Abstract
This unit describes a technique for the direct and quantitative measurement of the capacity of peptide ligands to bind Class I and Class II MHC molecules. The binding of a peptide of interest to MHC is assessed based on its ability to inhibit the binding of a radiolabeled probe peptide to purified MHC molecules. This unit includes protocols for the purification of Class I and Class II MHC molecules by affinity chromatography, and for the radiolabeling of peptides using the chloramine T method. An alternate protocol describes alterations in the basic protocol that are necessary when performing direct binding assays, which are required for (1) selecting appropriate high-affinity, assay-specific, radiolabeled ligands, and (2) determining the amount of MHC necessary to yield assays with the highest sensitivity. After a predetermined incubation period, dependent upon the allele under examination, the bound and unbound radiolabeled species are separated, and their relative amounts are determined. Three methods for separation are described, two utilizing size-exclusion gel-filtration chromatography and a third using monoclonal antibody capture of MHC. Data analysis for each method is also explained.
Keywords: MHC class I, MHC class II, T cell epitope, peptide ligand, binding affinity, CTL, epitope recognition
INTRODUCTION
This unit describes a technique for the direct and quantitative measurement of the capacity of peptide ligands to bind Class I and Class II MHC molecules. The binding of a peptide of interest to MHC is assessed based on its ability to inhibit the binding of a radiolabeled probe peptide to MHC molecules. MHC molecules are solubilized with detergents and purified by affinity chromatography. Purified MHC is then incubated with the inhibitor peptide and a high-affinity binding radiolabeled probe peptide, in the presence of a cocktail of protease inhibitors. Incubation is typically for 2 days at room temperature, although some alleles require incubation at higher temperatures (i.e., 37°C) and longer time periods. At the end of the incubation period, MHC-peptide complexes are separated from unbound radiolabeled peptide by (a) an antibody-capture phase or (b) size-exclusion gel-filtration chromatography. The percent of bound radioactivity is then determined. The binding affinity of a particular peptide for an MHC molecule may be determined by co-incubation of various doses of unlabeled competitor peptide with the MHC molecules and labeled probe peptide. The concentration of unlabeled peptide required to inhibit the binding of the labeled peptide by 50% (IC50) can be determined by plotting dose versus % inhibition. Under conditions where [label] < [MHC] and IC50 ≥ [MHC], the measured IC50 values are reasonable approximations of true Kd values (Cheng and Prusoff, 1973; Gulukota et al., 1997). To date, the methods described below have been utilized to establish over 130 binding assays for Class I and Class II molecules of human, mouse, macaque (Indian and Chinese Rhesus macaque, and pig-tailed macaque), chimpanzee, and gorilla origin (Tables 18.3.1, 18.3.2, and 18.3.3). Note that UNIT 18.4 describes an alternate method for measuring peptide-MHC interactions using a spin column gel filtration assay.
Table 18.3.1.
Human, Non-Human Primate, and Murine Class I MHC-Peptide Binding Assays Established Using Purified MHC Molecules and Radiolabeled Ligands
| Radiolabeled peptidea |
||||
|---|---|---|---|---|
| Organism | Allele | Preferred cell line | Sequence | Source |
| Human | A*0101 | STEINLIN | YTAVVPLVY | H. sapiens (J chain 102) |
| A*0201 | JY | FLPSDYFPSV | HBV (core 18) | |
| A*0202 | M7 | GLYSSTVPV | HBV (Pol 61) | |
| A*0203 | FUN | FVNYNFTLV | T. cruzi (trans-sialidase) | |
| A*0205 | DAH | FLPSDYFPSV | HBV (core 18) | |
| A*0206 | CLA | FLPSDYFPSV | HBV (core 18) | |
| A*0207 | AP | FLPSDYFPSV | HBV (core 18) | |
| A*0217 | AMALA | FLPSDYFPSV | HBV (core 18) | |
| A*0301 | GM3107 | YVFPVIFSK | H. sapiens (MAGE3 138) | |
| A*1101 | BVR | YVFPVIFSK | H. sapiens (MAGE3 138) | |
| A*2301 | WT51 | AYIDNYNKF | Non-natural (A24 consensus) | |
| A*2402 | KT3 | AYIDNYNKF | Non-natural (A24 consensus) | |
| A*2601 | QBL | ETFGFEIQSY | H. sapiens (leucine zipper 51) | |
| A*2902 | SWEIG | YTAVVPLVY | H. sapiens (J chain 102) | |
| A*3001 | RSH or S Buus | KTKDYVNGL | H. sapiens (F actin 235) | |
| A*3002 | DUCAF | RISGVDRYY | H. sapiens (NADH 53) | |
| A*3101 | SPACH | YVFPVIFSR | H. sapiens (MAGE3 138) | |
| A*3201 | WT47 | RILHNFAYSL | H. sapiens (Her2/neu 434) | |
| A*3301 | LWAGS | YVFPVIFSR | H. sapiens (MAGE3 138) | |
| A*6601 | TEM | YVFPVIFSR | H. sapiens (MAGE3 138) | |
| A*6801 | CIR | YVFPVIFSK | H. sapiens (MAGE3 138) | |
| A*6802 | AMAI | YVIKVSARV | H. sapiens (MAGE1 282) | |
| A*7401 | Pure Protein | YVFPVIFSR | H. sapiens (MAGE3 138) | |
| B*0702 | GM3107 | APRTLVYLL | H. sapiens (A2 signal seq 5) | |
| B*0801 | STEINLIN | FLRGRAYGI | HSV (EBNA 3 nuc) | |
| B*1402 | HO301 | DAYRRIHSL | H. sapiens (BRCA1/2) | |
| B*1501 | SPACH | AQIDNYNKF | Non-natural (A24 consensus) | |
| B*1503 | S Buus | YQAVVPLVY | H. sapiens (J chain 102) | |
| B*1801 | DUCAF | SEIDLILGY | H. sapiens (unknown) | |
| B*2705 | LG2 | FRYNGLIHR | H. sapiens (60s rL28 38) | |
| B*3501 | CIR | FPFKYAAAF | Non-natural (B35 consensus) | |
| B*3503 | KOSE | FPFKYAAAF | Non-natural (B35 consensus) | |
| B*3508 | TISI | FPFKYAAAF | Non-natural (B35 consensus) | |
| B*3701 | KAS011 | AEFKYIAAV | Non-natural (B4006 consensus) | |
| B*3801 | TEM | YHIPGDTLF | Variola virus (RNA-hel 346) | |
| B*4001 | 2F7 | YEFLQPILL | H. sapiens (XP090897) | |
| B*4002 | SWEIG | YEFLQPILL | H. sapiens (XP090897) | |
| B*4201 | RSH | FPFKYAAAF | Non-natural (B35 consensus) | |
| B*4402 | WT47 | SEIDLILGY | H. sapiens (unknown) | |
| B*4403 | PITOUT | SEIDLILGY | H. sapiens (unknown) | |
| B*4501 | OMW | AEFKYIAAV | Non-natural (B4006 consensus) | |
| B*5101 | KAS116 | FPYSTFPII | Non-natural (B51 consensus) | |
| B*5201 | HARA | YGFSDPLTF | Unknown (Mamu B39 ligand) | |
| B*5301 | AMAI | FPFKYAAAF | Non-natural (B35 consensus) | |
| B*5401 | KT3 | FPFKYAAAF | Non-natural (B35 consensus) | |
| B*5701 | DBB | KAGQYVTIW | H. sapiens (lamin C 490) | |
| B*5801 | AP | ISDSNPYLTQW | H. sapiens (E46 407) | |
| B*5802 | 35841 | GSVNVVYTF | H. sapiens (glucose trans 5 322) | |
| C*0401 | CIR | QYDDAVYKL | Non-natural (consensus) | |
| C*0602 | 721.221b | YRHDGGNVL | Rat (Ig variable 80) | |
| C*0702 | 721.221 | YRHDGGNVL | Rat (Ig variable 80) | |
| Macaque | Mamu A*01 | 721.221 | ATPYDINQML | SIV (Gag 181) |
| Mamu A*02 | 721.221 | YTAVVPLVY | H. sapiens (J chain 102) | |
| Mamu A*07 | 721.221 | YHSNVKEL | SIV (Pol 782) | |
| Mamu A*11 | 721.221 | GDYKLVEI | SIV (Env 497) | |
| Mamu A1*2201 | 721.221 | YVADALAAF | Non-natural (Mamu A26 consensus) | |
| Mamu A1*2601 | 721.221 | YLPTQQDVL | Non-natural (Mamu A26 consensus) | |
| Mamu B*01 | 721.221 | SDYLELDTI | Macaque (tumor reject gp96 235) | |
| Mamu B*03 | 721.221 | RRAARAEYL | Non-natural (consensus) | |
| Mamu B*08 | 721.221 | RRDYRRGL | Non-natural (Vif 172) | |
| Mamu B*17 | 721.221 | IRFPKTFGY | SIV (Nef 165) | |
| Mamu B*48 | 721.221 | AQFSPQYL | Non-natural | |
| Mamu B*52 | 721.221 | VGNVYVKF | H. sapiens (U2 RNA factor 1) | |
| Mamu B*8301 | 721.221 | KSINKVYGK | Vaccinia (B13R 64) | |
| Mane A*0301 | 721.221 | DHQAAFQYI | SIV (Gag analog 176) | |
| Mane A*0302 | 721.221 | DHQAAFQYI | SIV (Gag analog 176) | |
| Chimpanzee | Patr A*0101 | 721.221 | KVFPYALINK | Non-natural (A3 consensus) |
| Patr A*03 | 721.221 | KVFPYALINK | Non-natural (A3 consensus) | |
| Patr A*0401 | 721.221 | KFYGPFVDR | SARS (Orf 1ab 3420) | |
| Patr A*0701 | 721.221 | AYIDNYNKV | Non-natural (A24CON1) | |
| Patr A*0901 | 721.221 | AYISSEATTPV | HCV (NS4 1963) | |
| Patr B*0101 | 721.221 | YTGDFDSVI | HCV (NS3 1444) | |
| Patr B*1301 | 721.221 | FPFKYAAAF | Non-natural (B35 consensus) | |
| Patr B*2401 | 721.221 | SDYLELDTI | Macaque (tumor reject gp96 235) | |
| Mouse | Db | EL4 | SGPSNTYPEI | Adenovirus (EA1) |
| Dd | P815 | RGPYRAFVTI | HIV (Env 18) | |
| Kb | EL4 | RGYVFQGL | VSV (NP 52) | |
| Kd | P815 | KFNPMFTYI | Non-natural | |
| Kk | CH27 | SEAAYAKKI | Non-natural (Kk consensus) | |
| Ld | P815 | IPQSLDSYWTSL | HBV (Env 28) | |
All Class I assays are performed at pH 7.0, at room temperature, with a 48-hr incubation. With the exception of B*4402 and B*4403, which are captured with the B123.2 antibody, all primate Class I assays are captured for 3 hr with W6/32. Mouse Class I assays are captured using the antibodies as listed in Table 18.3.5.
721.221 refers to corresponding single allele transfected cell lines.
Table 18.3.2.
Human, Non-Human Primate, and Murine Class II MHC-Peptide Binding Assays Established Using Purified MHC Molecules and Radiolabeled Ligands: Cell Lines and Ligands for Class II Assays
| Radiolabeled peptide |
||||
|---|---|---|---|---|
| Organism | Allele(s) | Preferred cell line | Sequence | Source |
| Human | DPB1*0101 | VAVY | EKKYFAATQFEPLAA | H. sapiens (aminopeptidase 285) |
| DPB1*0201 | WT51 | KYFAATQFEPLAARL | H. sapiens (aminopeptidase 287) | |
| DPB1*0301 | COX | AFKVAATAANAAPAY | Non-natural (Phl p 5 analog 196) | |
| DPB1*0401 | PITOUT | KYFAATQFEPLAARL | H. sapiens (aminopeptidase 287) | |
| DPB1*0402 | AMAI | EKKYFAATQFEPLAA | H. sapiens (aminopeptidase 285) | |
| DPB1*0501 | HO301 | IGRIAETILGYNPSA | H. sapiens (chimeric protein) | |
| DPB1*1401 | KAS011 | AFKVAATAANAAPANY | Non-natural (Phl p 5 analog 196) | |
| DPB1*2001 | EBV-D8 | AFKVAATAANAAPAY | Non-natural (Phl p 5 analog 196) | |
| DQA1*0501/B1*0201 | VAVY | KPLLIIAEDVEGEY | M. tuberculosis (65 kDa hsp 32) | |
| DQA1*0201/B1*0202 | PITOUT | EEDIEIIPIQEEEY | H. sapiens (CD20 249) | |
| DQA1*0301/B1*0301 | PF | YAHAAHAAHAAHAAHAA | Non-natural (ROIV reiterative) | |
| DQA1*0501/B1*0301 | HERLUF | YAHAAHAAHAAHAAHAA | Non-natural (ROIV reiterative) | |
| DQA1*0505/B1*0301 | SWEIG | YAHAAHAAHAAHAAHAA | Non-natural (ROIV reiterative) | |
| DQA1*0301/B1*0302 | PRIESS | EEDIEIIPIQEEEY | H. sapiens (CD20 249) | |
| DQA1*0401/B1*0402 | OLL | EEDIEIIPIQEEEY | H. sapiens (CD20 249) | |
| DQA1*0101/B1*0501 | LG2 | AAHSAAFEDLRVSSY | Influenza (nucleoprotein 335) | |
| DQA1*0102/B1*0502 | KAS011 | AAHSAAFEDLRVSSY | Influenza (nucleoprotein 335) | |
| DQA1*0104/B1*0503 | TEM | AAHSAAFEDLRVSSY | Influenza (nucleoprotein 335) | |
| DQA1*0102/B1*0602 | MGAR | AAATAGTTVYGAFAA | Non-natural (GAD65 analog 334) | |
| DQA1*0103/B1*0603 | OMW | AAATAGTTVYGAFAA | Non-natural (GAD65 analog 334) | |
| DRB1*0101 | LG2 | YPKYVKQNTLKLAT | Influenza (HA 307) | |
| DRB1*0301 | MAT | YARIRRDGCLLRLVD | H. sapiens (telomerase 854) | |
| DRB1*0401 | PRIESS | PVVHFFKNIVTPRTPPY | H. sapiens (MBP 85) | |
| DRB1*0404 | BIN40 | PVVHFFKNIVTPRTPPY | H. sapiens (MBP 85) | |
| DRB1*0405 | KT3 | PVVHFFKNIVTPRTPPY | H. sapiens (MBP 85) | |
| DRB1*0701 | PITOUT | YATFFIKANSKFIGITE | C. tetani (tetanus toxin 828) | |
| DRB1*0802 | OLL | EVFFQRLGIASGRARY | H. sapiens (PSA 522) | |
| DRB1*0901 | HID | TLSVTFIGAAPLILSY | H. sapiens (PSA 9) | |
| DRB1*1001 | WTAIL | AFKVAATAANAAPAY | Non-natural (Phl p 5 analog 196) | |
| DRB1*1101 | SWEIG | YATFFIKANSKFIGITE | C. tetani (tetanus toxin 830) | |
| DRB1*1201 | HERLUF | EALIHQLKINPYVLS | H. sapiens (unknown) | |
| DRB1*1302 | H0301 | QYIKANAKFIGITE | C. tetani (tetanus toxin 830) | |
| DRB1*1501 | L466.1 | PVVHFFKNIVTPRTPPY | H. sapiens (MBP 78) | |
| DRB1*1602 | RML | EALIHQLKINPYVLS | H. sapiens (unknown) | |
| DRB3*0101 | MAT | YTVDFSLDPTFTIETT | HCV (polyprotein 1460) | |
| DRB3*0202 | HERLUF | VIDWLVSNQSVRNRQEGLY | Non-natural | |
| DRB4*0101 | L257.6 | QVPLVQQQQFLGQQQP | T. aestivum (alpha gliadin 41) | |
| DRB5*0101 | GM3107 | YATFFIKANSKFIGITE | C. tetani (tetanus toxin 828) | |
| Macaque | Mamu DRBw*20101 | RM3 | ELYKYKVVKIEPLGV | HIV (gp120 482) |
| Mamu DRB1*0406 | RM3 | THGIRPVVSTQLLLY | HIV (gp120 analog 248) | |
| Mouse | IAb | LB27.4 | YAHAAHAAHAAHAAHAA | Non-natural (ROIV reiterative) |
| IAd | A20/ LB27.4 | YAHAAHAAHAAHAAHAA | Non-natural (ROIV reiterative) | |
| IAk | CH12/CH27 | YNTDGSTDYGILQINSR | G. gallus (HEL 46) | |
| IAs | LS102.9 | YAHAAHAAHAAHAAHAA | Non-natural (ROIV reiterative) | |
| IAu | 91.7 | YAHAAHAAHAAHAAHAA | Non-natural (ROIV reiterative) | |
| IEd | A20/ LB27.4 | YRKILRQRKIDRLID | HIV (Vpu 30) | |
| IEk | CH12 | YLEDARRLKAIYEKKK | Bacteriophage (lambda rep. 12) | |
Table 18.3.3.
