the environment as a result of geological processes and incomplete combustion of biofuels. Although natural processes such as petroleum seepage and forest fires produce PAHs, anthropogenic processes such as coal burning and oil spills have led to environmental PAH concentrations significantly larger than naturally occurring levels (Zhang and Tao, 2009). Benzo[a]pyrene (BaP) and dibenzo[def,p]chrysene (DBC) are classified by the International Agency on Cancer Research (IARC) as known or probable human carcinogens, respectively, and induce carcinogenesis in lung, liver, thymus, prostate and skin (IARC, 2010). To date, more than 100 PAHs have been identified in the atmosphere (Schauer et al., 2003) with a wide range of carcinogenic potencies.
The first study linking PAH exposure with cancer was performed by Percival Pott in 1775, noting the prevalence of scrotal (skin) cancer in chimney sweeps. Although environmental PAH mixtures such as soot have long been considered carcinogenic, variability in PAH mixture composition, depending on the organic source, combustion temperatures, age, and surrounding environment (Moldoveanu, 2010), have made understanding and regulating PAH exposures difficult. Today skin cancer remains prevalent throughout society, being the highest diagnosed cancer in the United States2. Melanoma skin cancer was responsible for an estimated 8800 deaths in the US in 2011 (Siegel et al., 2011), and melanoma as well as non-melanoma skin cancer incidence is increasing both in the US and worldwide (Siegel et al., 2011; Lomas et al., 2012). A large percentage of these cancers are due to UV exposures but PAHs may play a contributory role (Burke and Wei, 2009; Toyooka et al., 2006).
Current methods for environmental PAH mixture risk assessment involve the calculation/estimation of relative potency factors (RPFs). Individual PAHs are assigned a potency value based on carcinogenic potential relative to the most well studied PAH, BaP, and the RPFs of all components (with a designated RPF) within a mixture are summed to determine the potency of the mixture as a whole (Damon, 1997). RPFs provide a clear, transparent method for risk assessment, at the cost of assuming a common mechanism of action (MOA) and no interactions between mixture components.
Recent studies in our laboratory utilizing a mouse two-stage skin tumor model have shown that RPFs grossly underestimate the carcinogenicity of dermal exposure to DBC or mixtures containing CTE (Siddens et al., 2012). These studies also raise questions regarding possible alternative MOAs for DBC exposures in contrast to BaP, and have demonstrated interactive effects occurring at the gene expression level for genes relevant to well-known BaP MOAs, including Cyp1b1.
BaP and many other high molecular weight PAHs have three well-known MOAs (Cavalieri and Rogan, 1995; Penning et al., 1999): 1) single electron oxidation by peroxidases, creating BaP radical cations capable of binding to and depurinating DNA, 2) Production of reactive oxygen species and electrophilic DNA-binding quinones through aldoketo reductase (AKR)-mediated catechol formation and subsequent electron redox cycling and 3) formation of (±)-syn/anti-BaP-7,8-dihydrodiol-9,10-epoxide (BaPDE) isomers that form covalent DNA adducts . CYPs in the 1 family (1A1, 1B1, 1A2) play a critical role in bioactivation of high molecular weight PAHs such as BaP: redox cycling and BaPDE pathways are dependent on CYPs for activation of the parent BaP compound through epoxygenation at the 7,8 position and further BaPDE formation by CYP epoxygenation across the 9,10-double bond.
mRNA expression of several CYP1 isoforms, including CYP1A1, CYP1A2, and CYP1B1, are induced by high molecular weight PAHs through aryl hydrocarbon receptor (AhR) activation (Fujii-Kuriyama and Mimura, 2005; Lin et al., 2003). The AhR pathway and corresponding CYP1B1 up-regulation requires a number of co-activators and co-repressors, including heat shock protein 90 (HSP90), aryl hydrocarbon receptor nuclear translocator (ARNT), and aryl hydrocarbon receptor repressor (AhRR), contributing to the regulation of AhR-mediated CYP1 expression (Hosoya et al., 2008; Hahn et al., 2009). Previous studies investigating exposures to environmental PAH mixtures and binary PAH combinations have demonstrated synergistic cyp1b1 and cyp1a1 induction in zebrafish (Billiard et al., 2006; Timme-Laragy et al., 2007) and Cyp1b1 and Cyp1a1 induction in dermally PAH-treated mice (Courter et al., 2007a). Our laboratory previously observed highly distinct gene signatures for several Phase I and II metabolizing enzymes, including differential regulation of Cyp1a1 and Cyp1b1, in mouse skin after initiation with PAHs and environmental PAH mixtures suggesting that these enzymes and associated pathways may be important for the carcinogenic potential of PAHs (Siddens et al, 2012). Despite the importance of CYP1 in PAH mixture carcinogenesis, currently accepted methods based on RPFs cannot predict PAH-mediated perturbations of CYP1 gene expression. The ToxCast and Tox21 programs were created by the Environmental Protection Agency, National Institutes of Health, and other federal agencies in recognition of the limitations of current RPF methods and the need to develop high throughput in vitro and in silico methods to predict potential toxicity of chemicals and chemical mixtures. ToxCast efforts have resulted in a number of first generation pathway-level predictive models for high-throughput screening of single chemical exposures (Abdelaziz et. al, 2012, Sipes et. al, 2011). Currently, however, there are still no known quantitative prediction models which can account for non-interactive effects within chemical mixtures (Altenburger et. al, 2012).
We created a fuzzy neural network (a neural network model with layers consisting of fuzzy logic mathematical operations) for utilizing gene expression patterns and applied it in predicting quantitative changes in CYP1B1 expression in human skin as a function of PAH mixture composition. The model was trained and evaluated using microarrays of dermal RNA from mice treated with PAH mixtures, in which the microarray data set was partitioned into training sets consisting of n-1 treatment groups and testing data sets consisting of the treatment groups excluded from the training process. Based on the initial findings, we conclude that neural network modeling, when coupled with fuzzy logic mathematics and constructed using logic functions, may be useful in predicting interactive effects of PAH, or other environmental, mixtures on gene expression.
Materials and methods
BaP and DBC were handled in accordance with National Cancer Institute (NCI) guidelines. All pure PAHs and mixtures were prepared under UV depleted light.
Chemicals
Coal tar extract (CTE) SRM 1597a was purchased from the National Institute of Standards and Technology (NIST) (Gaithersburg, MD), and was concentrated to 10 mg/ml by evaporation under nitrogen. Diesel exhaust particulate matter (SRM 1650b) was also purchased from the NIST. Two hundred mg of diesel particulate were extracted into 200 ml dichloromethane using a Soxhlet Apparatus at 40° C for 24 h. Dichloromethane extract was concentrated, exchanged into toluene, and evaporated under a stream of nitrogen gas to a final volume of 10 ml. Cigarette smoke condensate (CSC) (40 mg/ml in DMSO) was generously provided by Dr. Hollie Swanson (University of Kentucky, Lexington, KY). The CSC was evaporated using a speed vac centrifuge and diluted to 40 mg/ml and 5% DMSO in toluene. BaP and DBC were purchased from Midwest Research Institute, Kansas City, MO. Dichloromethane, toluene, acetone, and DMSO were purchased from Fisher Scientific (Pittsburgh, PA). All other reagents were purchased from Sigma-Aldrich (St. Louis, MO). An aliquot of diesel particulate extract (DPE) was diluted into vehicle (toluene containing 5% DMSO) to create a 5 mg/mL solution designated as PAH mixture 1 (Mix 1). PAH mixture 2 (Mix 2) consisted of 5 mg/ml DPE and 5 mg/ml CTE diluted in vehicle, while PAH mixture 3 (Mix 3) consisted of 5 mg/ml DPE, 5 mg/ml CTE, and 10 mg/ml CSC in vehicle.
Mouse dermal treatment
All procedures were conducted according to National Institutes of Health guidelines and were approved by the Oregon State University Animal Care and Use Committee. Six week old female FVB/N inbred mice were obtained from NCI-Frederick, Frederick, MD, and housed four mice per cage in micro ventilated racks. Mice were on a 12 h light/dark cycle, 22°C, 40–60% humidity and fed AIN93-G pellets (Research Diets, Inc., New Brunswick, NJ) for 10 days. At 7.5 weeks mice were shaved on the dorsal side from the front shoulders to tail and observed for 48 h in order to confirm hair of mice were in the resting phase of growth. Treatments were delivered by pipetting 200 µl of the designated treatment evenly over the shaved area according to the dosing scheme shown in Supplemental Table 1. Mice were euthanized 12 h post-treatment using CO2 and cervical dislocation. Dermal and epidermal layers of the shaved area were excised and snap frozen in liquid nitrogen for RNA extraction.
Dermal RNA extraction
One cm2 subsections of the frozen skin were homogenized in 2 ml Trizol® Reagent (Life Technologies, Carlsbad, CA), using 15 mL disposable conical homogenizers (VWR International, West Chester, PA). RNA isolation was performed according to commercial protocol. RNA was further purified using QIAGEN RNeasy mini prep kit according to protocols provided by the manufacturer (RNeasy Miniprep kit, Qiagen, Valencia, CA). Nucleic acid purity and concentrations were determined using Nanodrop spectrophotometry (Thermo Fisher Scientific, Waltham, MA), and Agilent Bioanalyzer (Santa Clara, CA) analysis, respectively. Samples with A260/280 ratios of 1.9–2.2 and RNA integrity numbers 6.5 or greater were selected for microarray analysis.
Microarray analysis
Individual mouse dermal samples were analyzed by Agilent microarray after initiation with PAHs (N=4 biological replicates, Supplemental Table 1) or toluene control (N=5 biological replicates) as previously described (Siddens et al, 2012). Briefly, the RNA was labeled with Agilent’s 2 color Quickamp kit for hybridization to the Agilent 8 X 60K mouse array. Raw intensity data were quantile normalized by RMA summarization (Bolstad et al., 2003) and subject to pairwise analysis of variance (Kerr et al., 2000) with Tukey's post hoc test and 5% false discovery rate calculation (Benjamini and Hochberg, 1995). Raw and normalized Agilent data files are available online at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE39455. Microarrays results were confirmed using RT-qPCR on a subset of genes with decreased, increased, and no significant change in expression levels relative to control (Supplemental Figure 1).
Model inputs
To develop a model for quantitative prediction of CYP1B1 expression in skin as a function of PAH mixture composition, a list of candidate input genes were identified by searching Pubmed and Google Scholar databases for genes involves in the epithelial perturbation of human/murine CYP1B1/Cyp1b1, and Cyp1a1/Cyp1a1. The list of genes was filtered to a short list of candidate input genes from the microarray analysis reported in Siddens et al., (2012) The 11 candidate genes from the microarray analysis selected for model inputs are summarized in Table 1.
TABLE 1.
Microarray Expression Levels of Genes Selected for FNN Inputsa (mean log2 fold change ± SE)
| Treatment | |||||||
|---|---|---|---|---|---|---|---|
| Gene Name |
GenBank ID: |
BaP | DBC | Mix 1 | Mix 2 | Mix 3 | |
| Input Gene | Ahr | NM_013464 | −0.23 ±0.29 | −0.01 ±0.22 | −0.50 ±0.03 | −0.83 ±0.08 | −0.76 ±0.10 |
| Hsp90a1 | NM_010480 | −0.22 ±0.37 | −0.58 ±0.11 | 0.32 ±0.23 | 0.18 ±0.15 | 0.07 ±0.14 | |
| Ahrr | NM_009644 | 2.00 ±0.44 | 0.25 ±0.37 | 1.39 ±0.14 | 3.05 ±0.04 | 2.62 ±0.35 | |
| Nfe2l3 | NM_010903 | −0.13 ±0.27 | −0.47 ±0.90 | −0.56 ±0.40 | −1.15 ±0.30 | −1.03 ±0.49 | |
| Tnfrsfl4 | NM_178931 | 1.12 ±0.07 | −0.53 ±0.93 | −0.36 ±0.35 | 0.99 ±0.46 | 1.30 ±0.24 | |
| Tnfrsfl9 | NM_001164155 | 0.24 ±0.54 | 0.57 ±1.10 | −1.09 ±0.24 | −1.47 ±0.53 | −1.51 ±0.15 | |
| Arntl2 | NM_172309 | 0.61 ±0.17 | 0.31 ±0.48 | −0.46 ±0.34 | −0.22 ±0.09 | −0.24 ±0.25 | |
| E2f6 | NM_033270 | −0.15 ±0.21 | −0.55 ±0.33 | −0.05 ±0.03 | 0.24 ±0.21 | 0.40 ±0.09 | |
| Rbbp4 | NM_009030 | 0.07 ±0.05 | −0.01 ±0.14 | −0.14 ±0.11 | −0.32 ±0.12 | −0.34 ±0.12 | |
| Rbbp8 | NM_001081223 | 0.15 ±0.19 | 0.24 ±0.41 | 0.15 ±0.07 | −0.06 ±0.15 | −0.44 ±0.01 | |
| Nfe2l2 | NM_010902 | 0.22 ±0.34 | −0.20 ±0.25 | 0.38 ±0.14 | 0.68 ±0.15 | 0.77 ±0.07 | |
Complete data set reported in Siddens et al., (2012)
Model structure
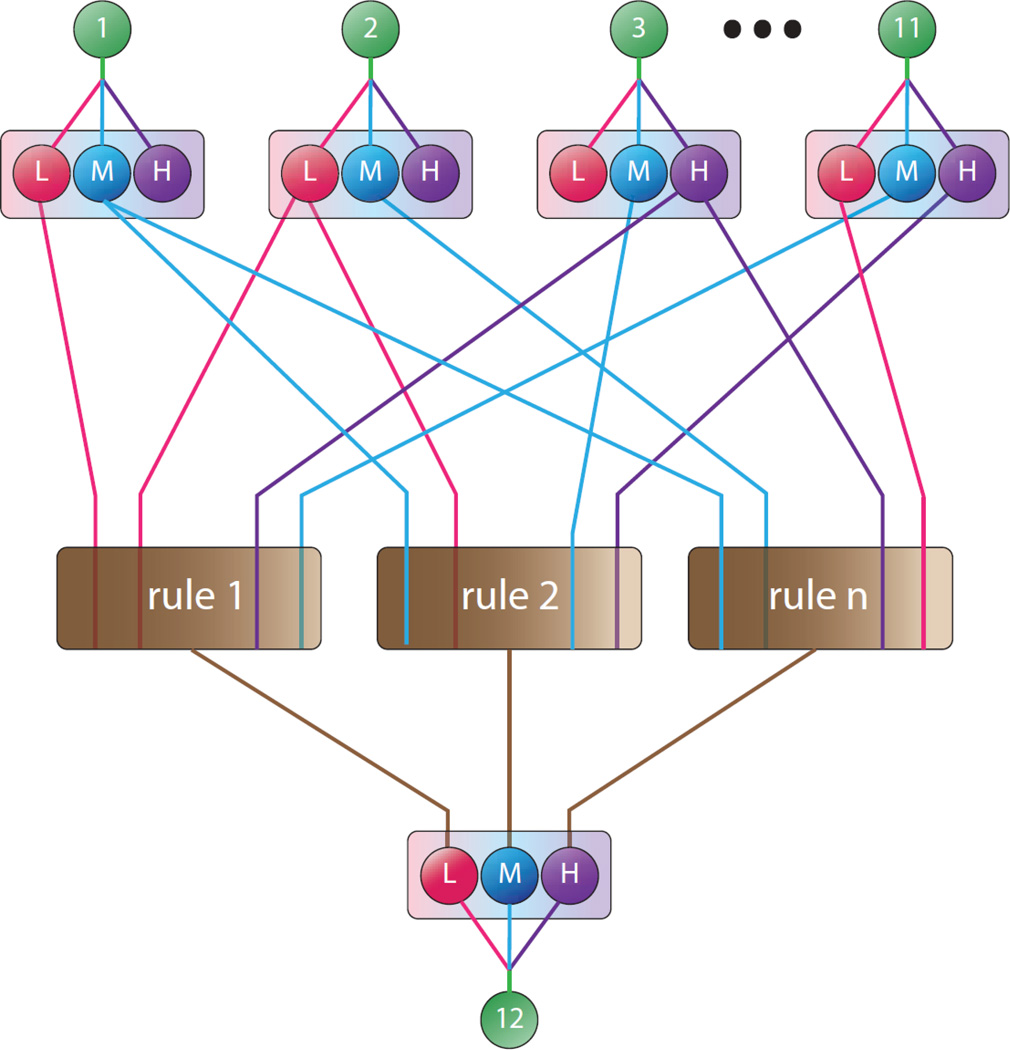
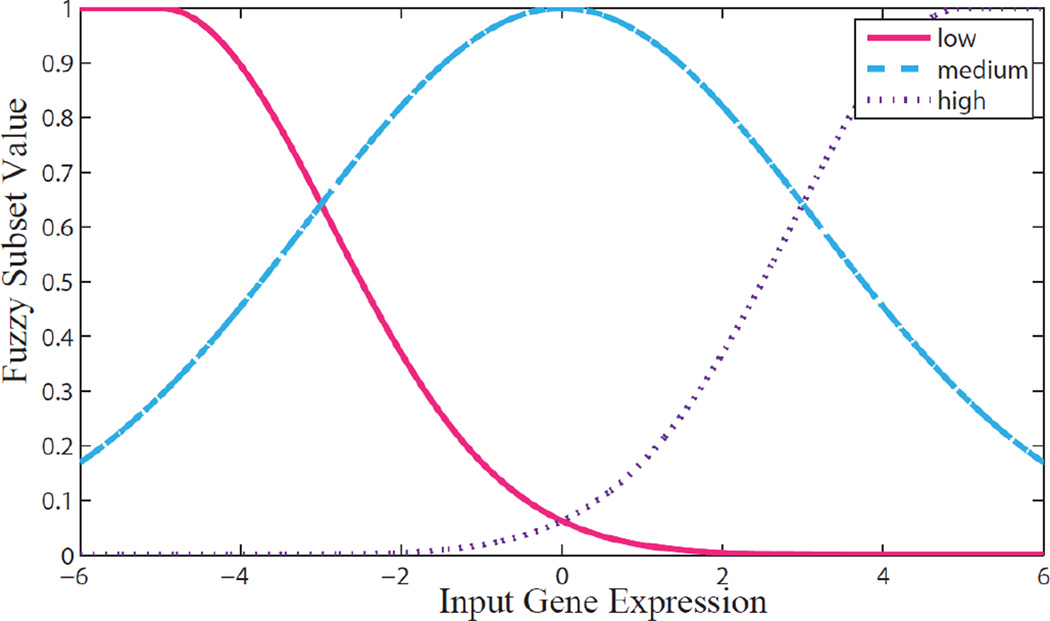