Human, Non-Human Primate, and Murine Class II MHC-Peptide Binding Assays Established Using Purified MHC Molecules and Radiolabeled Ligands: Conditions for Class II Assays and MHC Capture Steps
| Assay conditions |
Capture conditions |
|||||||
|---|---|---|---|---|---|---|---|---|
| Organism | Allele(s) | pH | NEM | Detergenta | Temp. | Incubation (hr) | Antibody | Time (hr) |
| Human | DPB1*0101 | 5.5 | Yes | 1.6% NP40 | RT | 72 | B7/21 | 24 |
| DPB1*0201 | 5.5 | Yes | 1.6% NP40 | RT | 48 | B7/21 | 24 | |
| DPB1*0301 | 7 | Yes | 1.6% NP40 | 37°C | 72 | B7/21 | 24 | |
| DPB1*0401 | 7 | Yes | 1.6% NP40 | RT | 48 | B7/21 | 24 | |
| DPB1*0402 | 7 | Yes | 1.6% NP40 | 37°C | 72 | B7/21 | 24 | |
| DPB1*0501 | 5.5 | Yes | 1.6% NP40 | 37°C | 72 | B7/21 | 24 | |
| DPB1*1401 | 7 | Yes | 1.6% NP40 | 37°C | 72 | B7/21 | 24 | |
| DPB1*2001 | 7 | Yes | 1.6% NP40 | RT | 72 | B7/21 | 24 | |
| DQA1*0501/B1*0201 | 5.5 | No | 0.82%Pluronic | 37°C | 72 | HB180 | 24 | |
| DQA1*0201/B1*0202 | 5 | No | 0.82% Pluronic | 37°C | 72 | HB180 | 24 | |
| DQA1*0301/B1*0301 | 7 | Yes | 0.82% Pluronic | RT | 72 | HB180 | 24 | |
| DQA1*0501/B1*0301 | 7 | Yes | 0.82% Pluronic | 37°C | 72 | HB180 | 24 | |
| DQA1*0505/B1*0301 | 7 | Yes | 0.82% Pluronic | 37°C | 72 | HB180 | 24 | |
| DQA1*0301/B1*0302 | 5 | Yes | 0.82% Pluronic | 37°C | 48 | HB180 | 24 | |
| DQA1*0401/B1*0402 | 5 | No | 0.82% Pluronic | 37°C | 72 | HB180 | 24 | |
| DQA1*0101/B1*0501 | 7 | No | 0.82% Pluronic | 37°C | 72 | HB180 | 24 | |
| DQA1*0102/B1*0502 | 7 | No | 0.82% Pluronic | 37°C | 72 | HB180 | 24 | |
| DQA1*0104/B1*0503 | 7 | No | 0.82% Pluronic | 37°C | 72 | HB180 | 24 | |
| DQA1*0102/B1*0602 | 5.5 | No | 0.82% Pluronic | 37°C | 72 | HB180 | 24 | |
| DQA1*0103/B1*0603 | 4.5 | No | 0.82% Pluronic | 37°C | 72 | HB180 | 24 | |
| DRB1*0101 | 7 | Yes | 1.6% NP40 | RT | 48 | LB3.1 | 3 | |
| DRB1*0301 | 4.5 | Yes | 1.6% NP40 | RT | 48 | LB3.1 | 3 | |
| DRB1*0401 | 7 | No | 1.6% NP40 | RT | 48 | LB3.1 | 3 | |
| DRB1*0404 | 7 | No | 1.6% NP40 | RT | 48 | LB3.1 | 3 | |
| DRB1*0405 | 7 | No | 1.6% NP40 | RT | 48 | LB3.1 | 3 | |
| DRB1*0701 | 7 | Yes | 1.6% NP40 | RT | 48 | LB3.1 | 3 | |
| DRB1*0802 | 7 | Yes | 1.6% NP40 | RT | 48 | LB3.1 | 3 | |
| DRB1*0901 | 7 | Yes | 1.6% NP40 | RT | 48 | LB3.1 | 3 | |
| DRB1*1001 | 7 | Yes | 1.6% NP40 | RT | 48 | LB3.1 | 3 | |
| DRB1*1101 | 7 | Yes | 1.6% NP40 | RT | 48 | LB3.1 | 3 | |
| DRB1*1201 | 7 | Yes | 1.6% NP40 | 37°C | 48 | LB3.1 | 3 | |
| DRB1*1302 | 7 | Yes | 1.6% NP40 | RT | 48 | LB3.1 | 3 | |
| DRB1*1501 | 7 | No | 1.6% NP40 | RT | 48 | LB3.1 | 3 | |
| DRB1*1602 | 7 | Yes | 1.6% NP40 | RT | 48 | LB3.1 | 3 | |
| DRB3*0101 | 7 | No | 1.6% NP40 | RT | 48 | LB3.1 | 3 | |
| DRB3*0202 | 7 | Yes | 1.6% NP40 | RT | 48 | LB3.1 | 3 | |
| DRB4*0101 | 5 | No | 1.6% NP40 | RT | 48 | LB3.1 | 3 | |
| DRB5*0101 | 7 | Yes | 1.6% NP40 | RT | 48 | LB3.1 | 3 | |
| Macaque | Mamu DRBw*20101 | 7 | Yes | 1.6% NP40 | RT | 48 | LB3.1 | 3 |
| Mamu DRB1*0406 | 5.5 | Yes | 1.6% NP40 | RT | 48 | LB3.1 | 3 | |
| Mouse | IAb | 7 | Yes | Digitonin 10% | RT | 48 | Y3JP | 3 |
| IAd | 7 | Yes | Digitonin 10% | RT | 48 | MKD6 | 3 | |
| IAk | 7 | Yes | 1.6% NP40 | RT | 48 | 10.3.6 | 3 | |
| IAs | 7 | Yes | 1.6% NP40 | RT | 48 | Y3JP | 3 | |
| IAu | 7 | Yes | 0.82% Pluronic | RT | 48 | Y3JP | 3 | |
| IEd | 4.5 | No | 0.82% NP40 | RT | 48 | 14.4.4 | 3 | |
| IEk | 5 | Yes | 1.6% NP40 | RT | 48 | 14.4.4 | 3 | |
Indicates the detergent utilized in the protease inhibitor cocktail, as well as added as a 1-μl spike to the reaction mix.
Because some residual proteolytic activity is evident in most preparations of purified MHC, the use of a cocktail of protease inhibitors in each assay is extremely critical. Many protease inhibitors are light-sensitive and very labile, so it is also very important that the cocktail be prepared fresh and used immediately (i.e., within minutes). Failure to use or prepare the protease inhibitor cocktail properly can result in assays with poor specificity and low sensitivity, and can give irreproducible results.
The establishment of an MHC/peptide binding assay, and its subsequent use in determining the MHC binding capacities of peptide ligands, requires sufficient stocks of purified MHC and both labeled and unlabeled peptides. Accordingly, this unit includes protocols for the purification of Class I and Class II MHC molecules by affinity chromatography (see Support Protocol 1) and for the radiolabeling of peptides using the chloramine T method (see Support Protocol 2). Peptides may be synthesized by a number of alternative methods described elsewhere (e.g., UNIT 9.1). The Alternate Protocol describes alterations in the Basic Protocol that are necessary when performing direct binding assays, which are required for (1) selecting appropriate high-affinity, assay-specific, radiolabeled ligands and (2) determining the amount of MHC necessary to yield assays with the highest sensitivity.
After a 2- to 3-day incubation, the bound and unbound radiolabeled species are separated, and their relative amounts are determined. Methods for separation by size-exclusion gelfiltration chromatography or antibody-based MHC capture are described in respective support protocols (see Support Protocols 3 and 4), along with data analysis. Support Protocol 5 describes the preparation of immunoaffinity columns for MHC purification.
DETERMINATION OF PEPTIDE BINDING TO AFFINITY-PURIFIED CLASS I AND CLASS II MHC MOLECULES
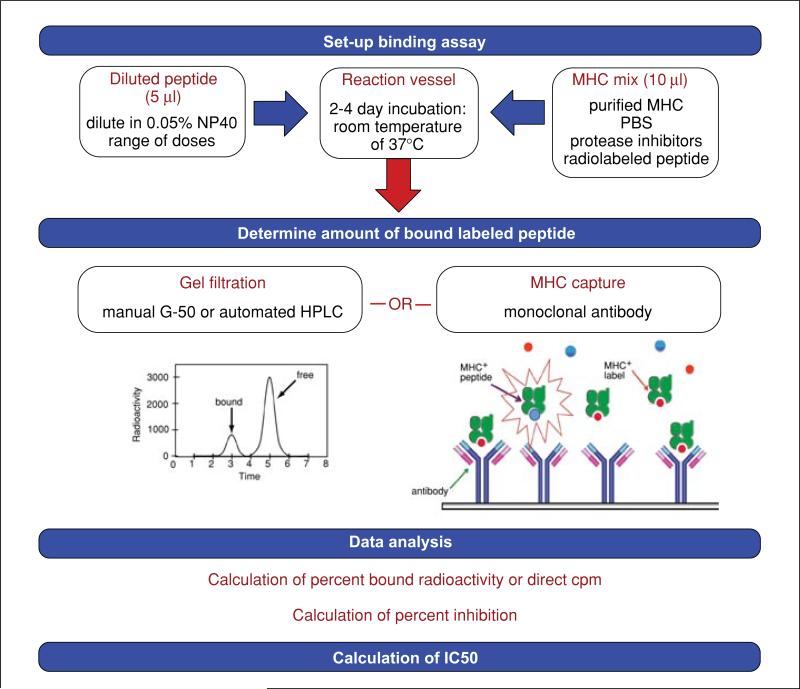
This protocol describes an assay in which the MHC binding capacity of peptides is determined by their ability to inhibit the binding of a high-affinity radiolabeled probe peptide to a specific MHC molecule. This assay is useful for screening synthetic peptides, or other materials, and the procedure can be used, with little variation, for either Class I or Class II MHC purified from a number of different sources (e.g., from human, mouse, or transfected Drosophila cell lines). The Alternate Protocol describes the minor changes required for conducting direct binding assays, which are used to establish binding assay conditions or perform MHC titrations. A flow chart schematizing the assay is presented in Figure 18.3.1.
Figure 18.3.1.
A schematic overview of the steps involved in performing an MHC-peptide binding assay.
With a few exceptions, Class I and Class II assays are largely performed in the same manner. These exceptions include (1) that N-ethylmaleimide (NEM) is not used in the protease inhibitor cocktail for Class I assays and (2) that human β2-microglobulin is included in Class I assays. These differences are also noted where appropriate in the protocol. Throughout the protocol we have indicated vendors for various reagents. These listings are by way of example, and other suppliers of comparable material may be utilized.
Materials
Inhibitor peptides
Phosphate-buffered saline (PBS; APPENDIX 2A), pH 7.2 (Invitrogen)
Dimethylsulfoxide (DMSO)
0.05% (v/v) Nonidet P-40 (NP-40; Fluka)/PBS, pH 7.2
Citrate/phosphate buffer (optional; see recipe)
MHC (see Support Protocol 1 and Alternate Protocol for preparation and titration, respectively)
Protease inhibitor cocktail (prepare at step indicated, not in advance; see recipe)
1 to 3 μM human β2-microglobulin (Class I only; Scripps Laboratories, cat. no. M0114)
1.6% (v/v) NP-40/PBS: PBS, pH 7.2 (Class II only)
0.82% Pluronic in PBS, pH 7.2
10% digitonin in water
Radiolabeled peptide (see Support Protocol 2)
Reaction vessels (e.g., 96-well polypropylene round-bottom plates from Costar, or 12 × 75–mm culture tubes)
Mylar film plate sealer with adhesive backing (ICN Biomedicals) or Costar storage mat III (Corning)
Additional reagents and equipment for gel filtration or MHC capture and analysis (see Support Protocols 3 and 4)
Prepare peptides
- Solubilize lyophilized inhibitor peptides in water, PBS, pH 7.2, or 100% DMSO. Serially dilute peptides to the desired concentrations in 0.05% (v/v) NP-40/PBS.While peptide solubility and stability are optimal in DMSO, MHC assays do not appear to tolerate more than 1% DMSO (Class I) or 5% DMSO (Class II) in the final assay. Therefore, DMSO stocks should be concentrated enough that the DMSO is sufficiently diluted over the range of doses tested. For most applications, 10 to 20 mg/ml is suitable for peptide stocks.
- Load 5 μl of each peptide dose to be tested into a reaction vessel using a micropipettor. For positive (i.e., no inhibitor peptide) and negative (no MHC) controls, load 5 μl 0.05% (v/v) NP-40/PBS in place of inhibitor peptide.Peptides are tested in a final assay volume of 15 μl, which contains inhibitor peptide, MHC, labeled peptide, and a protease inhibitor cocktail. If separations are to be performed in an automated gel filtration system (see Support Protocol 3) or capture-based system (see Support Protocol 4), the reaction is performed in 96-well polypropylene plates or other format as required by the specific system configuration. For manual separation by gel filtration, the reaction may be performed in 12 × 75–mm culture tubes, snap-cap vials, or other similar vessels, depending on the configuration of the separation scheme and radiodetection method utilized.It is recommended that a full titration of a standard reference peptide (typically unlabeled probe peptide) also be included. Examples of high-affinity standard peptides for various Class I and Class II assays may be found in Tables 18.3.1, 18.3.2, and 18.3.3.
Prepare MHC/labeled peptide reaction mix
-
3Prepare a mix of the remaining ingredients. Prepare sufficient reaction mix for all data points, scaling the ingredients for a total volume of 10 μl per well/data point. Take care to add ingredients in the following order, and to prepare the protease inhibitor cocktail immediately before adding MHC to PBS.
- PBS, pH 7.2: Enough to bring volume to 10 μl (for experiments to be performed at a pH other than 7.0, substitute 6 μl PBS with 6 μl citrate/phosphate buffer at a pH 0.5 points below the desired final pH in the mix).
- MHC: Enough to yield a signal-to-noise ratio (S/N; maximum bound counts/background cpm) of at least 3, but ideally 5 to 10, in the capture-based system, or ~15% binding of labeled peptide, as determined by previous titration (see Alternate Protocol), in the gel-filtration system.Support Protocol 1 describes the purification of MHC from cell lysates. However, the assay described here is fully amenable to MHC purified from other sources, such as the recombinant solubilized MHC produced in a hollow-fiber bioreactor, as described by Hildebrand (Buchli et al., 2004, 2005), or the E. coli expression system described by Buus (Ferre et al., 2003; Justesen et al., 2009; Harndahl et al., 2011).
- 2 μl protease inhibitor cocktail.
- 1 to 3 μl of 1 μM human β2-microglobulin (Class I assays only; in Class II this spike is replaced with 1.6% (v/v) NP-40/PBS, 0.82% Pluronic, or 10% digitonin; see Table 18.3.3).
- Radiolabeled peptide: Sufficient amount to give ~40,000 counts per well in the gel-filtration system, or ~8500 cpm in the capture system for Class I, and 15,000 for Class II.An example of a mix “recipe” is given in Table 18.3.4. Mix proportions should be calculated prior to beginning an assay setup. Note that the proportions of MHC, radiolabeled peptide, and PBS will vary for each assay, depending upon the activity of the MHC preparation and specific activity of the radiolabeled peptide.A small amount of mix is typically irretrievable. Therefore, it is prudent to prepare a small (10%) excess of mix to avoid running short.Protease inhibitor cocktail must be used immediately, ideally within 2 min of preparation. Therefore, it should be prepared when indicated above, and the reaction mix should be added to wells (step 4) immediately upon its completion.Examples of assay-specific peptides that may be radiolabeled are given in Tables 18.3.1, 18.3.2, 18.3.3.
Table 18.3.4.
Sample Reaction Mix Recipe
| Ingredient | Vol (ml) per 10 ml | Vol (ml) for 100 samples | Vol (ml) with 10% excess |
|---|---|---|---|
| PBS | 5.5 | 550 | 605 |
| MHC | 1.0 | 100 | 110 |
| Protease inhibitor cocktail | 2.0 | 200 | 220 |
| β2-microglobulin | 1.0 | 100 | 110 |
| Labeled peptide | 0.5 | 50 | 55 |
| Total | 10 | 1000 | 1100 |
Perform binding reaction
-
4Immediately add 10 μl reaction mix to all but the negative control well(s). For negative controls, add 2 μl protease inhibitor cocktail, 1 μl of 1 μM human β2-microglobulin (in Class I assays only), the appropriate amount of radiolabeled peptide, and enough PBS, pH 7.2, to bring the final volume to 15 μl.The mix may be loaded from a reagent reservoir by multichannel pipettor if 96-well plates are used.
-
5Seal the reaction vessel to prevent evaporation. Use mylar film plate sealer or Costar mats (preferable) to seal 96-well plates; tightly seal tubes or close with stoppers.To avoid problems with evaporation in the outer wells of the plate, the mylar should be trimmed neatly along the edge of the plate, and then taped down with general-purpose laboratory tape.
-
6Incubate 2 days (for most assays) in the dark at room temperature or 37°C moist incubator (assay dependent).Kinetic studies have shown that, under the conditions described here, peptide binding to Class I, and most Class II, molecules generally begins to plateau after ~36 hr (e.g., Buus et al., 1986; Sette et al., 1992). Accordingly, all Class I, and most Class II assays are performed at pH 7, at room temperature, with a 48-hr incubation. However, some Class II assays do require longer incubation times and somewhat different conditions. Optimal conditions for each Class II assay, from our experience, are summarized in Table 18.3.3. Gel filtration or Ab capture can begin after the minimum incubation time. Once formed, peptide-MHC complexes are relatively stable, and assays can still be analyzed after a 60- to 72-hr incubation.
Separate unbound peptide from peptide-MHC complexes
-
7Separation of peptide-MHC complexes from unbound peptide can be achieved using two distinct platforms:
- Gel-filtration chromatography.Two procedures for gel-filtration separation are described (see Support Protocol 3). Other alternatives (e.g., spin columns) may be used. Data analysis is also discussed in Support Protocol 3.Alternatively, Class II assay plates may be frozen (–20°C) and analyzed at a later time.
- Capture assay utilizing monoclonal antibody-coated plates.A detailed procedure for capture-based separation is described in Support Protocol 4.
MHC PURIFICATION
Establishing a binding assay requires appropriate reagents and protocols for production and isolation of purified MHC molecules. The procedure outlined below for the affinity purification of MHC can be used for the isolation of both Class I and Class II molecules. It has been used to purify Class I and Class II MHC from mouse, human, and primate origin, as well as from transfected Drosophila cells (Buus et al., 1986, 1987, 1988; O'Sullivan et al., 1990; Sette et al., 1994). Regardless of the source, it does not appear to be necessary to specifically generate empty MHC molecules, or to copurify accessory molecules such as the molecule that catalyzes MHC-II peptide loading in endosomal/lysosomal compartments. Scatchard analysis of both Class I (Olsen et al., 1994; Sette et al., 1994) and Class II (Sette et al., 1992) assay systems indicate that, in most cases, a sufficient pool of active receptor, ranging between 2% and 20% of MHC present, is available for peptide binding.
Materials
Cell line(s): examples include Epstein-Barr virus (EBV)–transformed human B cell lines; mouse B cell lymphomas or mastocytomas; singly transfected fibroblast, C1R, or 721.221 lines; or Drosophila cells (see Tables 18.3.1, 18.3.2, and 18.3.3 for specific lines that have been used). Cells should be checked for MHC expression prior to purification (or at harvest when freezing for later use).
Complete RPMI-10 (APPENDIX 2A)
Phosphate-buffered saline (PBS; APPENDIX 2A), pH 7.4
Lysis buffer (see recipe), ice cold
Washing buffer: 10 mM Tris·Cl, pH 8.0 (APPENDIX 2A) with 1% Nonidet P-40 (store up to 6 months at 4°C)
0.4% (w/v) octylglucoside in PBS
Elution buffer (see recipe)
2 M Tris·Cl, pH 6.8 (APPENDIX 2A)
2.5 M glycine, pH 2.5
225-cm2 tissue culture flasks or roller bottle apparatus
Refrigerated centrifuge
0.8-μm filter
Columns (50-ml borosilicate glass containing one each of the following): inactivated Sepharose CL-4B (10-ml bed volume), protein A–Sepharose CL-4B (5-ml bed volume), and protein A-Sepharose CL-4B conjugated to the appropriate anti-MHC antibody (Table 18.3.5; 10-ml bed volume; see Support Protocol 5 for conjugation)
Centriprep-30 concentrator (Amicon)
Additional reagents and equipment for counting cells (APPENDIX 3A)
Table 18.3.5.
Monoclonal Antibodies Used in MHC Purification or Capture
| Monoclonal antibody | Specificity | Sourcea |
|---|---|---|
| M1/42 | H-2 Class I | ATCC |
| 28-14-8S | H-2 Db and Ld | ATCC |
| 34-5-8S | H-2 Dd | ATCC |
| Y3JP | H-2 IAb,IAs,IAu | Janeway et al. (1994) |
| MKD6 | H-2 IAd | ATCC |
| 10.3.6 | H-2 IAk | ATCC |
| 14.4.4 | H-2 IEd, IEk | ATCC |
| B8-24-3 | H-2 Kb | ATCC |
| Y-3 | H-2 Kb, Kk | ATCC |
| SF1-1.1.1 | H-2 Kd | ATCC |
| B123.2 | HLA B and Cb | Rebai and Malissen (1983) |
| W6/32 | HLA Class I | ATCC |
| HB180 | HLA Class II | ATCC |
| B7/21 | HLA DP | ATCC |
| IVD12 | HLA DQ | ATCC |
| SPVL3 | HLA DQ | Nepom et al. (1996) |
| LB3.1 | HLA DR | ATCC |
ATCC, American Type Culture Collection.
The B123.2 antibody will also bind some HLA A molecules. To date, we have identified HLA A*2301, A*2601, A*2902, A*3001, A*3002, A*3101, A*3201 and A*3301 as B123.2 reactive.
We would presume that corresponding subtypes are also reactive.
Prepare lysate
- Grow cell line(s) in complete RPMI-10 in 225-cm2 tissue culture flasks or, for large-scale cultures, in roller bottle apparatus. Centrifuge cells 10 min at 500 to 800 × g, 4°C, and wash three times with PBS 7.4. Count cells with a hemacytometer or Coulter counter (APPENDIX 3A), and pellet ~1010 cells.Drosophila cells require Schneider medium (Invitrogen) supplemented with 10% FBS and 500 μg/ml Geneticin (Invitrogen).
Lyse cells by adding 100 ml ice-cold lysis buffer (i.e., 10 ml lysis buffer and 100 μl 200 mM PMSF per 1 ml pellet; see Reagents and Solutions) and incubating 30 min at 4°C.
Clear nuclei and other debris by centrifuging 30 min at 15,000 × g, 4°C. Decant the lysate and filter through a 0.8-μm filter. Store the lysate at 4°C or continue directly with the purification.
Purify MHCs
-
4Pass lysate by gravity flow twice through two 50-ml precolumns: an inactivated Sepharose CL-4B column (10-ml bed volume) followed by a protein-A Sepharose-CL-4B column (5-ml bed volume).Both of these columns are stable for ~6 months to 1 year, depending on usage. Prior to use, these columns should be stripped with elution buffer, and then neutralized with washing buffer.
-
5Capture MHC by two passages on a protein A–Sepharose CL-4B column conjugated with the appropriate anti-MHC antibody (10-ml bed volume).Monoclonal antibodies relevant for human and mouse Class I and Class II molecules, and their specificities, are listed in Table 18.3.5. Depending on the antibodies used, it may be necessary to cascade a set of columns of differing antibody specificity to capture the appropriate MHC molecule. For example, the purification of HLA-DR molecules may be performed by removing HLA-A, -B, and -C molecules from the lysate by passage over a W6/32 (anti-HLA-A, -B, and -C) column, and then capturing DR molecules with an LB3.1 (anti-HLA-DR alpha) column.
-
6
Wash column by gravity flow with 4 column volumes (200 ml) of washing buffer.
-
7
Wash column with 50 ml PBS/0.4% octylglucoside. Allow wash to proceed to dryness.
-
8Elute MHC from the column with 50 ml elution buffer. Immediately neutralize the eluate with 2 M Tris·Cl, pH 6.8, for Class I MHC or 2 M glycine, pH 2.5, for Class II MHC, until pH 7 to 7.5 is reached.The column should also be immediately neutralized using washing buffer.
-
9Concentrate MHC preparation to a final volume of ~500 μl in a Centriprep-30 concentrator, according to manufacturer's specifications.Protein content may be evaluated using a BCA protein assay kit (Pierce) and confirmed by SDS-PAGE (UNIT 8.4). MHC preparations should be stored at 4°C. Alternatively, they may be diluted 50% with glycerol, and stored at –20°C. Stability in storage varies for different preparations and cell lines. Most preparations are stable for years at 4°C; others degrade within weeks and must be kept at –20°C.
RADIOLABELING OF PEPTIDES BY THE CHLORAMINE T METHOD
Peptides to be used as radiolabeled probes are iodinated using the chloramine T method (UNIT 8.11; Greenwood et al., 1963; Bolton and Hunter, 1986).
Materials
Tyrosinated peptide (10 to 20 mg/ml)
Phosphate-buffered saline (PBS), pH 7.4 (APPENDIX 2A) with and without 0.05% Tween 20 (Sigma)
~40 μM [Na125]I (~100 μCi/μl; NEN Life Sciences, Perkin Elmer)
0.1 mg/ml chloramine T (Sigma) in PBS/0.05% Tween 20
0.1 mg/ml sodium metabisulfite (Fisher) in PBS/Tween
10% (w/v) sodium azide
Ethanol
0.82% NP-40
Sephadex G-10 column: multispin separation kits (Genesee Scientific) with a 0.8 ml bed volume suspended in PBS, pH 7.2
Microcentrifuge: Labnet International Spectrafuge 16M (cat. no. C0160-R; http://www.labnetinternational.com/)
Polyethylene storage vessel (e.g., 1.5-ml microcentrifuge tubes)
Label peptide
Dilute tyrosinated peptide at 1:600 for Class II, or 1:1200 for Class I, in PBS/0.05% Tween 20.
Decant excess PBS from a G-10 filled multispin column.
- Incubate 12.5 μl diluted peptide sequentially, for 60 sec per step, with each of the following:
- 100 μCi [Na125]I (~1 μl)
- 10 μl 0.1 mg/ml chloramine T (the reaction catalyst)
- 10 μl 0.1 mg/ml of sodium metabisulfite (to quench the reaction).That is, the peptide is incubated for 60 sec in the presence of Na[125I], and then, to catalyze the reaction, for an additional 60 sec after adding chloramine T. Lastly, to quench the reaction, the peptide is incubated for a final 60 sec in the presence of sodium metabisulfate. The radiolabeled peptides should be as hot as possible. Labels of low specific activity require the use of high amounts of radiolabeled peptide in the binding assay to achieve a significant signal. However, the use of too large an excess of peptide reduces the sensitivity of the assay substantially (see Troubleshooting, discussion of radiolabeled peptide). The proportions given here yield labeled peptide preparations of suitable specific activity for the binding assay; typically, between 0.01 and 0.1 μl of labeled peptide should be sufficient to give 10,000 counts per well.
Dilute the quenched reaction with 25 μl PBS/0.05% Tween 20.
- Separate labeled peptide from free iodine by passing the reaction over the G-10 multispin columns. Layer the newly labeled peptide on the G-10 beads so as not to disturb the bed, and spin in centrifuge for 2 min at 1 speed (see the note below). Add 50 μl PBS, pH 7.4, to the top of the spin column×and centrifuge for another 2 min at 1× speed. Next add 150 μl of PBS, 7.4, and centrifuge for an additional 2 min at 1× speed. Then, in a last rinse, add 75 μl PBS, pH 7.4, and centrifuge again for 2 min. Collect the labeled peptide in a 1.5-ml microcentrifuge tube containing 5 μl of 10% NaN3, 10 μl ethanol (as a free radical scavenger), and 25 μl 0.82% NP40/PBS. Store at 4°C.The centrifugal force of the specific microcentrifuge that we utilize (Labnet International Spectrafuge 16M, catalog #C0160-R) is 82 times the force of gravity when the centrifuge dial is set at its 1× speed. Because each make and model of microcentrifuge will be different in terms of rotor size and weight, etc., the appropriate speed adjustment necessary to obtain force appropriate to allow clear separation of free and bound iodine must be determined empirically for each specific instrument.It is crucial that these settings be optimized to enable consistent and sufficient separation of the radiolabeled peptide from the later-eluting free iodine. Besides ensuring collection of the maximal amount of radiolabeled peptide, establishing the optimal times for separation ensures that radiolabeled peptide preparations are free from contaminating amounts of free iodine, the presence of which would represent a major safety risk outside of an appropriate chemical hood.The length of time that a peptide remains usable as a label is dependent on the particular peptide. Typically, a label remains very active for 2 to 3 weeks. The integrity of a label may be checked periodically by HPLC. If any sign of degradation is detected, the peptide should be discarded, even if binding is observed. Otherwise, the quantitative aspects of the peptide-MHC assay could be seriously affected.
DIRECT BINDING ASSAYS TO IDENTIFY APPROPRIATE HIGH-AFFINITY LIGANDS
In the establishment of a binding assay, it is necessary to identify an appropriate high-affinity ligand that can be used as the radiolabeled probe in subsequent inhibition assays. A high-affinity ligand not only ensures that the assay will be as sensitive as possible, but also allows the most efficient use of a purified MHC preparation. The importance of identifying a high-affinity ligand cannot be overstated.
Additionally, it is necessary to perform complete titrations of MHC preparations. These titrations reveal the appropriate concentration of MHC for use in inhibition assays, which is also a critical factor in determining assay sensitivity. A high MHC concentration results in poor assay sensitivity, while a low concentration yields a signal that is difficult to distinguish from background. Under conditions where [label] < [MHC], and IC50 ≥ [MHC], measured IC50 values are reasonable approximations of true Kd values. With typical MHC preparations, these conditions are achieved when the percent bound radioactivity is ~15%, or with a signal to noise ratio of 5 to 10.
The identification of an appropriate high-affinity ligand and the determination of the optimal MHC concentration are both done using direct binding assays. These assays are performed essentially as described in the Basic Protocol, with the following variations at the indicated steps. MHC titrations and ligand screening can be performed simultaneously by combining these steps.
For materials, see Basic Protocol.
For MHC titrations
-
2a
Rather than loading 5 μl of an inhibitor peptide (see Basic Protocol, step 2), add 5 μl of various concentrations of MHC to the assay well. As in the Basic Protocol, load the negative control (no MHC) wells with 5 μl PBS/0.05% (v/v) NP-40.
-
3a
Omit MHC from the 10 μl reaction mix (see Basic Protocol, step 3), replacing it with an equal volume of PBS or citrate/phosphate buffer.
For screening ligands
-
3b
Perform multiple binding assays using a separate reaction mix (see Basic Protocol, step 3) for each labeled peptide.
SEPARATION OF MHC-PEPTIDE COMPLEXES BY SIZE-EXCLUSION GEL-FILTRATION CHROMATOGRAPHY
Filtration
The use of HPLC systems capable of automated loading, separation, radio-detection, and data analysis of samples following incubation offers the potential to analyze binding events literally around the clock. As a result, hundreds of data points may be generated daily. For automated separation, use a TosoHaas QC-PAK TSK GFC200 column (7.8 mm × 15 cm) with a particle size of 5 μm. Use PBS, pH 6.5 (APPENDIX 2A), containing 0.5% (v/v) Nonidet P-40 (NP-40) and 0.1% (w/v) sodium azide as the eluent, at a flow rate of 1.2 ml/min. In this configuration, one sample can be analyzed every 7 to 9 min, depending on the age of the column. Set up the HPLC hardware configuration to include an integrator (e.g., Hewlett-Packard 3396A), a disc drive (e.g., Hewlett-Packard 9114B), a dilutor (e.g., Gilson 401), a sample injector (e.g., Gilson 231), a radioisotope detector (e.g., Beckman 170), and a solvent delivery module (pump; e.g., Beckman 110B).
Alternatively, samples may be separated manually by gravity flow over medium-grade Sephadex G-50 (Buus et al., 1986; O'sullivan et al., 1990), or with spin columns (Boyd et al., 1992; Olsen et al., 1994). For Sephadex G-50 separation on a 22.5 × 1.5–cm borosilicate column, elute with PBS, pH 7.0 (APPENDIX 2A) containing 0.5% (v/v) NP-40 and 0.1% (w/v) sodium azide. Collect 1-ml fractions and determine the amount of radioactivity per fraction with a standard gamma counting apparatus. Although this method cannot achieve the high throughput of an automated system, its low cost may be attractive in situations where only a limited number of samples need to be analyzed.
Analysis
Chromatographic separation typically results in the identification of two prominent peaks, representing peptide-MHC complexes and free peptide, respectively (Fig. 18.3.1). Calculate percent binding, which is equal to the percent area of the first peak relative to the total area integrated:
Calculate IC50 values for each test peptide by plotting dose versus percent inhibition, where:
Use this plot to extrapolate the dosage yielding 50% inhibition (IC50). Assay sensitivity may vary somewhat from day to day, or between batches of purified MHC. To compare data obtained in different experiments and with different batches of MHC, normalize binding values for each peptide to an assay-specific positive control for inhibition (i.e., the assay standard peptide; see Basic Protocol, step 2). These relative binding values represent the ratio of the IC50 of the positive control for inhibition to the IC50 of the test peptide:
Relative binding values appear to be the most accurate and consistent for comparing peptides that have been tested on different days or with different lots of purified MHC. To convert back into nM IC50 values, divide the nM IC50 of the positive controls for inhibition by the relative binding of the peptide of interest.
Affinity/rate constant information may be obtained directly using the direct binding assay (see Alternate Protocol). In this case, multiple tubes/wells are set up, and % binding is determined at multiple time points starting from t = 0. Constants may then be calculated as described in any basic biochemistry text.