Traditional neural networks have unconstrained connections between nodes in adjacent layers. Unconstrained network structures allow for mathematical optimization during model training, but are considered to be “black boxes” due to the inability to interpret functionality of the network in a manner that provides insight into the process that is being modeled. The connections between nodes in our FNN were constrained so that each connection has a well-understood statistical or biological meaning related to the Cyp1b1 pathway. Model input genes and corresponding Cyp1b1 values (Table 1) for samples in each training data set were used to construct the neural network structure prior to model training and testing. The neural network structure consisted of 5 layers (Fig. 1). The first layer consisted of the model input gene values. The second layer consisted of gene expression values that were transformed using fuzzy logic into low, medium, and high fuzzy subsets with membership values defined by Gaussian membership functions:
| Eq. 1 |
| Eq. 2 |
| Eq. 3 |
where X is the calculated fuzzy subset value and input is the corresponding gene input value (Fig.2). The low, medium, and high fuzzy logic subsets are related to the likelihood that a gene expression level is below, at, or above a control treatment group given the observed expression level observed in the microarrays. Transforming gene expression levels into fuzzy membership subsets helps prevent model overtraining, reduces model sensitivity to inaccurate input expression levels, and allows the user to account for uncertainty by altering the scale parameter of the Gaussian distribution functions. A matrix consisting of all combinations of low, medium, and high fuzzy subsets with membership values greater than 0.1 for input genes and Cyp1b1 was constructed, and combinations were partitioned based upon the Cyp1b1 subset (matrices which contained fuzzy subsets with membership values less than 0.1 were not included to reduce processing time). Combinations within each partition were then sorted using k-means clustering (5 clusters per partition). Each cluster was then transformed into a Mamdani logic function, or “If-Then” rule (Mamdani, 1977) for predicting low, medium, or high Cyp1b1 expression levels. Rules were created using the most common fuzzy subset within a cluster for each input gene and Cyp1b1 gene The third layer of the FNN is comprised of the Mamdani If-Then rules, with one node for each rule. The fourth layer of the FNN consist of the low, medium, and high Cyp1b1 predicted expression levels predicted by the If-Then rules in the third layer of the network. The low, medium, and high Cyp1b1 predictions in the fourth layer were combined or defuzzified into a single quantitative value of Cyp1b1 expression by applying the inverse of the respective membership functions. The predicted Cyp1b1 expression is the fifth and final layer of the network. For a more thorough review of neural network structure design, see the review by Lee (1990).
Fig. 1.
Five layer fuzzy neural network structure for quantitative prediction of Cyp1b1 expression. Layer 1 consists of input gene expression values (input genes 1–11). Layer 2 fuzzifies input values into low, medium, and high fuzzy subsets. Layer 3 creates Mamdani “If-Then” rules. Layer 4 predicts low, medium, and high Cyp1b1 expression, and layer 5 (output) consists of a single quantitative prediction of Cyp1b1 mRNA expression (output gene 12).
Fig. 2.
Graph of membership functions used to fuzzify input genes. Note that the scale parameter of the medium Gaussian membership function is larger than low and high membership functions (7.5, 5, and 5, respectively), increasing bias towards control level predictions. Low, medium, and high membership values are related to the likelihood that a treatment group is expressed below, at, or above the control treatment group, respectively, given the observed expression level.
Model Training and evaluation
Model training was performed using MATLAB version 2011B with Neural Network Toolbox algorithms (“train” command using Quasi-Newton formula for minimizing means-squared error). Connections between the first and second and fourth and fifth layers were fixed with weight values equal to corresponding fuzzy subset membership values. Model training was performed using leave one out cross-validation (LOOCV). A data set is partitioned into a training data set consisting of n-1 treatment groups and a validation data set consisting of the treatment group excluded from the training data (Agrafiotis et al., 2002, Lek and Guégan, 1999). Percent relative contribution of each input gene, also referred to in neural network literature as relative importance or percent influence, towards prediction of low, medium, and high dermal Cyp1b1 expression was determined through back-calculation of the neural network connection weights followed by unbiased multi-model averaging and percent normalization to average (mean) gene contribution (9.09%) (Garson, 1991,Vasilakos et al., 2009).
The FNN was evaluated by first comparing the qualitative and quantitative predictive capabilities of the model to microarrays of treatment groups excluded from the training process using LOOCV. The structure of the model was then evaluated by comparing If-Then rules to Cyp1b1 expression levels in treatment groups included in the training data set: If-Then rules for predicting high and low Cyp1b1 expression would be ideally based upon samples in the training data set with Cyp1b1 expression levels above and below control, respectively. The model’s ability to identify and emphasize which input genes are most important for low, medium, and high Cyp1b1 expression was then evaluated by calculating the relative influence of each input gene on model predictions for low, medium, and high Cyp1b1 expression, and comparing the importance of the genes most influential in model predictions to their relative importance in the biological pathway of Cyp1b1 transcriptional regulation. Lastly, the efficacy of the genes selected for input in capturing Cyp1b1 transcriptional regulation was evaluated repeating the LOOCV studies using three permutations of randomly selected genes as model inputs for comparing predictive capabilities of models with an expert selection of inputs to models with randomly selected inputs.
Results and Discussion
This study develops an FNN model that can be applied to quantitative prediction of Cyp1b1 enzyme expression from environmental PAH mixtures in support of Tox21 and ToxCast efforts to create in vivo and in silico toxicological methods for the 21st century. Our lab previously observed complex effects of PAH mixture exposures in CYP activity and CYP1B1 expression (Courter et al, 2007b) in human cell cultures, as well as Cyp1a1 and Cyp1b1 protein and activity levels in dermally treated mice (Courter et al, 2007a). Recent studies in our lab have also demonstrated distinct differences in the carcinogenic potency of PAHs and environmental PAH mixtures in mouse skin that are not predicted by RPF values alone (Siddens et al, 2012). To further investigate the Cyp1b1 pathway, we applied a multi-layer fuzzy neural network model to predict quantitative changes in Cyp1b1 expression as a function of PAH mixture composition.
The primary purpose of fuzzy logic or fuzzy expert systems is to evaluate measurements or other quantitative values in a manner similar to the evaluation process performed by an individual with expert knowledge of the system of interest (Leondes, 1998). A fuzzy logic program evaluating the influence of co-activators or co-repressors on a transcription factor of interest, for example, would use documented network pathways, rules defined from observations of previous studies, and/or high quality data sets to predict how changes in the co-activators or co-repressors would change transcription factor activity (Pham and Liu, 1995). Fuzzy logic systems categorize, or fuzzify, quantitative values, similar to the categorization process performed by scientists when interpreting experimental results. Gene expression levels after chemical treatment, for example, are fuzzified into categories, called fuzzy subsets, such as low, medium, or high tumor potency for chemical mixtures relative to a control group.
Distribution functions such as the normal or Gaussian distribution are used to determine the value assigned to a fuzzy subset based upon the likelihood that a particular value for a given observation falls within the fuzzy subset category. Assigning values to fuzzy subsets allows the system to retain the quantitative information in a data set, which can be used to defuzzify system outputs (using the mathematical inverse of the distribution function used for fuzzifying inputs) in order to attain quantitative outputs. Incorporating fuzzy logic operations into neural networks allow network properties such as layer connections and weights to be defined based on known or observed correlations between selected variables rather than by mathematical optimization, which in turn facilitates comparisons between the behaviors of the neural network and the systems of interest (Halgamuge and Glesner, 1994).
Cyp1b1 prediction in individual treatment groups
Overall, the FNN model accurately predicted Cyp1b1 expression for each PAH treatment group, including the three environmental PAH mixtures. Model predictions, corresponding microarray observations, and the root mean squared error (RMSE, the square root of the sum of the difference between predicted and observed expression levels for all samples in a treatment group divided by the number of samples in the treatment group) are listed in Table 2. Average model predictions are within one log2- fold unit change of microarrays for all treatment groups except for DBC, where the model correctly predicted the unexpected result of DBC differing from the other treatment groups although to a greater extent (−1.34 fold-change (Log2) predicted compared to −0.28 fold-change (Log2) actual). RMSEs range from 0.36–1.16 log2-fold change units. Treatment groups with the smallest standard deviations in microarray expression levels are associated with the smallest RMSEs (Mix1 and Mix2, respectively), whereas treatment groups with the largest microarray SDs also produced the largest RMSEs (Mix3 and DBC, respectively). Model error appears to be associated with variances in treatment effects, and treatment groups which have large variances in dose-response may require larger training data sets.
TABLE 2.
Cyp1b1 Expression Levels (mean log2 fold change ± SE) and RMSE
| BaP | DBC | Mix 1 | Mix 2 | Mix 3 | |
|---|---|---|---|---|---|
| Model Predictiona | 2.32 ± 1.08 | −1.34 ± 0.16 | 0.92 ± 0.69 | 3.19 ± 0.08 | 3.02 ± 0.40 |
| Microarrays | 2.56 ± 0.37 | −0.28 ± 0.62 | 0.95 ± 0.28 | 3.85 ± 0.13 | 3.78 ± 0.51 |
| RMSE | 0.75 | 1.16 | 0.36 | 0.67 | 1.08 |
Model predications are based on 1000 replications for each treatment group.
The discrepancy in magnitude between model predictions and microarray observations for DBC treatment can be partly attributed to differences between induction, steady-state level, and repression of Cyp1b1 expression. Cyp1b1 transcriptional regulation is dynamic. The interactions between regulatory proteins change when Cyp1b1 transcriptional regulation shifts between repressed, steady-state, and induced expression. As a consequence regulation of Cyp1b1 gene expression should be viewed as a function with non-identical parameters for up and down-regulation. Using leave one out cross-validation, the FNN created rules for predicting Cyp1b1 following DBC treatment using BaP, Mix 1, Mix 2, and Mix 3 treatment groups, in which Cyp1b1 was induced. Therefore the FNN was unable to create If-Then rules for low Cyp1b1 prediction and rules for medium Cyp1b1 prediction were based exclusively on induced Cyp1b1 samples. Without gene expression profiles of treatment groups with steady-state or repressed Cyp1b1 expression, the FNN extrapolated based on induced Cyp1b1 samples, to predict the effects of DBC treatment on Cyp1b1 expression, levels lower but not significantly less than control (p=0.05). Successful quantitative prediction of induced Cyp1b1 along with unsuccessful prediction of non-induced Cyp1b1 expression suggests that Cyp1b1 induction and repression are not mirror images of each other. For that reason, quantitatively predicting the entire range of Cyp1b1 expression requires including samples with down-regulated and steady-state as well as up-regulated Cyp1b1 expression in the training set. The lack of If-Then rules for low Cyp1b1 prediction when DBC is excluded from the training data set and the abundance of If-Then rules for high Cyp1b1 prediction suggest that the model correctly identifies which samples are appropriate for developing rules to predict low, medium, or high Cyp1b1. Future models which include down-regulated and steady-state Cyp1b1 samples in the training data set are more likely to accurately predict Cyp1b1 expression after DBC treatment.
Model predictions for up-regulated treatments are all closer to control than microarray observations (bias towards the null hypothesis). Gaussian distribution functions with two or more parameters belong to the location-scale family of distribution functions. The scale parameter of the medium fuzzy logic membership function is larger than the low and high membership functions (7.5, 5, and 5, respectively). The larger scale parameter value adds bias towards greater influence from the medium subset. Similarly, defuzzifying low, medium, and high Cyp1b1 subsets is biased towards greater influence from the medium subset. Bias was intentionally added in order to prevent over-fitting during model training, as well as to structure model evaluation and interpret results from a null hypothesis paradigm. Adopting a null hypothesis viewpoint allows results to be better compared and integrated with scientific studies, and to have greater confidence in predictions of adverse biological responses at the cost of diverting from current risk assessment paradigms favoring the precautionary principle, or bias towards overestimation of responses for the purposes of protecting sensitive members of the population.