SEPARATION OF MHC-PEPTIDE COMPLEXES BY ANTIBODY-BASED MHC CAPTURE
Capture Assay
As a more efficient, higher-throughput alternative to the gel-filtration-based separation protocol described in Support Protocol 3, MHC-peptide complexes can be separated from unbound peptide using monoclonal antibody capture. In this approach, monoclonal antibodies specific for various MHC types are coated onto the wells of 96-well microplates. After an incubation phase to maximize MHC capture, the plates are washed and the amount of bound radioactivity is determined with a microscintillation counter.
The specific approach described here has been developed to use 96-well white polystyrene microtiter plates specifically designed for high-volume, in-plate, radiometric assays. Measurement of the 125I-labeled peptide bound to MHC is accomplished using the Topcount (PerkinElmer Instruments) benchtop microplate scintillation and luminescence counter, although other system configurations are feasible.
Capture assays are performed essentially as described in the Basic Protocol. In this section, however, we have provided additional details, including some variations, facilitating the use of the capture assay for the purpose of high-throughput screening. Accordingly, the present section details dilution and handling of peptides for compatibility with the 96-well format, preparation of antibody-coated plates, transfer of assay samples for the MHC-capture phase, and then final washing and counting of the plates. All of the steps described here can be performed with standard 8- or 12-channel pipettors, and most can take advantage of the high capacity of instrumentation such as automated plate washers and other liquid-handling devices. As described here, the protocol can essentially permit the analysis of about 150 plates, or 3600 samples, per Topcount instrument, per day.
Additional Materials (also see Basic Protocol)
Anti-MHC monoclonal antibody (see first annotation to step 9): 30 μg/ml in 0.1 M Tris·Cl, pH 8.0 (APPENDIX 2A; the same antibodies used for the affinity columns for MHC purification are used in the capture assay)
Blocking solution: 0.3% (v/v) Tween 20 in PBS or 1% (w/v) BSA in PBS
96-well round-bottom polystyrene microtiter plate (Greiner-bio-one, cat. no. 650201, http://www.greinerbioone.com/en/start/)
Mylar plate sealer with adhesive back (MP Biomedicals, LLC, cat. no. 76-402-05)
View Seal (Greiner-bio-one, cat. no. 676070, http://www.greinerbioone.com/en/start/)
Costar Sealing Mat (Corning, cat. no. 3080)
96-well flat-bottom white polystyrene Optiplate (Greiner-bio-one, cat. no. 655074, http://www.greinerbioone.com/en/start/)
TopSeal-A for 96-well microplates (PerkinElmer, cat. no. 6005185)
Microscint-20 (PerkinElmer; Cat. #6013621)
Topcount microscintillation counter (Perkin-Elmer Instruments)
Basic capture assay setup
Dilution of inhibitor peptides
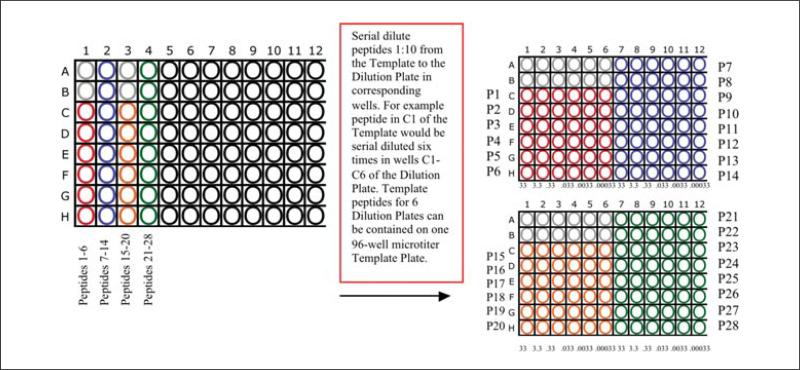
- Inhibitor peptides are first diluted 1:10 in a “Template Plate” (Fig. 18.3.2). Peptides then undergo six subsequent 1:10 serial dilutions in a “Dilution Plate.” Typically, the six serial dilutions in the “Dilution Plate” range from 100 μg/ml to 300 pg/ml. The six serial dilutions from the “Dilution Plate” are transferred at 5 μl/well into the “Assay Plate.” Since the final assay volume is 15 μl, this represents an assay range of 33 μg to 0.33 ng.
- Dilute inhibitor peptides 1:10 in 96-well round-bottom polystyrene microtiter plates (“Template Plate”) using 0.05% NP-40. Typically, 5.5 μl of 10 mg/ml peptide is diluted into 50 μl 0.05% NP-40, although larger (or smaller) volumes may be utilized if necessary.
- Further dilute inhibitor peptides in 0.05% NP-40 in additional 96-well round bottom polystyrene microtiter plates (“Dilution Plate”) six times in 10-fold serial dilutions.
- The “Dilution Plate” is typically laid out with peptide 1 in C1 to C6, peptide 2 in D1 to D6, etc., with peptides 7 to 12 in A7 to A12, B7 to B12, etc. Wells A1 to A6 and B1 to B6 are left blank such that standard and control peptides may be added to the “Assay Plate.”
- High-affinity peptides, such as those used as the assay standard (i.e., a positive control), may require additional dilution to ensure that an appropriate titration range is achieved to cover from 100% to 0% inhibition.
- Seal the “Template Plate” and the “Dilution Plate” with plate sealer (mylar or mat) for storage. Plates may be stored at 4°C for up to 4 weeks.
Figure 18.3.2.
Example layout for serial dilution of inhibitor peptides amenable to high throughput screening.
Assay plate set-up
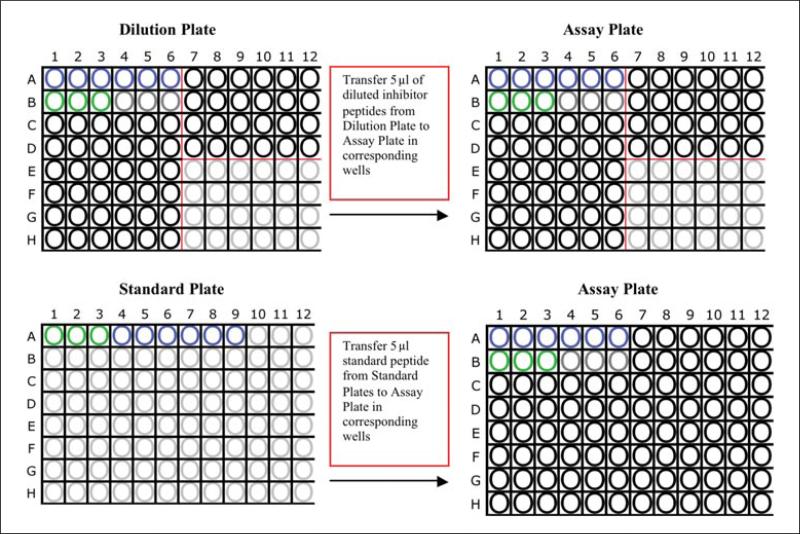
-
2Transfer 5 μl from each well of the “Dilution Plate” to the corresponding well in the “Assay Plate” (Fig. 18.3.3). For replicates, this may be done in triplicates, or other replicates.
- Load 5 μl of the six lower dilutions of standard peptide to the “Assay Plate” from the “Standard Plate” in wells A1 to A6.
- Load 5 μl of “high-concentration” standard peptide to wells B1 to B3. These wells are negative (background) control wells. “Cold Mix” is also loaded into these wells (see below).
- Wells B4 to B6 are not loaded with peptide as they are the no-inhibitor positive control wells. Only “Hot Mix” (see below) and 5 μl PBS NP-40 is loaded into these wells.
Figure 18.3.3.
Example layout for setting-up assays plates to screen serial dilutions covering a 6-log dose range.
Prepare radiolabeled peptide/MHC mix (Hot Mix)
-
3
The radiolabeled peptide/MHC mix is prepared as described in the Basic Protocol. Dispense the radiolabeled peptide/MHC mix at 10 μl/well into the “Assay Plate” containing the unlabeled inhibitor peptides. Do not add “Hot Mix” to the negative (background) control wells, B1 to B3.
Prepare protease inhibitors (PI Mix)
-
4
Protease inhibitors are prepared as described in Reagents and Solutions.
Plate out controls
-
5Add the No Inhibitor (+) into wells B4 to B6 of the 96-well round-bottom “Assay Plate.” Alternatively, these wells may be loaded when loading the inhibitor peptide wells. The no-inhibitor control contains the following ingredients.
- 10 μl/well “Hot Mix” (radiolabeled peptide/MHC mix).
- 5 μl/well 0.05% NP-40.
-
6Add the Cold (–) (Background) Control into wells B1 to B3 of the 96-well round bottom “Assay Plate.” This mix is the same as the Hot Mix, but without MHC. Enough mix should be prepared sufficient for all of the “Assay Plates.” These wells contain:
- 5 μl “high concentration” unlabeled high-affinity peptide.
- 10 μl “Cold Mix,” composed of:
- 2 μl protease inhibitor cocktail.
- X μl 125I-labeled peptide (enough to provide assay specific optimal cpm).
- Spike: 0.1 μl or 0.3 μl β2 microglobulin at 2 mg/ml or 1 μl detergent, as in “Hot Mix.”
- 6 μl pH buffer if other than pH 7.2 is used.
- X μl PBS, pH 7.2 (adjust volume for a total of 10 μl).In general, Class I assays have optimal sensitivity at 8,000 to 10,000 cpm of input radioactivity. Class II assay have optimal sensitivity at about 15,000 cpm, although in some cases higher counts (up to about 40,000 cpm) may be necessary.
Seal and incubate plates
-
7
Seal plates with Greiner bio-one View Seal. Alternatively, plates may also be sealed with a Costar Sealing Mat applied with a Costar Sealing Press. These sealing mats may be recycled once for room temperature assays (assays incubated at 37°C must be sealed with a new sealing mat).
-
8
Incubate plates in the dark, at room temperature or 37°C, for 48 to 120 hr. All Class I assays are incubated for 48 hr at room temperature. For Class II, see Tables 18.3.3 for assay-specific temperature and incubation times.
Coat capture plates
-
9Coat antibody capture plate(s) (Optiplates) with 125 μl/well of anti-MHC antibody.Anti-MHC antibodies, such as W6/32 or L243 (see Table 18.3.5), are common reagents available from a wide variety of commercial vendors (e.g., OneLambda, eBioscience, etc.). However, such sourcing can be prohibitively expensive, given the quantity of antibody required. Alternatively, it is recommended that antibodies be purified from hybridomas (see Table 18.3.5) following standard methods (see, e.g., Bonifacino et al., 2013, Chapter 16). Purification from hybridomas on a large scale can also be performed under contract from various vendors, such as Strategic Biosolutions (http://www.sdix.com/) or Maine Biotechnology Services, Inc. (http://www.mainebiotechnology.com/).Antibody stocks must be kept frozen until needed. Thawed aliquots of purified antibody are added to 0.1 M Tris·Cl, pH 8.0, to make a working antibody solution with a final concentration of 30 μg/ml. Because the working solution is at 30 μg/ml, each well is effectively coated with 3.75 μg of antibody.Plates are incubated at room temperature, in the dark, for 24 hr. With the exception of B*4402 and B*4403, which are captured with the B123.2 antibody, all primate Class I assays are captured using the W6/32 antibody. Mouse Class I assays are captured using the antibodies as listed in Table 18.3.5. See Table 18.3.2 to determine the specific antibody used to capture a specific Class II MHC.
Block coated plates
-
10
Antibody solution is removed from the 96-well flat-bottom Optiplate and discarded, or collected for recycling (retrieved antibody solution may be used for one additional coating).
-
11Blocking solution (0.3% Tween 20 /PBS or 1% BSA/PBS) is added to each well, at 180 μl for short-term storage, or 200 μl/well for long-term storage. Typically, Class I antibody capture plates are blocked with 0.3% Tween 20/PBS and Class II antibody capture plates are blocked with 1% BSA to reduce background counts.96-well flat bottom Optiplates pre-coated with anti-MHC antibody can be used for the capture assay after incubating with blocking buffer at least 3 hr. Plates can be stored up to 5 days at room temperature, or stored for up to 3 months at 4°C with blocking solution remaining in the wells. If stored, plates must be covered with mylar adhesive seals.
Prepare capture plates
-
12Antibody pre-coated and blocked Optiplates (see above) must be prepared before the “Assay Plate” is transferred into them. To prepare the Optiplate:
- Remove blocking buffer.
- Wash plates with 200 μl of 0.05% (v/v) Tween 20/PBS two times.
- Transfer the “Assay Plate” into the antibody plate as soon as possible to prevent the antibody from drying inside the plate.
Transfer binding assay
-
13Peptide/MHC complexes from binding “Assay Plates” are transferred to anti-MHC antibody coated and blocked plates.
- Add 100 μl of 0.05% NP-40/PBS to each well of the 96-well round-bottom microtiter assay plate. The entire plate can be transferred at one time using the Costar 96-well pipettor and the Costar 96-well pipet cartridge.
- Using a 12-channel pipettor set at 125 to 150 μl or a Costar 96-well pipettor, transfer each well from the assay plate to the precoated/blocked and washed antibody plate.
- If using the Costar 96-well pipettor and the Costar 96-well pipet cartridge, change the cartridge after approximately nine plates, or when transferring different alleles.
- The antibody plate is then sealed with a mylar adhesive seal and incubated at room temperature for > 3 hr for Class I and most Class II assays. Some Class II assays are incubated for up to 24 hr (see Table 18.3.3).
Wash and count capture assay plate
-
14
Once the capture step (step 13) is completed, the liquid contents of the plate are discarded into an appropriate radioactive liquid waste container.
-
15
Wash plates with 200 μl of 0.05% Tween 20/PBS two times for Class I assays, and four times for Class II assays.
-
16
Dispense Microscint-20 scintillation fluid at 125 μl to each well, then seal the plates with a TopSeal adhesive cover sheet. Wipe plates with antistatic wipes.
-
17
Radiolabeled peptide binding to MHC is then determined by counting cpm for 1 min for each plate in the TOPCOUNT microscintillation and luminescence counter.
Notes on capture assay variations
Some assays require specific conditions and reagents. Essentially, however, all Class I and most Class II assays are performed at pH 7 and incubated at room temperature for 48 hr, followed by a 3-hr antibody plate capture incubation. However, all HLA-DQ assays, and some DP and DR assays, must be incubated at 37°C for 72 hr, followed by a 24-hr antibody plate capture incubation.
Class I assays always have a β2-microglobulin spike, whereas Class II assays require different detergent spikes. 1.6% NP-40 is generally used for DR and DP assays, while 0.82% Pluronic or 1.6% Pluronic is used for DQ assays. Additionally, some H-2 Class II assays require a 10% digitonin spike. Assays may also differ in their final pH. These assay variations for Class II are indicated in Table 18.3.3.
Analysis
In principle, analysis of data generated from the capture assay is performed essentially as described in Support Protocol 3 for the gel filtration–based assay. The primary difference is that while the chromatographic separation results in the identification the percent of input radioactivity associated with the free peptide and peptide-MHC complex species, the capture assay provides a direct count of cpm associated with radiolabeled peptide bound to the MHC. As with the gel-filtration system, the percent inhibition of binding of the radiolabeled peptide attributed to the specific dose of the input unlabeled inhibitor peptide can be determined by comparison with the positive control wells (i.e., those with no inhibitor peptide added). Then, the IC50 value of a test inhibitor peptide can be determined with a dose-response curve (dose versus percent inhibition).
PREPARATION OF IMMUNOAFFINITY COLUMNS
Anti-MHC antibodies, such as W6/32 or L243 (see Table 18.3.5), are common reagents available from a wide variety of commercial vendors (e.g., OneLambda, eBioscience, etc.). However, such sourcing can be prohibitively expensive, given the quantity of antibody required. Alternatively, it is recommended that antibodies be purified from hybridomas (see Table 18.3.5) following standard methods (see, e.g., Bonifacino et al., 2013, Chapter 16). Purification from hybridomas on a large scale can also be performed under contract from various vendors, such as Strategic Biosolutions (http://www.sdix.com/) or Maine Biotechnology Services, Inc. (http://www.mainebiotechnology.com/). A typical purification column will have a bed volume of 10 ml. Antibody is used at a ratio of 2 to 3 mg per ml of column bed.
Triethanolamine, DMP, and ethanolamine solutions should all be made immediately before use.