Cyp1b1 prediction in adding multiple PAH sources
Comparing the addition of PAH mixtures, in which Mix 2 contains CTE added to Mix 1, and Mix 3 contains CSC added to Mix 2, model predictions are in agreement with microarray results, in which adding CTE increases and adding CSC does not increase Cyp1b1 expression (Table 2). The inverse also applies, in which model predictions of subtracting CSC from Mix 3 and subtracting CTE from Mix 2 are in agreement with microarrays. Model predictions of adding or subtracting PAH sources can supplement current statistical efforts of capturing PAH effects by establishing PAH gene expression signatures of complex mixtures, then using sufficient similarity to predict how deviations in composition from the well-studied mixtures correlate to differences in biological responses.
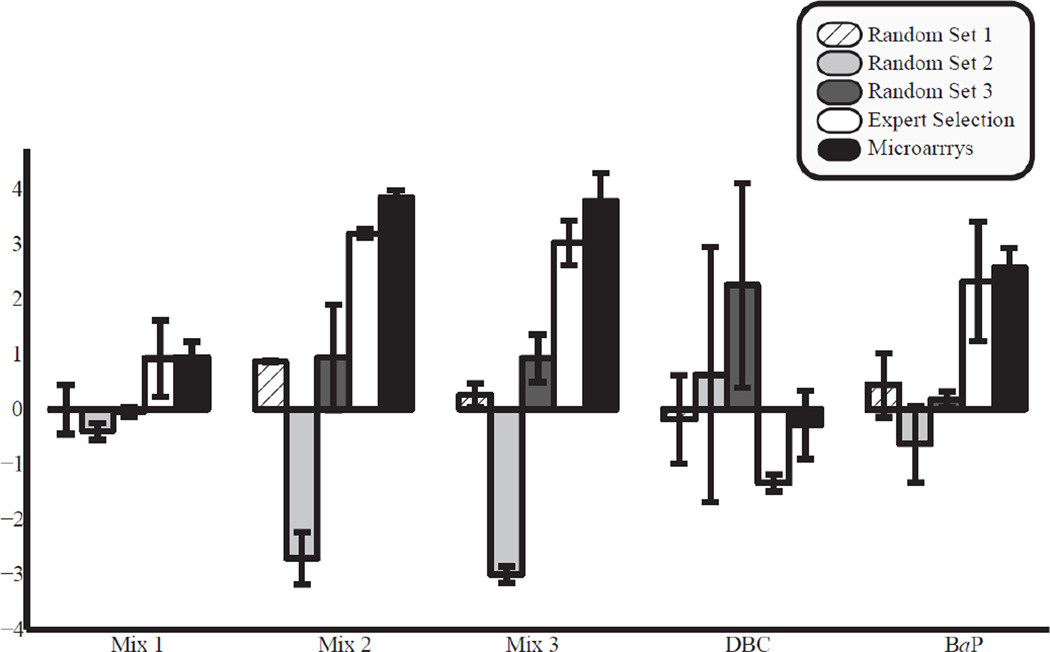
Comparison between expertly selected and randomly selected gene inputs
As mentioned above, expert systems are designed to evaluate measurements in a manner similar to an individual with expert knowledge in the subject. As described in the methods section, the structure of our expert system is derived from the relationship between genes involved in epithelial Cyp1b1 transcriptional regulation and Cyp1b1 gene expression. If the model is properly structured, connections between network layers and network nodes will capture the relationships between input genes and Cyp1b1 expression. A properly structured expert model should therefore provide more accurate predictions of Cyp1b1 expression compared to models that are structured and trained with a random selection of genes. Figure 3 shows Cyp1b1 expression predicted from models with random gene inputs (described in Supplemental Table 2) compared to those with expertly selected gene inputs (from Table 1). For most of the PAH treatment scenarios, the Cyp1b1 expression predicted by the expertly selected gene inputs most closely followed the actual Cyp1b1 expression patterns from the microarray analysis providing validation for the expert-driven approach. Differences between prediction and microarray expression levels were greater for models with random inputs for all treatment groups with the exception of DBC, where two of the three models with random inputs predicted mean Cyp1b1 levels closer to microarrays than the expert selection model, albeit with large standard errors in model predictions. As discussed above, this is likely due to the lack of data available for prediction of low Cyp1b1 expression (or down-regulation) in the testing and training of the FNN model.
Fig. 3.
Comparison of randomly selected versus expertly selected gene lists as input for the FNN model and prediction of Cyp1b1 expression. Differences between predicted and observed Cyp1b1 expression levels were greater for models with randomly selected gene inputs (described in Supplemental Table 2) than for models with an expert selection of gene inputs (Table 1) for all treatment groups with the exception of DBC.
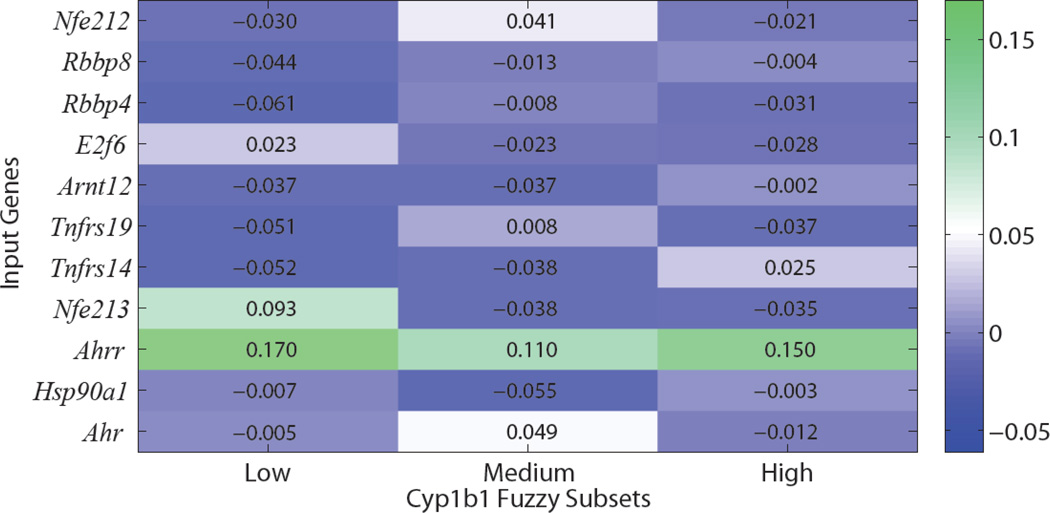
Relative percent weights of input genes
Relative percent weights of input genes are shown in Figure 4 for low, medium, and high Cyp1b1 prediction. Ahrr has the largest relative percent weight across all three Cyp1b1 fuzzy subsets, suggesting that Ahrr is a better indicator than Ahr of changes in Cyp1b1 expression at the sample time point (12 h). Cyp1b1 levels are more strongly correlated with Ahrr than Ahr in the microarrays (0.92 and −0.74, respectively), supporting the model’s decision to rely on Ahrr more than Ahr for predicting Cyp1b1 expression. Understanding the importance of selecting Ahrr over Ahr from a biological context requires a closer look at the AhR signaling pathway. As described in the introduction section, Ahrr gene expression is induced by Ahr/Arnt binding to xenobiotic response elements (XREs), and Ahrr heterodimerizes with Arnt, competitively inhibiting Ahr/Arnt XRE activation (Kawajiri and Fujii-Kuriyama, 2007). By emphasizing Ahrr over Ahr, the FNN is hypothesizing that at 12 hours post-initiation Cyp1b1 expression is more closely related to Ahrr than Ahr, further hypothesizing that Cyp1b1 inhibition at 12h is predominant over Cyp1b1 induction, and Cyp1b1 levels are in general decreasing. Studies investigating AhR induction in cultured cells have demonstrated peak nuclear localization of Ahr 1–2 h after TCDD exposure (Pollenz, 2002) and peak expression levels of reporter genes (100-fold induction) 5 h post TCDD exposure (Fujii-Kuriyama and Mimura, 2005), supporting the model-generated hypothesis of Cyp1b1 transcriptional regulatory network status. Ahr microarray expression levels are lower than control for all treatment groups with up-regulated Cyp1b1, again in agreement with the model-generated hypothesis. The importance of Ahrr in regulating Ahr-mediated Cyp1 expression has been previously shown using knockout mice as Ahrr-deficient mice exhibited higher levels of Cyp1a1 expression compared to wild type after exposure to BaP (Hosoya et al., 2008).
Fig. 4.
Heatmap of percent gene input weights for low, medium, and high Cyp1b1 prediction in the 1st, 2nd and 3rd columns, respectively. Weights are normalized relative to the average (mean) input weight (9.09%). Dark green indicates an above average (higher) relative input weight, whereas dark blue indicates a below average (lower) relative input.
Applications toward Risk Assessment
Risk assessment methods using RPFs assume interactions between mixture components are strictly additive. The published RPFs for the individual PAHs and mixtures used in this study are 100, 36, 0.004, 0.34 and 0.47 for BaP (100 µg dose), DBC, and mixtures 1, 2 and 3, respectively (Supplemental Table 1 and Siddens et al., 2012). Comparing the effects of BaP, DBC, Mix 2 and Mix 3, it is apparent that the RPF but does not correlate will with Cyp1b1 expression. RPFs do not have a well-defined method for including effects of non-PAH components present within a mixture, such as heavy metals, non-PAH urban air particulate matter, or for the effects of PAH components that are non-carcinogenic but may impact the potency of carcinogenic PAHs. Sufficient similarity is another potential method that has been proposed for risk assessment. Sufficient similarity involves evaluating the toxicity of mixtures with complex but well-defined compositions, and using the well-defined mixtures to predict toxic effects of other mixtures with similar compositions. FNN modeling can enhance both RPF and sufficient similarity risk assessment methods by predicting the effects of mixture components suspected to have non-additive interactions or predicting combinatory effects with metals and other non-PAH components and PAH mixtures. In the case of sufficient similarity, FNNs can consider a well-defined mixture as a single mixture and evaluate how adding or subtracting other components will change mixture effects, similar to comparing the effects of adding/subtracting CSC to/from Mix 2/Mix 3 or adding/subtracting CTE to/from Mix 1/Mix 2 from the microarray data mentioned above.
FNNs can be used to identify the best selection of genes to include in Tox screens for quantitative MOA modeling approaches. Our FNN model hypothesizes that Ahrr is a greater predictor than Ahr of PAH mediated Cyp1b1 induction in mouse skin at 12h post-exposure. ToxCast reference databases include a number of assays which screen for human AhR, but as of yet none of these high-throughput assays include AhRR. If the role of human AhRR is similar to Ahrr in mice, then the inclusion of AhRR in future ToxCast assays may be necessary for quantitatively predicting the effects of chemical combinations that perturb pathways associated with AhR activation.
Model limitations
Expert systems such as FNN, are dependent on accurate, previously obtained knowledge of the system of interest, and are consequently limited in scope compared to other array approaches such as modern quantitative structure activity relationship (QSAR) models. The advantage to using expert systems is the ability to predict outcomes at a quantitative level, which is an essential component for a high-throughput in silico based approach for predicting interactive effects of numerous chemical mixture combinations, as desired by the ToxCast and Tox21 programs. Expert systems are therefore not a replacement for transcriptome-wide pathway approaches, but rather a complement which allow scientists to quantitatively model known MOAs.
Current model predictions are based on microarray data from whole dermal samples collected from mice. Mouse and human skin tissues have several distinct morphological differences, including differences in epidermal thickness, densities of follicular hairs cells as well as melanocytes and inter-species differences in enzymatic activities. Nevertheless, the two-stage mouse skin tumor model has been used extensively for a number of years as a model for human skin cancer (reviewed in Di Giovanni, 1992; Yuspa and Poirier, 1988). Mouse microarrays were used for model assessment because the additive PAH dosing scheme provided the opportunity to compare model predictions and microarray measurements of gene expression for adding PAH mixtures. The model structure is not species-specific and can be adapted to other model systems such as zebrafish, human cell cultures, and human skin.
Our microarray data and model predictions evaluated Cyp1b1 expression at a single time point post treatment, 12 h. This time point was selected based on previous studies suggesting DNA adducts in murine epithelial tissue peaks at 12 h (Marsten et al., 2001). Time intervals for peak PAH metabolism and CYP1B1 expression in human keratinocytes and other human skin cells are currently unknown and could vary depending on exposure conditions, such as single vs. repeated dosing interactions or non-PAH components during co-exposures (Courter et al., 2007b). Previous work by our laboratory and others suggests, that Cyp1b1 expression is important in PAH (including DBC) carcinogenesis (Buters et al., 1999; 2002; Castro et al., 2008; Uno et al., 2006). The potency of DBC as a skin carcinogen in mouse seems to be inconsistent with DBC down-regulation of Cyp1b1 mRNA. Tumorigenesis is a complex process, and we agree that analysis of a single pathway or mechanism of action is insufficient to predict all of the ways in which a chemical or chemical mixture may contribute to tumor initiation, promotion, and progression. FNN analysis is not limited solely to the Cyp1b1 pathway. This model can be used to predict responses that may occur in numerous pathways or mechanisms of interest. The reason for focusing on a single transcriptional regulatory network in the paper is to provide a clear example of FNN analysis in a pathway that is relevant to PAH-mediated carcinogenesis and is well-known to most readers. Future studies including multiple time points would capture the temporal nature of gene regulatory processes and strengthen the predictions made by the algorithm.
The model has only been tested with mixtures that follow an S-shaped dose response curve, in which increased PAH concentrations produced non-linear increases in gene expression. The advantage to using a FNN in contrast to a traditional regression model is the ability to capture complex behavior such as feedback loops. FNNs therefore have the potential to model complex dose responses such as U-shaped dose response curves, but this ability has not yet been tested. Predicting complex dose-responses such as U-shaped curves would require larger training data sets than the data set used in this paper for predicting S-shaped dose responses.
As with any computational or mathematical model, the accuracy of model predictions are limited by the ability of a data set to capture the relevant information about the population of interest: models based on incomplete data, such as missing time points or concentrations, or data which has lost relevant information during sample and/or data processing are less likely to accurately simulate or predict conditions in the system of interest.
Conclusions
A Fuzzy Neural Network model was developed and evaluated for predicting PAH mixture-mediated perturbations of the dermal CYP1B1 transcriptional regulatory network. The model was evaluated with microarrays of RNA from FVB/N mice dermally treated with environmental PAH mixtures using leave one out cross-validation. Model predictions were within 1 log2 fold change unit of the microarrays for all treatment groups with the exception of DBC, where the unexpected down-regulation was predicted but failed to reach statistical significance on the microarray. Model predictions of adding coal tar extract (increase in Cyp1b1) or cigarette smoke condensate (no increase in Cyp1b1) to existing PAH mixtures were in agreement with microarray data. Ahrr greatly influenced model predictions. Further development of Fuzzy Neural Networks can supplement sufficient similarity or component-based risk assessment methods by integrating early, sensitive, and robust biological responses capable of capturing PAH interactive effects into the risk assessment process.