Materials
Protein A–Sepharose CL4 B (Sigma, cat. no. P-3391)
Sepharose CL4 B (Sigma, cat. no. CL4B-200; for use in uncoupled pre-columns)
100 mM borate buffer, pH 8.2: dissolve 6.18 g boric acid (Sigma, cat no. B-7660)/9.54 g borax (Sigma, cat. no. B-0127)/4.38 g NaCl in H2O
Monoclonal antibody (approximately 20 to 30 mg; see annotation to step 3) in PBS, pH 7.2 (see below) at a concentration of about 2 mg/ml or higher
PBS, pH 7.2: 20 mM Na2HPO4/150 mM NaCl/0.05% NaN3
200 mM triethanolamine, pH 8.2 (Sigma, cat. no. T-1377)
20 mM dimethyl pimelimidate (DMP; Pierce, cat. no. 21667) in 200 mM triethanolamine, pH 8.2
Phosphate-buffered saline (PBS, Invitrogen, cat. no. 10010-023) containing 0.02% sodium azide (NaN3; Fisher Scientific, cat. no. S227-500)
20 mM ethanolamine pH 8.2 (Sigma, cat. no. E-9508)
0.02% sodium azide (NaN3) (Fisher Scientific, cat. no. S227-500) in PBS, pH 7.2 (Invitrogen, cat. no. 10010-023)
Elution buffer (see recipe)
2 M glycine, pH 2.5
Washing buffer: 10 mM Tris·Cl, pH 8.0 (APPENDIX 2A) with 1% Nonidet P-40 (store up to 6 months at 4°C)
50-ml borosilicate glass column with stopcock
Rotator
Spectrophotometer
- Swell Protein A–Sepharose-CL4B (or Sepharose-CL4B for columns without antibody) in a column with 40 ml of borate buffer for 1 hr, while gently rotating the column.Approximately 2.5 mg of Protein A Sepharose-CL4B is used per 10 ml of column bed. A 10-ml column bed is the most commonly used column.
After swelling, drain, and then wash the column twice with 10 ml borate buffer.
Add 2 to 3 mg of antibody for each ml of bed volume, bring the volume to 40 ml with borate buffer, and incubate for 1 hr at room temperature on a rotator.
Collect the flowthrough and read optical density at 280 nm. If the reading is more than 20% of the initial reading, incubate for another hour.
Wash the column with 50 ml of borate buffer. Collect the flowthrough in 10-ml fractions and measure OD at 280 nm. Continue washing until the reading is less than 0.020.
Wash the column with 20 ml of 200 mM triethanolamine pH 8.2.
- Add 40 ml of 20 mM dimethyl pimelimidate (DMP) in 200 mM triethanolamine and incubate for 45 min at room temperature on a rotator.For this and the remaining steps, always save the column flowthrough in case the antibody did not cross-link.
Wash twice with 10 ml of 20 mM ethanolamine, pH 8.2, leaving the column on the rotator for 5 min between washes.
Wash the column with 200 ml of borate buffer, then with 100 ml PBS containing 0.02% sodium azide.
Take a final OD280 reading of flowthrough to completion of coupling. Store the column upright at 4°C in borate buffer.
To verify the column is not leaking antibody, perform a sham elution by passing 30 ml of elution buffer through the column and collect the flowthrough. Neutralize the flowthrough to pH 7.0 to 8.0 with 2 M glycine, pH 2.5. At the same time, neutralize the column to pH 8.0 using washing buffer, pH 8.0. Concentrate the flowthrough and run a gel to confirm that the column is not leaking antibody (no bands should appear on the gel).
REAGENTS AND SOLUTIONS
Use deionized, distilled water in all recipes and protocol steps. For common stock solutions, see APPENDIX 2A; for suppliers, see APPENDIX 5.
Citrate/phosphate buffer
Prepare stock solutions of 0.1 M citric acid (19.21 g/liter) and 0.2 M dibasic sodium phosphate (e.g., 53.65 g/liter Na2HPO4 ·7H2O). To prepare a working buffer at the desired pH, mix as shown in Table 18.3.6, and dilute to 100 ml with water. Store both stock and working solutions up to 6 months at 4°C.
Table 18.3.6.
Preparation of Citrate/Phosphate Buffer Working Solutions from Stock Solutionsa
| Citric acid (ml) | Phosphate (ml) | pH |
|---|---|---|
| 44.6 | 5.4 | 2.6 |
| 42.2 | 7.8 | 2.8 |
| 39.8 | 10.2 | 3.0 |
| 37.7 | 12.3 | 3.2 |
| 35.9 | 14.1 | 3.4 |
| 33.9 | 16.1 | 3.6 |
| 32.3 | 17.7 | 3.8 |
| 30.7 | 19.3 | 4.0 |
| 29.4 | 20.6 | 4.2 |
| 27.8 | 22.2 | 4.4 |
| 26.7 | 23.3 | 4.6 |
| 25.2 | 24.8 | 4.8 |
| 24.3 | 25.7 | 5.0 |
| 23.3 | 26.7 | 5.2 |
| 22.2 | 27.3 | 5.4 |
| 21.0 | 29.0 | 5.6 |
| 19.7 | 30.3 | 5.8 |
| 17.9 | 32.1 | 6.0 |
| 16.9 | 33.1 | 6.2 |
| 15.4 | 34.6 | 6.4 |
| 13.6 | 36.4 | 6.6 |
| 9.1 | 40.9 | 6.8 |
| 6.5 | 43.6 | 7.0 |
See recipe for citrate/phosphate buffer in Reagents and Solutions.
Elution buffer
0.15 M NaCl
50 mM diethylamine
1% (w/v) octylglucoside
0.02% (w/v) sodium azide
Adjust pH to 11.5
Store up to 6 months at 4°C
Ethylenediaminetetraacetic acid (EDTA), 86 mg/ml
Prepare 86 mg/ml tetrasodium EDTA tetrahydrate (Calbiochem, cat. no. 34103) in PBS (APPENDIX 2A). Warm gently in a water bath if EDTA does not go into solution easily. Adjust pH to 7.0 with 10 N NaOH. Store up to 6 months at –20°C.
Lysis buffer
20 mM Tris·Cl, pH 8.5 (APPENDIX 2A)
1% (v/v) Nonidet P-40 (NP-40; Fluka)
150 mM NaCl
2 mM phenylmethylsulfonyl fluoride (PMSF)
Adjust pH before adding detergent. Store without PMSF up to 6 months at 4°C. Immediately before use, add PMSF from a 40 mg/ml stock in isopropanol (store PMSF up to 6 months at –20°C).
Protease inhibitor cocktail
61 μl PBS/NP-40: PBS, pH 7.2 (APPENDIX 2A) with 0.82% (v/v) Nonidet P-40 (NP-40)
12 μl 8 mM tetrasodium EDTA (tetrasodium ethylenediaminetetraacetic acid tetrahydrate; 86 mg/ml in PBS, pH 7.2; Calbiochem, cat .no. 34103)
12 μl 73 μM pepstatin A (5 mg/ml in methanol; EMD, cat no. 516481)
12 μl 0.8 M N-ethylmaleimide (NEM; 100 mg/ml in isopropanol; Class II assays; Sigma, cat. no. E-1271)
12 μl 1.3 nM 1,10-phenanthroline (26 mg/ml in ethanol; Sigma, cat. no. P-9375)
6 μl 1 mM phenylmethylsulfonyl fluoride (PMSF; 40 mg/ml in isopropanol; Sigma, cat. no. P-7626)
3 μl 200 μM Nα-p-tosyl-L-lysine chloromethyl ketone (TLCK) hydrochloride (20 mg/ml in PBS; Sigma, cat. no. T-7254)
- Prepare fresh, immediately before use (e.g., within 2 min)NEM is not used in Class I assays; replace NEM with 12 μl isopropanol.For murine IA assays only, replace PBS/NP-40 with 61 μl of 20% digitonin (Fluka, Wako) or 61 μl Pluronic (PBS, pH 7.2, with 0.82% (w/v) Pluronic).Stock solutions can be stored up to 6 months at –20°C. It may be necessary to resolubilize the PMSF by gently warming before use.Because of water/alcohol mixing and alcohol evaporation, each batch yields ~100 μl of retrievable cocktail. A single large batch that is sufficient for the entire experiment should be made at the start.
COMMENTARY
Background Information
The induction and functionality of both helper and cytotoxic T cells are dependent upon the formation of a trimolecular complex between antigenic peptides, Class I or Class II MHC molecules, and the antigen-specific TCR of CD4+ or CD8+ T cells. As such, this event represents one of the most crucial elements in the generation of immune responses. Indeed, both the normal functions of the immune response—e.g., induction of delayed-type hypersensitivity (DTH) responses against parasites, induction of cytotoxic T lymphocyte (CTL) responses against tumors and viruses, and induction of specific antibodies against bacteria—as well as pathological reactions—e.g, autoimmunity and allergy—are ultimately dependent on the formation of such trimolecular complexes. To gain insight into the very basis of the functioning of the immune system, it is evident that the accurate and quantitative measurement of the capacity of peptides to bind MHC is essential.
Recognition of the vital role of peptide-MHC complexes in the immune response has generated considerable interest over the years in understanding the biology of peptide-MHC binding (for early, but still very relevant, reviews see, for example, Sette and Grey, 1992; Barber and Parham, 1993; Germain, 1993, 1994; Germain and Margulies, 1993; Engelhard, 1994; Joyce and Nathenson, 1994; Rothbard, 1994; Rötzschke and Falk, 1994; Sinigaglia and Hammer, 1994; Stern and Wiley, 1994; Madden, 1995; Rammensee et al., 1995). X-ray crystallographic structures of dozens of MHC molecules and MHC-peptide complexes have been reported in the literature, and new binding motifs still appear regularly. A considerable amount of data is now also available regarding the processing of peptides and the recognition of peptide-MHC complexes by T cell receptors.
Various methods, encompassing both whole-cell and cell-free systems, have been employed to characterize peptide binding to MHC molecules. Assay systems that are powerful in characterizing MHC peptide binding motifs or in identifying individual bound peptides include the use of phage-display libraries (Hammer et al., 1993), the sequencing of naturally processed peptides, either individually or in pools (van Bleek and Nathenson, 1990; Falk et al., 1991; Harris et al., 1993), and tandem mass spectrometry (Hunt et al., 1992; Cox et al., 1994).
Other assay systems are more amenable to the quantitation of the binding of individual peptides. A number of these systems use live-cell assays (Ceppellini et al., 1989; Busch et al., 1990; Christnick et al., 1991; Hill et al., 1991; del Guercio et al., 1995). Cell-free systems utilize detergent lysates (e.g., Cerundolo et al., 1991), biotinylated and europium-labeled ligand binding to immobilized purified MHC (Hill et al. 1994; Marshall et al., 1994), enzyme-linked immunosorbent assays (ELISAs; Reay et al., 1992; Sylvester-Hrid, 2002, 2004), surface plasmon resonance (Khilko et al., 1993), and a high-flux soluble-phase assay using both radiolabeled peptides and biotinylated antibodies (Hammer et al., 1994). Assays specific for Class I MHC based on stabilization or assembly are also commonly utilized (Ljunggren et al., 1990; Schumacher et al., 1990; Townsend et al., 1990; Parker et al., 1992). Buus and co-workers have developed high-throughput nonradioactive binding assays based on Luminescent Oxygen Channeling (LOCI) for both Class I (Harndahl et al., 2009) and Class II molecules (Justesen et al., 2009), as well as an essentially label-free scintillation proximity assay allowing real-time measurement of peptide-MHC Class I dissociation (Harndahl et al., 2011). Dedier and co-workers, as well as Hildebrand's group, have used fluorescence polarization and solubilized MHC for real-time measurement of peptide binding (Dedier et al., 2001; Buchli et al., 2004, 2005). The assay system described in this unit utilizes 125I-labeled peptide ligands and detergent-solubilized, affinity-purified MHC molecules. Similar procedures using solubilized MHC have also been reported (Roche and Cresswell, 1991; Boyd et al., 1992; Olsen et al., 1994).
Each method, including the one described in this unit, has distinctive advantages and disadvantages. Many of these aspects have been reviewed elsewhere (Engelhard 1994; Joyce and Nathenson, 1994; Hammer, 1995; Rammensee et al., 1995). The procedure described here is recommended because of its overall adaptability and very high sensitivity (typically in the 1 to 5 nM range). The techniques, reagents, and materials required are readily available to most immunology laboratories, and are relatively inexpensive. The method is highly quantifiable and very reproducible, and specifically addresses the capacity of individual peptides to bind MHC, rather than their capacity, for example, to survive processing in a live-cell environment. The method's high-throughput capacity enables screening of large peptide libraries in a reasonable period of time. As a result, detailed peptide binding motifs can be ascertained while simultaneously characterizing the binding of individual peptides. This particular utility has been demonstrated in a number of instances for both Class I (see, for example, Ruppert et al., 1993; Kondo et al., 1995, 1997; Sidney et al., 1996a,b; 2008a; for review see, e.g., Sidney et al., 2008b) and Class II (see, for example, O'sullivan et al., 1990, 1991; Sette et al., 1990, 1993; Sidney et al., 1992, 1994; 2001a,b; Alexander et al., 1994; Geluk et al., 1994, Greenbaum et al., 2011) molecules. Besides the identification of anchor residues, motifs generated by this procedure can also precisely address the facilitative or deleterious effects of specific amino acids at specific secondary positions.
Critical Parameters
Identification of a high-affinity ligand
The identification of a high-affinity ligand that can be radiolabeled and gives a clearly detectable signal is perhaps the single most challenging problem in establishing an MHC binding assay. Until such a ligand is identified and some binding is detected, it is not possible to optimize the experimental conditions and establish a final protocol.
This hurdle may often be bypassed by using peptides restricted to the MHC molecule of interest, or by using natural or engineered peptides known to have degenerate MHC binding (e.g., Sinigaglia et al., 1988; Ceppellini et al., 1989; Panina-Bordignon et al., 1989; Busch et al., 1990; Sette et al., 1990, 1994; Hill et al., 1991; Roche and Cresswell, 1991; Ruppert et al., 1993; Alexander et al., 1994; Kondo et al., 1995; Sidney et al., 1996a,b). In some cases, if the tyrosine residue required for iodination is not present in the parent sequence, it may be necessary to synthesize analogs. In these instances, a tyrosine residue should be substituted for a noncritical residue, if possible. Thus, for Class I ligands, the substitution is typically made at positions other than position 2 and the C-terminal main anchor. For Class II ligands, suitable peptides may be synthesized by adding an N- or C-terminal tyrosine. In both instances, if internal tyrosine residues are present in the natural sequence, analogs in which they have been replaced with phenylalanine should also be synthesized. This precaution reduces the possibility of labeling a potential anchor residue, or of producing doubly labeled peptides.
Sensitivity to size- and allele-specific motifs is also important. It has been widely reported in the literature that Class I ligands are of very specific size, and bear allele-specific motifs (for reviews see Sette and Grey, 1992; Barber and Parham, 1993; Germain, 1993; Germain and Margulies, 1993; Engelhard, 1994; Stern and Wiley, 1994; Madden, 1995; Rammensee et al., 1995). Peptides chosen for use as radiolabeled ligands for Class I assays should strictly adhere to these specifications. For Class II radiolabeled ligands, it has been the authors’ experience that long peptides (i.e., >18 residues in length) are often difficult to use from a separation standpoint, in the context of gel filtration assays, although they may still be suitable in the context of MHC capture assays. Ideally, peptides to be used as potential radiolabeled probes for Class II assays should be between 13 and 18 amino acids in length. Thus, it may be necessary to synthesize truncated analogs of known Class II–binding peptides.
Once a peptide that yields a detectable signal is identified, a larger panel of unlabeled peptides may be screened without having to label large numbers of peptides. From these panels, it may be possible to identify other ligands of even higher affinity, which may then be radiolabeled.
Assay validation
MHC molecules, in some situations, can yield low-affinity and artifactual binding. Thus, in the course of developing peptide-MHC binding assays, it is essential to validate them at both the biochemical and biological levels. Validity at the biochemical level is demonstrated by the specificity of the assay: the interaction should be inhibitable by an excess of unlabeled peptide ligand, and the peptide ligand should be capable of binding to some but not all MHC types. Conversely, it should also be demonstrated that the MHC molecule of interest binds some but not all peptide ligands.
The biological relevance of an assay may be established by assessing the correlation between binding specificity and MHC restriction. In most cases, peptides capable of eliciting a response restricted to a given MHC molecule will bind with relatively high affinity to that MHC molecule. For Class I alleles, this high-affinity binding has a threshold that appears to be ~500 nM (Ruppert et al., 1993; Sette et al., 1994; Sidney et al., 1995; Assarasson et al., 2007). For Class II, the threshold appears to be ~1 μM (Sidney et al., 1992, 1994; 2001a,b; Alexander et al., 1994; Southwood et al., 1998; Oseroff et al., 2010). Conversely, the failure to detect binding with a given MHC-peptide combination should correlate with a failure to detect a T cell response in the same context.
As an alternative approach, biological validity can be determined by measuring the correlation between the capacity of unrelated peptides to compete with antigen for MHC-restricted antigen presentation and their direct MHC binding capacity as measured in the binding assay. If the binding site involved in the interaction measured by the binding assay is the same site involved in the presentation of antigenic fragments to the T cell receptor, a good correlation should be observed (Buus et al., 1987, 1988; Lamont et al., 1990).
Troubleshooting
The assay system described in this unit is usually trouble free as long as fresh and high-quality reagents are used. However, there are some areas where problems may be encountered. Specific precautions and recommendations regarding these areas of concern are discussed briefly below.