Supplementary Material
Highlights.
Tested a model to predict PAH mixtures-mediated changes in Cyp1b1 expression
Quantitative predictions in agreement with microarrays for Cyp1b1 induction
Unexpected difference in expression between DBC and other treatments predicted
Model predictions for combining PAH mixtures in agreement with microarrays
Predictions highly dependent on aryl hydrocarbon receptor repressor expression
Acknowledgements
The Authors thank Dr. Hollie Swanson for providing cigarette smoke condensate and Laboratory Animal Resources Center at Oregon State University for help with the animal studies. Microarrays were processed by Bradley Stewart at the University of Wisconsin EDGE3 Core Facility. Part of the research described in this manuscript was presented at the 2011 Superfund Research Program annual meeting and 2012 Society of Toxicology annual meeting. Pacific Northwest National Laboratory is a multi-program national laboratory operated by Battelle Memorial Institute for the DOE under contract number DEAC05-76RLO1830.
Funding
This study was funded by the National Institute of Environmental Health Sciences (grants P42ES016465 and P42ES016465-S1).
Abbreviations
- PAH
polycyclic aromatic hydrocarbon
- CYP
cytochrome P450
- FNN
fuzzy neural network
- DBC
dibenzo[def,p]chrysene
- BaP
benzo[a]pyrene
- DPE
diesel particulate extract
- CTE
coal tar extract
- CSC
cigarette smoke condensate
- AhR
aryl hydrocarbon receptor
- AhRR
aryl hydrocarbon receptor repressor
- ARNT
aryl hydrocarbon receptor nuclear translocator
- BaPDE
B[a]P-7,8-dihydrodiol-9,10-epoxide
- HSP90
heat shock protein90
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reports of skin cancer incidence do not distinguish between UV-induced and chemical-induced carcinogenesis
Conflict of Interest Statement
None of the authors of this manuscript have any conflicts of interest associated with this work.
References
- Abdelaziz A, Shushko I, Wolfram T, Körner R, Novotarskyi S, Tetko IV. QSAR modeling for In vitro assays: linking ToxCast™ database to the integrated modeling framework “OCHEM”. J Chem. Inf. 2012;4:62. [Google Scholar]
- Agrafiotis DK, Cedeno W, Lobanov VS. On the use of neural network ensembles in QSAR and QSPR. J. Chem. Inf. Comput. Sci. 2002;42:903–911. doi: 10.1021/ci0203702. [DOI] [PubMed] [Google Scholar]
- Altenburger R, Scholz S, Mechthild S, Busch W, Escher BI. Mixture toxicity revisisted from a toxicogenomic perspective. Environ. Sci. Technol. 2012;46:2508–2522. doi: 10.1021/es2038036. [DOI] [PubMed] [Google Scholar]
- Baird WM, Hooven LA, Mahadevan B. Carcinogenic aromatic hydrocarbon-DNA adducts and mechanism of action. Environ. Mol. Mutagen. 2005;45:106–114. doi: 10.1002/em.20095. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. Roy. Statist. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- Billiard SM, Timme-Laragy AR, Wassenberg DM, Cockman C, Di Giulio RT. The role of the aryl hydrocarbon receptor pathway in mediating synergistic developmental toxicity of polycyclic aromatic hydrocarbons to zebrafish. Toxicol. Sci. 2006;92:526–536. doi: 10.1093/toxsci/kfl011. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Åstrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Burke KE, Wei H. Synergistic damage by UVA radiation and pollutants. Toxicol. Ind. Hlth. 2009;25(4–5):219–224. doi: 10.1177/0748233709106067. [DOI] [PubMed] [Google Scholar]
- Buters JT, Mahadevan B, Quintanilla-Martinez L, Gonzalez FJ, Greim H, Baird WM, Luch A. Cytochrome P450 1B1 determines susceptibility to dibenzo[a,l]pyrene-induced tumor formation. Chem. Res. Toxicol. 2002;15:1127–1135. doi: 10.1021/tx020017q. [DOI] [PubMed] [Google Scholar]
- Buters JT, Sakai S, Richter T, Pineau T, Alexander DL, Savas U, Doehmer J, Ward JM, Jefcoate CR, Gonzalez FJ. Cytochrome P450 CYP1B1 determines susceptibility to 7,12-dimethylbenz[a]thracene-induced lymphomas. Proc. Natl. Acad. Sci. (USA) 1999;96:1977–1982. doi: 10.1073/pnas.96.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro DJ, Baird WM, Pereira CB, Giovanini J, Löhr CV, Fischer KA, Yu Z, Gonzalez FJ, Krueger SK, Williams DE. Fetal mouse Cyp1b1 and transplacental carcinogenesis from maternal exposure to dibenzo(a,l)pyrene. Cancer Prev. Res. 2008;1:128–134. doi: 10.1158/1940-6207.CAPR-07-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalieri EL, Rogan EG. Central role of radical cations in metabolic activation of polycyclic aromatic hydrocarbons. Xenobiotica. 1995;25:677–688. doi: 10.3109/00498259509061885. [DOI] [PubMed] [Google Scholar]
- Courter LA, Musafia-Jeknic T, Fischer K, Bildfell R, Giovanini J, Pereira C, Baird WM. Urban dust particulate matter alters PAH-induced carcinogenesis by inhibition of CYP1A1 and CYP1B1. Toxicol. Sci. 2007a;95:63–73. doi: 10.1093/toxsci/kfl137. [DOI] [PubMed] [Google Scholar]
- Courter LA, Pereira C, Baird WM. Diesel exhaust influences carcinogenic PAH-indued genotoxicity and gene expression in human breast epithelial cells in culture. Mut. Res. 2007b;625:72–82. doi: 10.1016/j.mrfmmm.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damon DA. Toxic equivalency factor approach for assessment of polycyclic aromatic hydrocarbons. Toxicol. Environ. Chem. 1997;64:81–108. [Google Scholar]
- DiGiovanni J. Multistage carcinogenesis in mouse skin. Pharmacol. Ther. 1992;54:63–128. doi: 10.1016/0163-7258(92)90051-z. [DOI] [PubMed] [Google Scholar]
- Fujii-Kuriyama Y, Mimura J. Molecular mechanisms of AhR functions in the regulation of cytochrome P450 genes. Biochem. Biophys. Res. Commun. 2005;338:311–317. doi: 10.1016/j.bbrc.2005.08.162. [DOI] [PubMed] [Google Scholar]
- Garson GD. Interpreting neural-network connection weights. AI expert. 1991;6:46–51. [Google Scholar]
- Hahn ME, Allan LL, Sherr DH. Regulation of constitutive and inducible AHR signaling: complex interactions involving the AHR repressor. Biochem. Pharmacol. 2009;77:485–497. doi: 10.1016/j.bcp.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halgamuge SK, Glesner M. Neural networks in designing fuzzy systems for real world applications. Fuzzy Set. Syst. 1994;65:1–12. [Google Scholar]
- Hosoya T, Harada N, Mimura J, Motohashi H, Takahashi S, Nakajima O, Morita M, Kawauchi S, Yamamoto M, Fujii-Kuriyama Y. Inducibility of cytochrome P450 1A1 and chemical carcinogenesis by benzo[a]pyrene in AhR repressor-deficient mice. Biochem. Biophys. Res. Commun. 2008;365:562–567. doi: 10.1016/j.bbrc.2007.11.016. [DOI] [PubMed] [Google Scholar]