MHC
While MHC can be purified from many different types of cell lines, EBV-transformed homozygous cell lines are usually the best first choice for human MHC. These lines are typically easy to grow, and have high expression of MHC. However, because MHC expression varies between cell lines, it is prudent to have more than one cell line available that expresses the desired molecule. Single-allele-transfected, HLA-deficient, 721.221 or RM3 lines, for Class I and Class II, respectively, are also excellent for MHC cultivation and purification. The purity and concentration of MHC preparations should also be monitored, thereby guaranteeing that each assay is as specific and sensitive as possible.
Support Protocol 1 describes the purification of MHC from cell lysates. However, the assay described here is fully amenable to MHC purified from other sources, such as the recombinant solubilized MHC produced in a hollow-fiber bioreactor, as described by Hildebrand (Buchli et al., 2004, 2005), or the E. coli expression system described by Buus (Ferre et al., 2003; Justesen et al., 2009; Harndahl et al., 2011). This type of material may also be purchased from Pure Protein, LLC.
Radiolabeled peptide
As with all other reagents used, fresh material presents the fewest problems. Na[125I] stocks less than 6 weeks old generally give the most reliable labels. While some labels will last up to a month, in most cases peptides should be relabeled after 2 weeks. Old and degraded labels often show HPLC profiles with wide or multiple peaks on gel filtration, or poor signal in the capture assay, and will decrease the sensitivity of the assay, in general.
The hydrophobicity of some peptides causes them to form high–molecular weight “clumps” in solution. Because these clumps are often large enough to elute with the MHC-peptide complexes during gel filtration separation, resulting in either an uninhibitable front peak or poor separation between the first and second peaks, peptides known to be severely hydrophobic should not be used as radiolabeled ligands. In the gel filtration separation, this is particularly exacerbated in the case of Class II molecules, where larger ligands are typically used. Similarly, in the capture-based assay, the use of excessively hydrophobic peptides as radiolabeled ligands may result in reduced signal.
Poor separation of the peaks may also occur if the peptide is too large. In general, for Class II assays, peptides between 13 and 18 residues in length have proven to be the least problematic radiolabeled probes. Because of the specific requirement for peptides of small size, this is typically not an issue for Class I assays. Peptide length is also not an apparent problem for the capture assay.
Finally, because the chloramine T procedure attaches the 125I to tyrosine, it is essential that each peptide contain at least one, and preferably only one, tyrosine. In instances where no tyrosine residues are present, tyrosines may be added to either the N or C terminus of the peptide for Class II probes, or substituted for a nonanchor residue for Class I probes. For peptides bearing multiple tyrosines, tyrosine residues hypothesized to not be involved in peptide binding may be, in most instances, substituted with phenylalanine.
Protease inhibitors
A small amount of protease activity is evident in most MHC preparations, even those of considerably high purity. The degree, and classes, of protease activity present vary as a function of the particular cell line or MHC preparation utilized. For this reason, it is important that fresh protease inhibitor stocks be used in every assay. The protease inhibitor cocktail described for this unit (see Reagents and Solutions) contains broad-specificity inhibitors, which together allow redundant inhibition of most classes of proteases that may be present in an MHC preparation.
Some protease inhibitors are light sensitive and extremely labile in aqueous solutions. Therefore, the protease inhibitor cocktail should be prepared immediately prior to use in the binding assay. Proper storage and handling of these reagents is important to maintain their viability. The importance of the fresh preparation and immediate use (ideally within 2 min, or so) of the cocktail at the point indicated in the protocol cannot be overemphasized.
Detergent
To remain in solution, purified MHC molecules require detergent concentrations above the critical micelle formation level. The detergents and detergent concentrations used in this assay system have been worked out over a number of years, and appear to be optimal. MHC solubility problems can usually be alleviated by increasing the purity of the detergent stocks. This is especially evident with digitonin, which is used in the protease inhibitor cocktail for murine IA assays. As is the rule with most assay systems, fewer problems will be encountered when the highest grade reagents are used.
Temperature
The best and most reliable assay results for most purified MHC molecules are obtained when the 2- to 4-day incubation is performed at room temperature, although in some case incubation at 37°C is necessary. If there are significant temperature fluctuations in the surrounding environment, the assay should be placed in a temperature-controlled setting. Once complexes are formed, however, Class II assays may be placed in the freezer (–20°C) and analyzed at a later date (up to 1 week). The effects of temperature on the assay have been discussed elsewhere (Sette et al., 1992). In general, HLA- DP and DQ prefer 37°C humid conditions for a 48- to 72-hr incubation, with an additional antibody capture period of 24 hr at room temperature.
Evaporation
Because the small (15-μl) final assay volume is susceptible to evaporation, proper precautions must be made to seal the assay wells or tubes. In general, the smaller the vessel for incubation, the better. Sufficient sealing of 96-well plates may be achieved by overlaying a sheet of mylar film and taping down the edges. Costar storage mats are also only reused a maximum of one time or, if the assay is performed at 37°C, only new seals are used.
Assay pH
All Class I assays and the majority of Class II assays are performed at neutral pH. However, some Class II assays are optimal in acidic conditions (Table 18.3.1). For these situations, the pH of the assay is adjusted by preparing MHC/labeled peptide reaction mix with 6 μl of a citrate/phosphate buffer that is 0.5 pH points below the desired final pH. When developing Class II assays, it is prudent to perform direct binding titrations at varying pHs to find the optimal conditions.
Anticipated Results
In the automated separation system specified in Support Protocol 3, the first peak appears between 2.5 and 3.5 min after injection (Fig. 18.3.1). The second peak follows at 4.5 to 6.0 min, depending on the molecular weight of the peptide. In manually separated systems, the first peak will appear between the tenth and eighteenth 1-ml fractions.
If assays are performed under conditions where [label] < [MHC], and IC50 ≥ [MHC], the measured IC50 values are reasonable approximations of true Kd values. MHC-peptide interactions typically have affinities in the range of 1 nM to 50 μM at 23° to 37°C. Association rate constants are relatively low (1 to 1 × 103 M–1sec–1). Once formed, the complexes tend to be extremely stable, with half-lives at 37°C ranging between a few hours and several days (Buus et al., 1986; Sette et al., 1992; Assarsson et al., 2007). The expected affinity of the radiolabeled probe for most assays is in the 1-to-50-nM range. In only a very few cases has it not been possible to identify radiolabeled probes with affinities better than 100 nM.
Time Considerations
The amount of time required to set up an assay is almost entirely dependent upon the number of peptides tested. In fact, most of the time involved in performing an assay is the time required to dilute peptides to the desired concentrations. Performing steps 2 to 5 (see Basic Protocol) can be accomplished in as little as 15 min. Assays testing a few peptides may be set up in as little as 30 min, but even assays involving the testing of hundreds of peptides may be managed in 2 to 3 hr.
Kinetic data obtained using the protocol described in this unit has indicated that 36 to 48 hr is required to establish equilibrium for most peptide-MHC interactions (Buus et al., 1986; Sette et al., 1992, 1994). Thus, it is not advised to shorten the 2-day incubation. However, once formed, peptide-MHC complexes are relatively stable. As a result, assays may still be analyzed without any loss of information after a 60- to 72-hr incubation. Additionally, HLA-DR Class II binding assays may be frozen after incubation for up to 1 week.
Example Applications
Epitope identification
T cells recognize a complex formed between a major histocompatibility complex (MHC) molecule and an antigenic peptide, or epitope. The identification of T cell epitopes is crucial to facilitate the study of the correlates of immunity. MHC-peptide binding assays have proven to be valuable tools for screening targets of immunological interest for the purpose of identifying and characterizing candidate epitopes of diverse origin. In specific cases, known epitope regions can be further probed using truncated or frame-shifted peptides to identify the optimal responding peptide, providing information crucial for the generation of MHC-peptide tetramers, which in turn have proven invaluable for characterizing in detail the T cell response to various targets.
The ability of the assay described herein to be adapted to high-throughput instrumentation has made it feasible to probe large protein antigens in detail. Furthermore, when coupled with various bioinformatics approaches, the availability of high-throughput binding as-says makes it possible to probe large genomes, such as vaccinia virus (Pasquetto et al., 2005; Oseroff et al., 2005, 2008; Moutaftsi et al., 2006) or Mycobacterium tuberculosis (Arlehamn et al., 2012), or even families of related pathogens, such as arenaviruses (Kotturi et al., 2007, 2009, 2010), dengue viruses (Weiskopf et al., 2012), or influenza (Assarsson et al., 2008). These types of studies, combining bioinformatics with high-throughput determination of MHC binding capacity, can be undertaken with the expectation of identifying a large fraction of the antigen specific response (see, e.g., Oseroff et al., 2010; Kotturi et al., 2007, 2009, 2010; Moutaftsi et al., 2006; Arlehamn et al., 2012; Weiskopf et al., 2012).
Bioinformatic predictions
Similarly, the high-throughput capacity of the approach described here enables the testing of large panels of peptides to establish datasets sufficient for the generation of efficient bioinformatics tools. These tools, as noted above, have in turn proven invaluable for several large-scale epitope identification projects. Recent advances in bioinformatic techniques have been able to mine available MHC-peptide binding data to produce tools that can efficiently predict peptide binding to several hundred different MHC Class I and II specificities (see, e.g., Peters et al., 2006; Lin et al., 2008a,b; Wang et al., 2008, 2010, 2011; Hoof et al., 2009; Lundegaard et al., 2010; Nielsen et al., 2010a,b), making these tools valuable resources for the immunological community (see, for example, the prediction tools available at the Immune Epitope Database, http://www.iedb.org; Wulf et al., 2009; Vita et al., 2010).
Motif definition
Different MHC molecules are associated with different peptide-binding specificities, usually referred to as MHC peptide binding motifs. A large body of literature relates to the definition of MHC binding motifs for Class I and Class II molecules of several different species, including humans, mice, chimpanzees, and macaques (for review see, e.g., Rammensee et al., 1995, 1999; McFarland and Beeson, 2002; see also the “browse by allele” compilations at http://www.iedb.org).
A variety of different methods are available to define MHC peptide binding motifs, but the most common methods involve the pool sequencing of naturally presented MHC ligands or the evaluation of the binding capacity of individual peptide libraries. The binding assay approach described here has been utilized to screen peptide libraries for the purpose of defining binding motifs for almost a hundred different Class I and II specificities. Peptide libraries used for defining motifs may be composed of single substitution analogs of known high-affinity binding epitopes or ligands, or large libraries of unrelated peptides. Affinity data from individual peptides can be analyzed with different computational approaches to derive quantitative motifs that elucidate both primary and secondary influences on binding capacity with great detail. The most significant drawback of this approach is that it is dependent upon the availability of panels of several hundreds of allele-specific peptides. As a result, this approach can be relatively labor intensive and expensive. Also, the selection of peptide sequences can introduce biases into the training data, for example by over- or under-representing residues at specific sequence positions.
An alternative approach to characterize the binding specificity of MHC molecules is based on the use of positional scanning combinatorial peptide libraries (PSCL). PSCLs consist of combinatorial mixtures of large numbers of different peptides all sharing a single residue at a certain position (Pinilla et al., 1992). Measuring the affinity of such a library effectively evaluates the average influence of the shared residue on binding in a diverse set of surrounding sequences. Thus, measuring the affinity of a set of just 180 mixtures can derive an estimate of the binding contribution of all 20 naturally occurring amino acid residues in a 9-mer peptide. The use of PSCLs for the purpose of MHC binding motif studies was pioneered by Buus and co-workers (Stryhn et al., 1996; Lauemoller et al., 2001) and Udaka (Udaka et al., 1995, 2000), and has led to the definition of motifs for several dozen Class I and Class II alleles of mouse, human, chimpanzee and macaque origin (see, e.g., Sidney et al., 2007, 2008a, 2010a,b; Loffredo et al., 2009).
Like the single-substitution or peptide library approaches (see, e.g., Parker et al., 1994; Sidney et al., 1996a, 2003; Allen et al., 1998; Bui et al., 2005), data generated from positional scanning combinatorial library studies provide quantitative motifs. Matrices derived from PSCL analyses have been found to perform well in the prediction of peptides with high MHC binding affinity (see, e.g., Stryhn et al., 1996; Udaka et al., 2000; Lauemoller et al., 2001; Sidney et al., 2007, 2008a; Loffredo et al., 2009). The unique advantage of using positional scanning combinatorial libraries is that they can be reused for every allele, representing potentially very significant cost savings. Retesting the same probes for each allele also removes the risk of introducing bias into the set of tested ligands. In utilizing combinatorial libraries to characterize MHC specificity and identify binders, the approach is computationally simple, based on determining relative binding values for each residue/position coordinate. To predict binders, the independent binding of peptide side chains may be assumed, and the predicted binding propensity may be represented as a simple product of each coordinate.
Key References.
Buchli et al., 2004, 2005. See above.
Two papers from William Hildebrand's group demonstrating the use of fluorescence polarization techniques and soluble MHC for developing sensitive binding assays. Willie's group has done pioneering work in developing expression systems for the production of soluble MHC molecules.
Harndahl et al. 2009, 2011. See above
Justesen et al. 2009. See above.
Soren Buus has been a leader for over two decades in the MHC binding field, and the steady stream of excellent work from his laboratory is always creative, interesting and definitely relevant to the present context. His work covers a number of novel assay approaches, as well as related bioinformatics-based tools.
Germain and Margulies, 1993. See above.
A well-written review of antigen processing, which gives an appropriate backdrop for understanding MHC-peptide binding.
Greenbaum et al., 2011. See above.
A recent effort towards the classification of HLA Class II binding specificities into supertypes, defining sets of molecules with shared or largely overlapping repertoires.
Madden, 1995. See above.
Very detailed and clearly presented review of the structural aspects of MHC function.
McFarland and Beeson, 2002. See above.
Very detailed review of peptide binding to MHC Class II.
Rammensee et al., 1995, 1999. See above.
A large listing of MHC-peptide binding motifs, mostly as defined using MHC-peptide elution methodology, is available at the web site described in these references.
Sidney et al., 2008b. See above.
A recent analysis and compilation of HLA Class I binding specificities to define supertypes describing sets of molecules with shared or largely overlapping repertoires.
Internet Resources.
http://www.ebi.ac.uk/imgt/hla/
The ImMunoGeneTics Database: The IMGT/HLA Database, initiated by Marie-Paule Lefranc, provides a specialist database for sequences of the human major histocompatibility complex (HLA) and includes the official sequences for the WHO Nomenclature Committee For Factors of the HLA System. The IMGT/HLA Database is part of the international ImMunoGeneTics project (IMGT), and is hosted by the European Bioinformatics Institute (EBI).
http://www-bimas.cit.nih.gov/molbio/hla bind/
Kenneth Parker provides predictions for peptide binding to a number of different HLA Class I alleles based on pioneering matrices published in 1994.
SYFPEITHI is a pioneering database developed under the direction of Hans-Georg Rammensee. It comprises more than 7000 peptide sequences known to bind Class I and Class II MHC molecules. The entries are compiled from published reports only.
This is the Web site for the American Society of Histocompatibility and Immunogenetics.
The Immune Epitope Database and Analysis Resource (IEDB) contains data related to antibody and T cell epitopes for humans, nonhuman primates, rodents, and other animal species. Curation of peptidic and nonpeptidic epitope data relating to all infectious diseases (including NIAID Category A, B, and C priority pathogens and NIAID Emerging and Re-emerging infectious diseases), allergens, autoimmune diseases, and transplant/alloantigens is current and constantly being updated. The database also contains MHC binding data from a variety of different antigenic sources and immune epitope data from the FIMM (Brusic), HLA Ligand (Hildebrand), TopBank (Sette), and MHC binding (Buus) databases.
http://www.cbs.dtu.dk/services/NetMHC/
The NetMHC 3.2 server, developed by Morten Nielsen, Soren Buus, and co-workers, and hosted by the Center for Biological Sequence Analysis at the Technical University of Denmark, predicts binding of peptides to a number of different HLA alleles using artificial neural networks (ANNs) and weight matrices. Also available is a graphic MHC-peptide motif viewer.
Acknowledgments
This work was supported by funds through the following NI-NIAID contracts and grants (to AS): HHSN 272200900044C, HHSN 27220070048C, NIH U19 A110275, and HHSN 272200900042C.
Literature Cited
- Alexander J, Sidney J, Southwood S, Ruppert J, Oseroff C, Maewal A, Snoke K, Serra HM, Kubo RT, Sette A, Grey HM. Development of high potency universal DR-restricted helper epitopes by modification of high affinity DR-blocking peptides. Immunity. 1994;1:751–761. doi: 10.1016/s1074-7613(94)80017-0. [DOI] [PubMed] [Google Scholar]
- Allen TM, Sidney J, del Guercio MF, Glickman RL, Lensmeyer GL, Wiebe DA, De-Mars R, Pauza CD, Johnson RP, Sette A, Watkins DI. Characterization of the peptide binding motif of a rhesus MHC class I molecule (Mamu-A*01) that binds an immunodominant CTL epitope from simian immunodeficiency virus. J. Immunol. 1998;160:6062–6071. [PubMed] [Google Scholar]
- Arlehamn CS, Sidney J, Henderson R, Greenbaum JA, James EA, Moutaftsi M, Coler R, McKinney DM, Park D, Taplitz R, Kwok WW, Grey H, Peters B, Sette A. Dissecting mechanisms of immunodominance to the common tuberculosis antigens ESAT-6, CFP10, Rv2031c (hspX), Rv2654c (TB7.7), and Rv1038c (EsxJ). J. Immunol. 2012 doi: 10.4049/jimmunol.1103556. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assarsson E, Sidney J, Oseroff C, Pasquetto V, Bui H-H, Frahm N, Brander C, Peters B, Grey H, Sette A. A quantitative analysis of the variables affecting the repertoire of T cell specificities recognized after vaccinia virus infection. J. Immunol. 2007;178:7890–7901. doi: 10.4049/jimmunol.178.12.7890. [DOI] [PubMed] [Google Scholar]
- Assarsson E, Bui H-H, Sidney J, Zhang Q, Glenn J, Oseroff C, Mbawuike IN, Alexander J, Newman MJ, Grey H, Sette A. Immunomic analysis of the repertoire of T-cell specificities for influenza A virus in humans. J. Virol. 2008;82:12241–12251. doi: 10.1128/JVI.01563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber LD, Parham P. Peptide binding to major histocompatibility complex molecules. Annu. Rev. Cell Biol. 1993;9:163–206. doi: 10.1146/annurev.cb.09.110193.001115. [DOI] [PubMed] [Google Scholar]
- Bolton AE, Hunter WM. Radioimmunoassay and related methods. In: Weir DM, Herzenberg LA, Blackwell C, Herzenberg LA, editors. Handbook of Experimental Immunology, Vol. 1: Immunochemistry. Blackwell Scientific; Oxford: 1986. pp. 26.1–26.56. [Google Scholar]
- Bonifacino A, Dasso M, Harford JB, Lippincott-Schwartz J, Yamada KM. Current Protocols in Cell Biology. John Wiley & Sons; Hoboken, N.J.: 2013. [Google Scholar]
- Boyd LF, Kozlowski S, Margulies DH. Solution binding of an antigenic peptide to a major histocompatibility complex class I molecule and the role of β2-microglobulin. Proc. Natl. Acad. Sci. U.S.A. 1992;89:2242–2246. doi: 10.1073/pnas.89.6.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchli R, VanGundy RS, Hickman-Miller HD, Giberson CF, Bardet W, Hildebrand WH. Real-time measurement of in vitro peptide binding to soluble HLA-A*0201 by fluorescence polarization. Biochemistry. 2004;43:14852–14863. doi: 10.1021/bi048580q. [DOI] [PubMed] [Google Scholar]
- Buchli R, VanGundy RS, Hickman-Miller HD, Giberson CF, Bardet W, Hildebrand WH. Development and validation of a fluorescence polarization-based competitive peptide-binding assay for HLA-A*0201-a new tool for epitope discovery. Biochemistry. 2005;44:12491–12507. doi: 10.1021/bi050255v. [DOI] [PubMed] [Google Scholar]
- Bui HH, Sidney J, Peters B, Sathiamurthy M, Sinichi A, Purton KA, Mothe BR, Chisari FV, Watkins DI, Sette A. Automated generation and evaluation of specific MHC binding predictive tools: ARB matrix applications. Immunogenetics. 2005;57:304–314. doi: 10.1007/s00251-005-0798-y. [DOI] [PubMed] [Google Scholar]
- Busch R, Strang G, Howland K, Rothbard JB. Degenerate binding of immunogenic peptides to HLA-DR proteins on B cell surfaces. Int. Immunol. 1990;2:443–451. doi: 10.1093/intimm/2.5.443. [DOI] [PubMed] [Google Scholar]
- Buus S, Sette A, Colon SM, Jenis DM, Grey HM. Isolation and characterization of antigen-Ia complexes involved in T cell recognition. Cell. 1986;47:1071–1077. doi: 10.1016/0092-8674(86)90822-6. [DOI] [PubMed] [Google Scholar]
- Buus S, Sette A, Colon SM, Miles C, Grey HM. The relation between major histocompatibility complex (MHC) restriction and the capacity of Ia to bind immunogenic peptides. Science. 1987;235:1353–1358. doi: 10.1126/science.2435001. [DOI] [PubMed] [Google Scholar]
- Buus S, Sette A, Colon SM, Grey HM. Autologous peptides constitutively occupy the antigen binding site on Ia. Science. 1988;242:1045–1047. doi: 10.1126/science.3194755. [DOI] [PubMed] [Google Scholar]
- Buus S, Stryhn A, Winther K, Kirkby N, Pedersen LO. Receptor-ligand interactions measured by an improved spun column chromatography technique. A high efficiency and high throughput size separation method. Biochim. Biophys. Acta. 1995;1243:453–460. doi: 10.1016/0304-4165(94)00172-t. [DOI] [PubMed] [Google Scholar]
- Ceppellini R, Frumento G, Ferrara GB, Tosi R, Chersi A, Pernis B. Binding of labelled influenza matrix peptide to HLA DR in living B lymphoid cells. Nature. 1989;339:392–394. doi: 10.1038/339392a0. [DOI] [PubMed] [Google Scholar]
- Cerundolo V, Elliott T, Elvin J, Bastin J, Rammensee H-G, Townsend A. The binding affinity and dissociation rates of peptides for class I major histocompatibility complex molecules. J. Immunol. 1991;21:2069–2075. doi: 10.1002/eji.1830210915. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- Christnick ER, Luscher MA, Barber BH, Williams DB. Peptide binding to class I MHC on living cells and quantitation of complexes required for CTL lysis. Nature. 1991;352:67–70. doi: 10.1038/352067a0. [DOI] [PubMed] [Google Scholar]
- Cox AL, Skipper J, Chen Y, Henderson RA, Darrow TL, Shabanowitz J, Engelhard VH, Hunt DF, Slingluff CL., Jr. Identification of a peptide recognized by five melanoma-specific human cytotoxic T cell lines. Science. 1994;264:716–719. doi: 10.1126/science.7513441. [DOI] [PubMed] [Google Scholar]
- Dedier S, Reinelt S, Rion S, Folkers G, Rognan D. Use of fluorescence polarization to monitor MHC-peptide interactions in solution. J. Immunol. Methods. 2001;255:57–66. doi: 10.1016/s0022-1759(01)00423-9. [DOI] [PubMed] [Google Scholar]
- del Guercio M-F, Sidney J, Hermanson G, Perez C, Grey HM, Kubo RT, Sette A. Binding of a peptide antigen to multiple HLA alleles allows definition of an A2-like supertype. J. Immunol. 1995;154:685–693. [PubMed] [Google Scholar]
- Engelhard VH. Structure of peptides associated with class I and class II MHC molecules. Annu. Rev. Immunol. 1994;12:181–207. doi: 10.1146/annurev.iy.12.040194.001145. [DOI] [PubMed] [Google Scholar]
- Falk K, Rötzschke O, Stevanovic S, Jung G, Rammensee HG. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991;351:290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- Ferre H, Ruffet E, Blicher T, Sylvester-Hvid C, Nielsen LL, Hobley TJ, Thomas OR, Buus S. Purification of correctly oxidized MHC class I heavy-chain molecules under denaturing conditions: A novel strategy exploiting disulfide assisted protein folding. Protein Science. 2003;12:551–559. doi: 10.1110/ps.0233003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geluk A, van Meijgaarden KE, Southwood S, Oseroff C, Wouter Drijfhout J, de Vries RRP, Ottenhoff THM, Sette A. HLA-DR3 molecules can bind peptides carrying two alternative specific submotifs. J. Immunol. 1994;152:5742–5748. [PubMed] [Google Scholar]
- Germain RN. Antigen processing and presentation. In: Paul WE, editor. Fundamental Immunology. 3rd ed. Raven Press; New York: 1993. pp. 629–676. [Google Scholar]
- Germain RN. MHC-dependent antigen processing and peptide presentation: Providing ligands for T lymphocyte activation. Cell. 1994;76:287–299. doi: 10.1016/0092-8674(94)90336-0. [DOI] [PubMed] [Google Scholar]
- Germain RN, Margulies DH. The biochemistry and cell biology of antigen processing and presentation. Annu. Rev. Immunol. 1993;11:403–450. doi: 10.1146/annurev.iy.11.040193.002155. [DOI] [PubMed] [Google Scholar]
- Greenbaum J, Sidney J, Chung J, Brander C, Peters B, Sette A. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics. 2011;63:325–335. doi: 10.1007/s00251-011-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood F, Hunter W, Glover J. The preparation of 131I-labeled human growth hormone of high specific radioactivity. Biochem. J. 1963;89:114–123. doi: 10.1042/bj0890114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulukota K, Sidney J, Sette A, DeLisi C. Two complementary methods for predicting peptides binding major histocompatibility complex molecules. J. Mol. Biol. 1997;267:1258–1267. doi: 10.1006/jmbi.1997.0937. [DOI] [PubMed] [Google Scholar]
- Hammer J. New methods to predict MHC-binding sequences within protein antigens. Curr. Opin. Immunol. 1995;7:263–269. doi: 10.1016/0952-7915(95)80013-1. [DOI] [PubMed] [Google Scholar]
- Hammer J, Valsasnini P, Tolba K, Bolin D, Higelin J, Takacs B, Sinigaglia F. Promiscuous and allele-specific anchors in HLA-DR-binding peptides. Cell. 1993;74:197–203. doi: 10.1016/0092-8674(93)90306-b. [DOI] [PubMed] [Google Scholar]
- Hammer J, Bono E, Gallazzi F, Belunis C, Nagy Z, Sinigaglia F. Precise prediction of major histocompatibility complex class II-peptide interaction based on peptide side chain scanning. J. Exp. Med. 1994;180:2353–2358. doi: 10.1084/jem.180.6.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harndahl M, Justesen S, Lamberth K, Roder G, Nielsen M, Buus S. Peptide binding to HLA class I molecules: Homogenous, high-throughput screening, and affinity assays. J. Biomol. Screen. 2009;14:173–180. doi: 10.1177/1087057108329453. [DOI] [PubMed] [Google Scholar]
- Harndahl M, Rasmussen M, Roder G, Buus S. Real-time, high-throughput measurements of peptide-MHC-I dissociation using a scintillation proximity assay. J. Immunol. Methods. 2011;374:5–12. doi: 10.1016/j.jim.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris PE, Colovai A, Liu Z, Dalla Favera R, Suciu-Foca N. Naturally processed HLA class I bound peptides from c-myctransfected cells reveal allele-specific motifs. J. Immunol. 1993;151:5966–5974. [PubMed] [Google Scholar]
- Hill CM, Hayball JD, Allison AA, Roth-bard JB. Conformational and structural characteristics of peptide binding to HLA-DR molecules. J. Immunol. 1991;147:189–197. [PubMed] [Google Scholar]
- Hill CM, Liu A, Marshall KW, Mayer J, Jorgensen B, Yuan B, Cubbon RM, Nichols EA, Wicker LS, Rothbard JB. Exploration of requirements for peptide binding to HLA DRB1*0101 and DRB1*0401. J. Immunol. 1994;152:2890–2898. [PubMed] [Google Scholar]
- Hoof I, Peters B, Sidney J, Pedersen LE, Sette A, Lund O, Buus S, Nielsen M. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics. 2009;61:1–13. doi: 10.1007/s00251-008-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt DF, Henderson RA, Shabanowitz J, Sakaguchi K, Michel H, Sevilir N, Cox AL, Appella E, Engelhard VH. Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science. 1992;255:1261–1266. doi: 10.1126/science.1546328. [DOI] [PubMed] [Google Scholar]
- Janeway CA, Conrad PJ, Lerner EA, Babich J, Wettstein P, Murphy DB. Monoclonal antibodies specific for Ia glycoproteins raised by immunization with activated T cells: Possible role of T cell bound Ia antigens as targets of immunoregulatory T cells. J. Immunol. 1984;132:662–667. [PubMed] [Google Scholar]
- Joyce S, Nathenson SG. Methods to study peptides associated with MHC class I molecules. Curr. Opin. Immunol. 1994;6:24–31. doi: 10.1016/0952-7915(94)90029-9. [DOI] [PubMed] [Google Scholar]
- Justesen S, Harndahl M, Lamberth K, Nielsen LL, Buus S. Functional recombinant MHC class II molecules and high-throughput peptide-binding assays. Immunome Res. 2009;5:2. doi: 10.1186/1745-7580-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khilko SN, Corr M, Boyd LF, Lees A, Inman JK, Margulies DH. Direct detection of major histocompatibility complex class I binding to antigenic peptides using surface plasmon resonance. J. Biol. Chem. 1993;268:15425–15434. [PubMed] [Google Scholar]
- Kondo A, Sidney J, Southwood S, del Guercio M-F, Appella E, Sakamoto J, Celis E, Grey HM, Chesnut RW, Kubo RT, Sette A. Prominent roles of secondary anchor residues in peptide binding to HLA-A24 human class I molecules. J. Immunol. 1995;155:4307–4312. [PubMed] [Google Scholar]
- Kondo A, Sidney J, Southwood S, del Guercio M-F, Appella E, Sakamoto J, Celis E, Grey HM, Celis E, Chesnut RW, Kubo RT, Sette A. Two distinct HLA-A*0101-specific submotifs illustrate alternative peptide binding modes. Immunogenetics. 1997;45:249–258. doi: 10.1007/s002510050200. [DOI] [PubMed] [Google Scholar]
- Kotturi M, Peters B, Buendia-Laysa F, Sidney J, Oseroff C, Botten J, Grey H, Buchmeier M, Sette A. The CD8+ T-cell response to lymphocytic choriomeningitis virus involves the L antigen: Uncovering new tricks for an old virus. J. Virol. 2007;81:4928–4940. doi: 10.1128/JVI.02632-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotturi MF, Botten J, Sidney J, Bui H-H, Giancola L, Maybeno M, Babin J, Oseroff C, Pasquetto V, Greenbaum JA, Peters B, Ting J, Do D, Vang L, Alexander J, Grey H, Buchmeier MJ, Sette A. A multivalent and cross-protective vaccine strategy against Arenaviruses associated with human disease. PLoS Pathog. 2009;5:e1000695. doi: 10.1371/journal.ppat.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotturi M, Botten J, Maybeno M, Sidney J, Glenn J, Bui H-H, Oseroff C, Crotty S, Peters B, Grey H, Altmann D, Buchmeier M, Sette A. Polyfunctional CD4+ T cell responses to a set of pathogenic arenaviruses provide broad population coverage. Immunome Res. 2010;6:4. doi: 10.1186/1745-7580-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont AG, Powell MF, Colon SM, Miles C, Grey HM, Sette A. The use of peptide analogs with improved stability and MHC binding capacity to inhibit antigen presentation in vitro and in vivo. J. Immunol. 1990;144:2493–2498. [PubMed] [Google Scholar]
- Lauemoller SL, Holm A, Hilden J, Brunak S, Holst Nissen M, Stryhn A, Ostergaard Pedersen L, Buus S. Quantitative predictions of peptide binding to MHC class I molecules using specificity matrices and anchor-stratified calibrations. Tissue Antigens. 2001;57:405–414. doi: 10.1034/j.1399-0039.2001.057005405.x. [DOI] [PubMed] [Google Scholar]
- Lin HH, Zhang GL, Tongchusak S, Reinherz EL, Brusic V. Evaluation of MHC-II peptide binding prediction servers: Applications for vaccine research. BMC Bioinformatics. 2008a;9:S22. doi: 10.1186/1471-2105-9-S12-S22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HH, Ray S, Tongchusak S, Reinherz EL, Brusic V. Evaluation of MHC class I peptide binding prediction servers: Applications for vaccine research. BMC Immunol. 2008b;9:8. doi: 10.1186/1471-2172-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljunggren HG, Stam NJ, Ohlen C, Neefjes JJ, Hoglund P, Heemels MT, Bastin J, Schumacher TNM, Townsend A, Karre K, Ploegh HL. Empty MHC Class I molecules come out in the cold. Nature. 1990;346:476–480. doi: 10.1038/346476a0. [DOI] [PubMed] [Google Scholar]
- Loffredo JT, Sidney J, Bean AT, Beal DR, Bardet W, Wahl A, Hawkins OE, Piaskowski S, Wilson NA, Hildebrand WH, Watkins DI, Sette A. Two MHC class I molecules associated with elite control of immunodeficiency virus replication, Mamu-B*08 and HLA-B*2705, bind peptides with sequence similarity. J. Immunol. 2009;182:7763–7775. doi: 10.4049/jimmunol.0900111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden DR. The three-dimensional structure of peptide-MHC complexes. Annu. Rev. Immunol. 1995;13:587–622. doi: 10.1146/annurev.iy.13.040195.003103. [DOI] [PubMed] [Google Scholar]