- IARC. Some non-heterocyclic polycyclicaromatic hydrocarbons and some related exposures. Monographs on the evaluation of carcinogenic risks to humans Lyon, France. 2010 [ http://monographs.iarc.fr/ENG/Monographs/vol92/mono92.pdf]. [PMC free article] [PubMed]
- Kawajiri K, Fujii-Kuriyama Y. Cytochrome P450 gene regulation and physiological functions mediated by the aryl hydrocarbon receptor. Arch. Biochem. Biophys. 2007;2:207–212. doi: 10.1016/j.abb.2007.03.038. [DOI] [PubMed] [Google Scholar]
- Kerr MK, Martin M, Churchill GA. Analysis of variance for gene expression microarray data. J. Comput. Biol. 2000;7:819–837. doi: 10.1089/10665270050514954. [DOI] [PubMed] [Google Scholar]
- International Agency for Research on Cancer. Some non-heterocyclic polycyclic aromatic hydrocarbons and some related exposures. IARC Monographs. 2010;92:7–465. [PMC free article] [PubMed] [Google Scholar]
- Lee CC. Fuzzy logic in control systems: fuzzy logic controller. IEEE. Trans. Syst. Man. Cybern. 1990;20:404–418. [Google Scholar]
- Lek S, Delacoste M, Baran P, Dimopoulos I, Lauga J, Aulagnier S. Application of neural networks to modelling nonlinear relationships in ecology. Ecol. Model. 1996;90:39–52. [Google Scholar]
- Lek S, Guégan JF. Artificial neural networks as a tool in ecological modelling, an introduction. Ecol. Modell. 1999;120:65–73. [Google Scholar]
- Leondes CT. Fuzzy logic and expert systems applications. San Diego: Academic Press; 1998. [Google Scholar]
- Lin P, Hu SW, Chang TH. Correlation between gene expression of aryl hydrocarbon receptor (AhR), hydrocarbon receptor nuclear translocator (Arnt), cytochromes P4501A1 (CYP1A1) and 1B1 (CYP1B1), and inducibility of CYP1A1 and CYP1B1 in human lymphocytes. Toxicol. Sci. 2003;71:20–26. doi: 10.1093/toxsci/71.1.20. [DOI] [PubMed] [Google Scholar]
- Lomas A, Leonardi-Bee J, Bath-Hextall F. A Systematic Review of worldwide incidence of non-melanoma skin cancer. Br. J. Dermatol. 2012;166:1069–1080. doi: 10.1111/j.1365-2133.2012.10830.x. [DOI] [PubMed] [Google Scholar]
- Mamdani EH. Application of fuzzy logic to approximate reasoning using linguistic synthesis. IEEE. Trans. Comput. 1977;100:1182–1191. [Google Scholar]
- Marston CP, Pereira C, Ferguson J, Fischer K, Hedstrom O, Dashwood WM, Baird WM. Effect of a complex environmental mixture from coal tar containing polycyclic aromatic hydrocarbons (PAH) on the tumor initiation, PAH–DNA binding and metabolic activation of carcinogenic PAH in mouse epidermis. Carcinogenesis. 2001;22:1077–1086. doi: 10.1093/carcin/22.7.1077. [DOI] [PubMed] [Google Scholar]
- Moldoveanu S. Toxicological and environmental aspects of polycyclic aromatic hydrocarbons (PAHs) and related compounds. Tech. Instrum. Anal. Chem. 2010;28:693–699. [Google Scholar]
- Penning TM, Burczynski ME, Hung C-F, McCoull KD, Palackal NT, Tsuruda LS. Dihydrodiol dehydrogenases and polycyclic aromatic hydrocarbon activation: generation of reactive and redox-active o-quinones. Chem. Res. Toxicol. 1999;12:1–18. doi: 10.1021/tx980143n. [DOI] [PubMed] [Google Scholar]
- Pham D, Liu X. Neural Networks for Identification, Prediction, and Control. London: Springer-Verlag; 1995. [Google Scholar]
- Pollenz RS. The mechanism of AH receptor protein down-regulation (degradation) and its impact on AH receptor-mediated gene regulation. Chem. Biol. Interact. 2002;141:41–61. doi: 10.1016/s0009-2797(02)00065-0. [DOI] [PubMed] [Google Scholar]
- Schauer C, Niessner R, Pöschl U. Polycyclic aromatic hydrocarbons in urban air particulate matter: decadal and seasonal trends, chemical degradation, and sampling artifacts. Environ. Sci. Technol. 2003;37:2861–2868. doi: 10.1021/es034059s. [DOI] [PubMed] [Google Scholar]
- Siddens LK, Larkin A, Krueger SK, Bradfield CA, Waters KM, Tilton SC, Pereira CB, Löjr CV, Arlt VM, Phillips DH, Williams DE, Baird WM. Polycyclic aromatic hydrocarbons as skin carcinogens: comparison of benzo[a]pyrene, dibenzo[def,p]chrysene and three environmental mixtures in the FVB/N mouse. Toxicol. Appl. Pharmacol. 2012;264:377–386. doi: 10.1016/j.taap.2012.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R, Ward E, Brawley O, Jeimal A. Cancer Statistics, 2011. The impact of eliminating socieoeconomic and racial disparities on premature cancer deaths. CA Cancer J. Clin. 2011;61:212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- Sipes NS, Martin MT, Reif DM, Kleinstreuer NC, Judson RS, Singh AV, Chandler KJ, Dix DJ, Kavlock RJ, Knudsen TB. Predictive models of prenatal developmental toxicity from ToxCast high-throughput screening data. Toxicol. Sci. 2011;124:109–127. doi: 10.1093/toxsci/kfr220. [DOI] [PubMed] [Google Scholar]
- Timme-Laragy AR, Cockman CJ, Matson CW, Di Giulio RT. Synergistic induction of AHR regulated genes in developmental toxicity from co-exposure to two model PAHs in zebrafish. Aquat. Toxicol. 2007;85:241–250. doi: 10.1016/j.aquatox.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka T, Ibuki Y, Takabayashi F, Goto R. Coexposure to benzo[a]pyrene and UVA induces DNA damage: first proof of double-strand breaks in a cell-free system. Environ. Molec. Mutagen. 2006;47:38–47. doi: 10.1002/em.20166. [DOI] [PubMed] [Google Scholar]
- Uno S, Dalton TP, Dragin N, Curran CP, Derkenne S, Miller ML, Shertzer HG, Gonzalez FJ, Nebert DW. Oral benzo[a]pyrene in Cyp1 knockout mouse lines: CYP1A1 important in detoxication, CYP1B1 metabolism required for immune damage independent of total-body burden and clearance rate. Molec. Pharmacol. 2006;69:1103–1114. doi: 10.1124/mol.105.021501. [DOI] [PubMed] [Google Scholar]
- Van den Berg M, Birnbaum L, Bosveld A, Brunström B, Cook P, Feeley M, Giesy JP, Hanberg A, Hasegawa R, Kennedy SW. Toxic equivalency factors (TEFs) for PCBs, PCDDs, PCDFs for humans and wildlife. Environ. Health Perspect. 1998;106:775–792. doi: 10.1289/ehp.98106775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilakos C, Kalabokidis K, Hatzopoulos J, Matsinos I. Identifying wildland fire ignition factors through sensitivity analysis of a neural network. Nat. Hazards. 2009;50:125–143. [Google Scholar]
- Yuspa SH, Poirier MC. Chemical carcinogenesis: from animal models to molecular models in one decade. Adv. Cancer Res. 1988;50:25–70. doi: 10.1016/s0065-230x(08)60434-0. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Tao S. Global atmospheric emission inventory of polycyclic aromatic hydrocarbons (PAHs) for 2004. Atmos. Environ. 2009;43:812–819. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.