- Marshall KW, Liu AF, Canales J, Perahia B, Jorgensen B, Gantzos RD, Aguilar B, Devaux B, Rothbard JB. Role of the polymorphic residues in HLA-DR molecules in allele-specific binding of peptide ligands. J. Immunol. 1994;152:4946–4957. [PubMed] [Google Scholar]
- McFarland BJ, Beeson C. Binding interactions between peptides and proteins of the class II Major Histocompatibility Complex. Med. Res. Rev. 2002;22:168–203. doi: 10.1002/med.10006. [DOI] [PubMed] [Google Scholar]
- Moutaftsi M, Peters B, Pasquetto V, Tscharke DC, Sidney J, Bui HH, Grey H, Sette A. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nat. Biotechnol. 2006;24:817–819. doi: 10.1038/nbt1215. [DOI] [PubMed] [Google Scholar]
- Nepom BS, Nepom GT, Coleman M, Kwok WW. Critical contribution of beta chain residue 57 in peptide binding ability of both HLA-DR and -DQ molecules. Proc. Natl. Acad. Sci. U.S.A. 1996;93:7202–7206. doi: 10.1073/pnas.93.14.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M, Justesen S, Lund O, Lundegaard C, Buus S. NetMHCIIpan-2.0—Improved pan-specific HLA-DR predictions using a novel concurrent alignment and weight optimization training procedure. Immunome Res. 2010a;6:9. doi: 10.1186/1745-7580-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M, Lund O, Buus S, Lundegaard C. MHC Class II epitope predictive algorithms. Immunology. 2010b;130:319–328. doi: 10.1111/j.1365-2567.2010.03268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen AC, Pedersen LO, Hansen AS, Nissen MH, Olsen M, Hansen PR, Holm A, Buus S. A quantitative assay to measure the interaction between immunogenic peptides and purified class I major histocompatibility complex molecules. Eur. J. Immunol. 1994;24:385–392. doi: 10.1002/eji.1830240218. [DOI] [PubMed] [Google Scholar]
- Oseroff C, Kos F, Bui HH, Peters B, Pasquetto V, Glenn J, Palmore T, Sidney J, Tscharke DC, Bennink JR, Southwood S, Grey HM, Yewdell JW, Sette A. HLA class I-restricted responses to vaccinia recognize a broad array of proteins mainly involved in virulence and viral gene regulation. Proc. Natl. Acad. Sci. U.S.A. 2005;102:13980–13985. doi: 10.1073/pnas.0506768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oseroff C, Peters B, Pasquetto V, Moutaftsi M, Sidney J, Panchanathan V, Tscharke DC, Maillere B, Grey H, Sette A. Dissociation between epitope hierarchy and immunoprevalence in CD8 responses to vaccinia virus western reserve. J. Immunol. 2008;180:7193–7202. doi: 10.4049/jimmunol.180.11.7193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oseroff C, Sidney J, Kotturi MF, Kolla R, Alam R, Broide DH, Wasserman SI, Weiskopf D, McKinney DM, Chung JL, Petersen A, Grey H, Peters B, Sette A. Molecular determinants of T cell epitope recognition to the common Timothy grass allergen. J. Immunol. 2010;185:943–955. doi: 10.4049/jimmunol.1000405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan D, Sidney J, Appella E, Walker L, Phillips L, Colon SM, Miles C, Chesnut RW, Sette A. Characterization of the specificity of peptide binding to four DR haplotypes. J. Immunol. 1990;149:1799–1808. [PubMed] [Google Scholar]
- O'Sullivan D, Arrhenius T, Sidney J, del Guercio M-F, Albertson M, Wall M, Oseroff C, Southwood S, Colon SM, Gaeta FCA, Sette A. On the interaction of promiscuous antigenic peptides with different DR alleles: Identification of common structural motifs. J. Immunol. 1991;147:2663–2669. [PubMed] [Google Scholar]
- Panina-Bordignon P, Tan A, Termijtelen A, Demotz S, Corradin G, Lanzavecchia A. Universally immunogenic T cell epitopes: Promiscuous binding to human MHC class II and promiscuous recognition by T cells. Eur. J. Immunol. 1989;19:2237–2242. doi: 10.1002/eji.1830191209. [DOI] [PubMed] [Google Scholar]
- Parker KC, DiBrino M, Hull L, Coligan JE. The β2-micro-globulin dissociation rate is an accurate measure of the stability of MHC class I heterotrimers and depends on which peptide is found. J. Immunol. 1992;149:1896–1904. [PubMed] [Google Scholar]
- Parker KC, Bednarek MA, Coligan JE. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J. Immunol. 1994;152:163–175. [PubMed] [Google Scholar]
- Pasquetto V, Bui HH, Giannino R, Banh C, Mirza F, Sidney J, Oseroff C, Tscharke DC, Irvine K, Bennink JR, Peters B, Southwood S, Cerundolo V, Grey H, Yewdell JW, Sette A. HLA-A*0201, HLA-A*1101, and HLA-B*0702 transgenic mice recognize numerous poxvirus determinants from a wide variety of viral gene products. J. Immunol. 2005;175:5504–5515. doi: 10.4049/jimmunol.175.8.5504. [DOI] [PubMed] [Google Scholar]
- Peters B, Bui H-H, Frankild S, Nielsen M, Lundegaard C, Kostem E, Basch D, Lamberth K, Harndahl M, Fleri W, Wilson SS, Sidney J, Lund O, Buus S, Sette A. A community resource benchmarking predictions of peptide binding to MHC-I molecules. PLoS Comput. Biol. 2006;2:e65. doi: 10.1371/journal.pcbi.0020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinilla C, Appel JR, Blanc P, Houghten RA. Rapid identification of high affinity peptide ligands using positional scanning synthetic peptide combinatorial libraries. Biotechniques. 1992;13:901–905. [PubMed] [Google Scholar]
- Rammensee H-G, Friede T, Stevanovic S. MHC ligands and peptide motifs: First listing. Immunogenetics. 1995;41:178–228. doi: 10.1007/BF00172063. [DOI] [PubMed] [Google Scholar]
- Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: Database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- Reay PA, Wettstein DA, Davis MM. pH dependence and exchange of high and low responder peptides binding to a class II MHC molecule. EMBO J. 1992;11:2829–2839. doi: 10.1002/j.1460-2075.1992.tb05350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebai N, Malissen B. Structural and genetic analyses of HLA class I molecules using monoclonal xenoantibodies. Tissue Antigens. 1983;22:107–117. doi: 10.1111/j.1399-0039.1983.tb01176.x. [DOI] [PubMed] [Google Scholar]
- Roche PA, Cresswell P. High-affinity binding of an influenza hemagglutinin-derived peptide to purified HLA-DR. J. Immunol. 1991;144:1849–1856. [PubMed] [Google Scholar]
- Rothbard JB. One size fits all. Curr. Biol. 1994;4:653–655. doi: 10.1016/s0960-9822(00)00146-9. [DOI] [PubMed] [Google Scholar]
- Rötzschke O, Falk K. Origin, structure and motifs of naturally processed MHC class II ligands. Curr. Opin. Immunol. 1994;6:45–51. doi: 10.1016/0952-7915(94)90032-9. [DOI] [PubMed] [Google Scholar]
- Ruppert J, Sidney J, Celis E, Kubo RT, Grey HM, Sette A. Prominent role of secondary anchor residues in peptide binding to HLA-A2.1 molecules. Cell. 1993;74:929–937. doi: 10.1016/0092-8674(93)90472-3. [DOI] [PubMed] [Google Scholar]
- Schumacher TN, Heemels MT, Neefjes JJ, Kast WM, Melief CJ, Ploegh HL. Direct binding of peptide to empty MHC Class I molecules on intact cells and in vitro. Cell. 1990;62:563–567. doi: 10.1016/0092-8674(90)90020-f. [DOI] [PubMed] [Google Scholar]
- Sette A, Grey HM. Chemistry of peptide interactions with MHC proteins. Curr. Opin. Immunol. 1992;4:79–86. doi: 10.1016/0952-7915(92)90130-7. [DOI] [PubMed] [Google Scholar]
- Sette A, Sidney J, Albertson M, Miles C, Colon SM, Pedrazzini T, Lamont AG, Grey HM. A novel approach to the generation of high affinity class II binding peptides. J. Immunol. 1990;145:1809–1813. [PubMed] [Google Scholar]
- Sette A, Southwood S, O'Sullivan D, Gaeta FCA, Sidney J, Grey HM. Effect of pH on MHC class II-peptide interactions. J. Immunol. 1992;148:844–851. [PubMed] [Google Scholar]
- Sette A, Sidney J, Oseroff C, del Guercio M-F, Southwood S, Arrhenius T, Powell MF, Colon SM, Gaeta FCA, Grey HM. HLA DR4w4-binding motifs illustrate the biochemical basis of degeneracy and specificity in peptide-DR interactions. J. Immunol. 1993;151:3163–3170. [PubMed] [Google Scholar]
- Sette A, Sidney J, del Guercio M-F, Southwood S, Ruppert J, Dahlberg C, Grey HM, Kubo RT. Peptide binding to the most frequent HLA-A class I alleles measured by quantitative molecular binding assays. Mol. Immunol. 1994;31:813–822. doi: 10.1016/0161-5890(94)90019-1. [DOI] [PubMed] [Google Scholar]
- Sidney J, Oseroff C, Southwood S, Wall M, Ishioka G, Koning F, Sette A. DRB1*0301 molecules recognize a structural motif distinct from the one recognized by most DRb1 alleles. J. Immunol. 1992;149:2634–2640. [PubMed] [Google Scholar]
- Sidney J, Oseroff C, del Guercio M-F, Southwood S, Krieger JI, Ishioka GY, Sakaguchi K, Appella E, Sette A. Definition of a DQ3.1-specific binding motif. J. Immunol. 1994;152:4516–4525. [PubMed] [Google Scholar]
- Sidney J, del Guercio M-F, Southwood S, Engelhard VS, Apella E, Rammensee HG, Falk K, Rötzschke O, Takiguchi M, Kubo RT, Grey HM, Sette A. Several HLA alleles share overlapping peptide specificities. J. Immunol. 1995;154:247–259. [PubMed] [Google Scholar]
- Sidney J, Grey HM, Southwood S, Celis E, Wentworth PA, del Guercio M-F, Kubo RT, Chesnut RW, Sette A. Definition of an HLA-A3-like supermotif demonstrates the overlapping peptide binding repertoires of common HLA molecules. Hum. Immunol. 1996a;45:79–93. doi: 10.1016/0198-8859(95)00173-5. [DOI] [PubMed] [Google Scholar]
- Sidney J, Southwood S, del Guercio M-F, Grey HM, Chesnut RW, Kubo RT, Sette A. Specificity and degeneracy in peptide binding to HLA-B7-like class I molecules. J. Immunol. 1996b;157:3480–3490. [PubMed] [Google Scholar]
- Sidney J, Southwood S, Pasquetto V, Sette A. Simultaneous prediction of binding capacity for multiple molecules of the HLA B44-supertype. J. Immunol. 2003;171:5964–5974. doi: 10.4049/jimmunol.171.11.5964. [DOI] [PubMed] [Google Scholar]
- Sidney J, Peters B, Moore C, Pencille TJ, Ngo S, Masterman KA, Asabe S, Pinilla C, Chisari FV, Sette A. Characterization of the peptide-binding specificity of the chimpanzee class I alleles A*0301 and A*0401 using a combinatorial peptide library. Immunogenetics. 2007;59:745–751. doi: 10.1007/s00251-007-0243-5. [DOI] [PubMed] [Google Scholar]
- Sidney J, Assarsson E, Moore C, Ngo S, Pinilla C, Sette A, Peters B. Quantitative peptide binding motifs for 19 human and mouse MHC class I molecules derived using positional scanning combinatorial peptide libraries. Immunome Res. 2008a;4:2. doi: 10.1186/1745-7580-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidney J, Peters B, Frahm N, Brander C, Sette A. HLA class I supertypes: A revised and updated classification. BMC Immunol. 2008b;9:1. doi: 10.1186/1471-2172-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidney J, Steen A, Moore C, Ngo S, Chung J, Peters B, Sette A. Divergent motifs but overlapping binding repertoires of six HLA-DQ molecules frequently expressed in the worldwide human population. J. Immunol. 2010a;185:4189–4198. doi: 10.4049/jimmunol.1001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidney J, Steen A, Moore C, Ngo S, Chung J, Peters B, Sette A. Five HLA-DP molecules frequently expressed in the worldwide human population share a common HLA supertypic binding specificity. J. Immunol. 2010b;184:2492–2503. doi: 10.4049/jimmunol.0903655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinigaglia F, Hammer J. Defining rules for the peptide-MHC class II interaction. Curr. Opin. Immunol. 1994;6:52–56. doi: 10.1016/0952-7915(94)90033-7. [DOI] [PubMed] [Google Scholar]
- Sinigaglia F, Guttinger M, Kilgus J, Doran DM, Matile H, Etlinger H, Trzeciak A, Gillessen D, Pink JRL. A malaria T-cell epitope recognized in association with most mouse and human MHC class II molecules. Nature. 1988;336:778–780. doi: 10.1038/336778a0. [DOI] [PubMed] [Google Scholar]
- Southwood S, Sidney J, Kondo A, del Guercio M-F, Appella E, Hoffman S, Kubo RT, Chesnut RW, Grey HM, Sette A. Several common HLA-DR types share largely overlapping peptide binding repertoires. J. Immunol. 1998;160:3363–3373. [PubMed] [Google Scholar]
- Stern LJ, Wiley DC. Antigenic peptide binding by class I and class II histocompatibility proteins. Structure. 1994;2:245–251. doi: 10.1016/s0969-2126(00)00026-5. [DOI] [PubMed] [Google Scholar]
- Stryhn A, Pedersen LO, Romme T, Holm CB, Holm A, Buus S. Peptide binding specificity of major histocompatibility complex class I resolved into an array of apparently independent subspecificities: Quantitation by peptide libraries and improved prediction of binding. Eur. J. Immunol. 1996;26:1911–1918. doi: 10.1002/eji.1830260836. [DOI] [PubMed] [Google Scholar]
- Sylvester-Hvid C, Kristensen N, Blicher T, Ferre H, Lauemoller SL, Wolf XA, Lamberth K, Nissen MH, Pedersen LO, Buus S. Establishment of a quantitative ELISA capable of determining peptide: MHC class I interaction. Tissue Antigens. 2002;59:251–258. doi: 10.1034/j.1399-0039.2002.590402.x. [DOI] [PubMed] [Google Scholar]
- Sylvester-Hvid C, Nielsen M, Lamberth K, Roder G, Justesen S, Lundegaard C, Worning P, Thomadsen H, Lund O, Brunak S, Buus S. SARS CTL vaccine candidates: HLA supertype-, genome-wide scanning and biochemical validation. Tissue Antigens. 2004;63:395–400. doi: 10.1111/j.0001-2815.2004.00221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend A, Elliott T, Cerundolo V, Foster L, Barber B, Tse A. Assembly of MHC class I molecules analyzed in vitro. Cell. 1990;62:285–295. doi: 10.1016/0092-8674(90)90366-m. [DOI] [PubMed] [Google Scholar]
- Udaka K, Wiesmuller KH, Kienle S, Jung G, Walden P. Decrypting the structure of major histocompatibility complex class I-restricted cytotoxic T lymphocyte epitopes with complex peptide libraries. J. Exp. Med. 1995;181:2097–2108. doi: 10.1084/jem.181.6.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udaka K, Wiesmuller KH, Kienle S, Jung G, Tamamura H, Yamagishi H, Okumura K, Walden P, Suto T, Kawasaki T. An automated prediction of MHC class I-binding peptides based on positional scanning with peptide libraries. Immunogenetics. 2000;51:816–828. doi: 10.1007/s002510000217. [DOI] [PubMed] [Google Scholar]
- van Bleek GM, Nathenson SG. Isolation of an endogenously processed immunodominant viral peptide from the class I H-2Kb molecule. Nature. 1990;348:213–216. doi: 10.1038/348213a0. [DOI] [PubMed] [Google Scholar]
- Vita R, Zarebski L, Greenbaum JA, Emami H, Hoof I, Salimi N, Damle R, Sette A, Peters B. The immune epitope database 2.0. Nucleic Acids Res. 2010;38:D854–D862. doi: 10.1093/nar/gkp1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Sidney J, Dow C, Mothe B, Sette A, Peters B. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput. Biol. 2008;4:e1000048. doi: 10.1371/journal.pcbi.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Sidney J, Kim Y, Sette A, Lund O, Nielsen M, Peters B. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics. 2010;11:568. doi: 10.1186/1471-2105-11-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Sidney J, Sette A, Peters B. A computational pipeline to generate MHC binding motifs. Immunome Res. 2011;7:3. doi: 10.4172/1745-7580.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D, Yauch LE, Angelo MA, John DV, Greenbaum JA, Sidney J, Kolla RV, De Silva AD, de Silva AM, Grey H, Peters B, Shresta S, Sette A. Insights into HLA-restricted T cell responses in a novel mouse model of dengue virus infection point toward new implications for vaccine design. J. Immunol. 2011;187:4268–4279. doi: 10.4049/jimmunol.1101970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulf M, Hoehn P, Trinder P. Identification of human MHC class I binding peptides using the iTOPIA-epitope discovery system. Methods Mol. Biol. 2009;524:361–367. doi: 10.1007/978-1-59745-450-6_26. [DOI] [PubMed] [Google Scholar]