Abstract
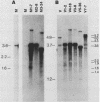
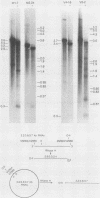
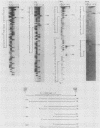
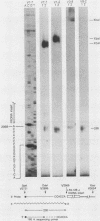
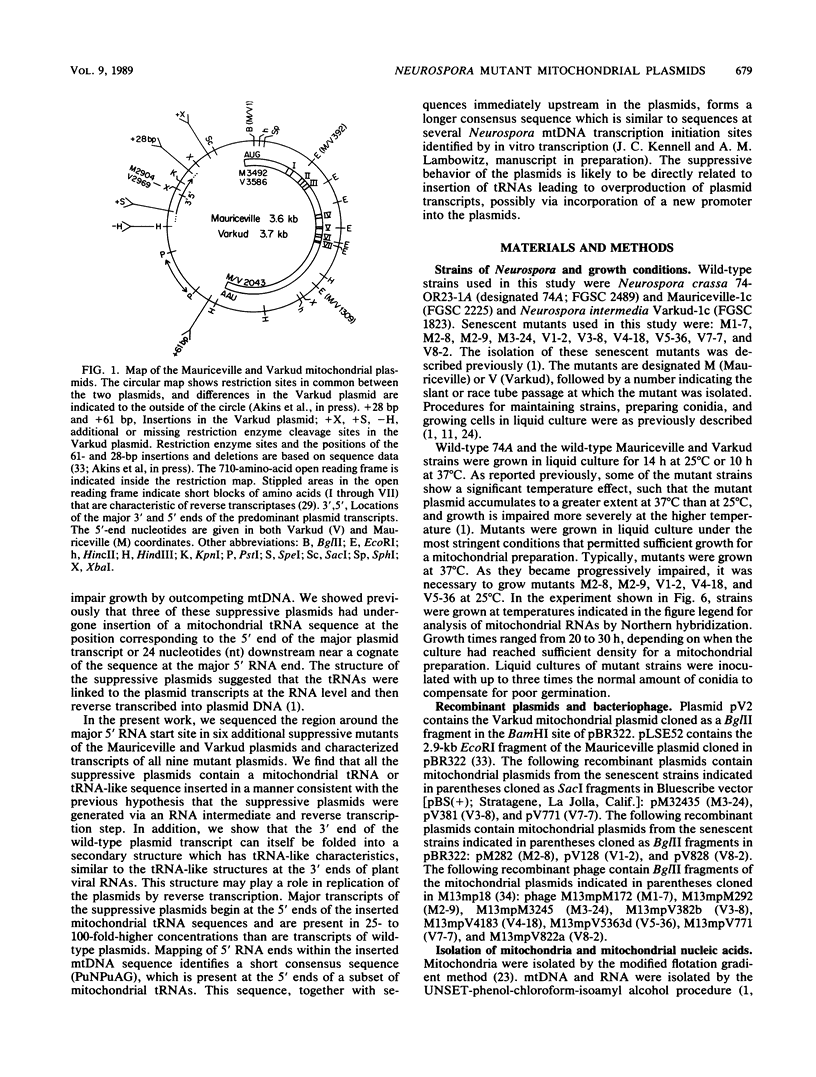
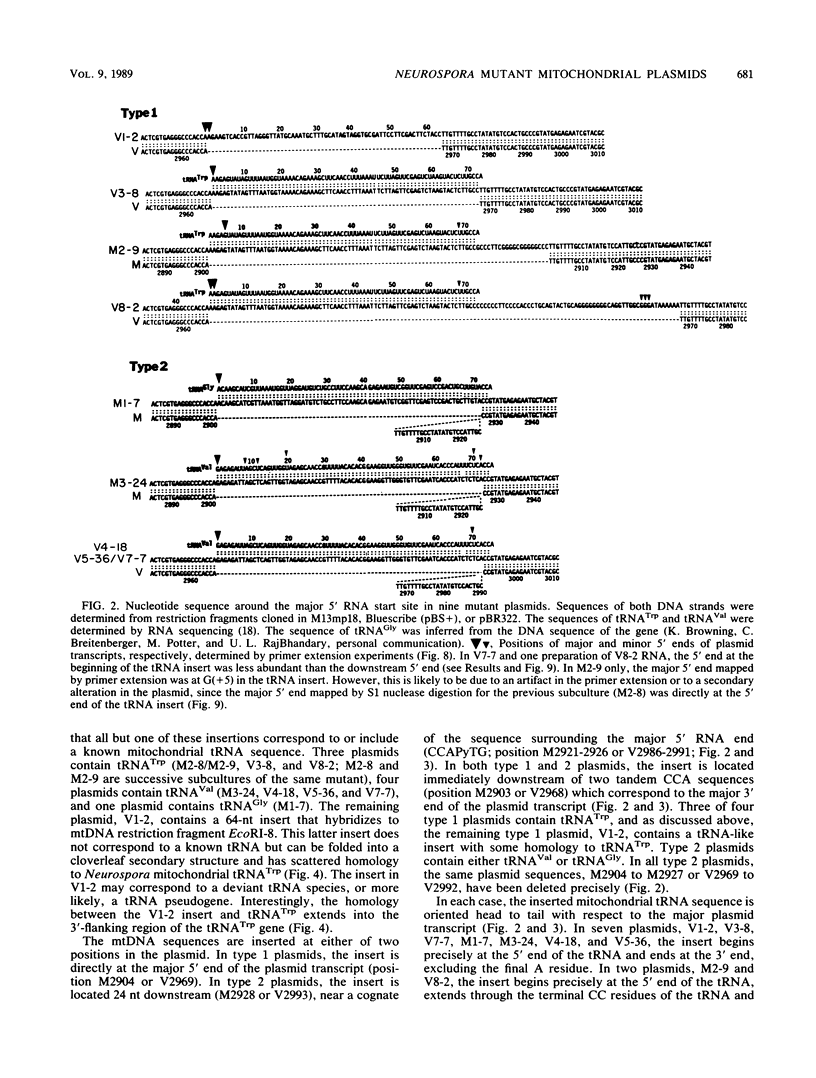
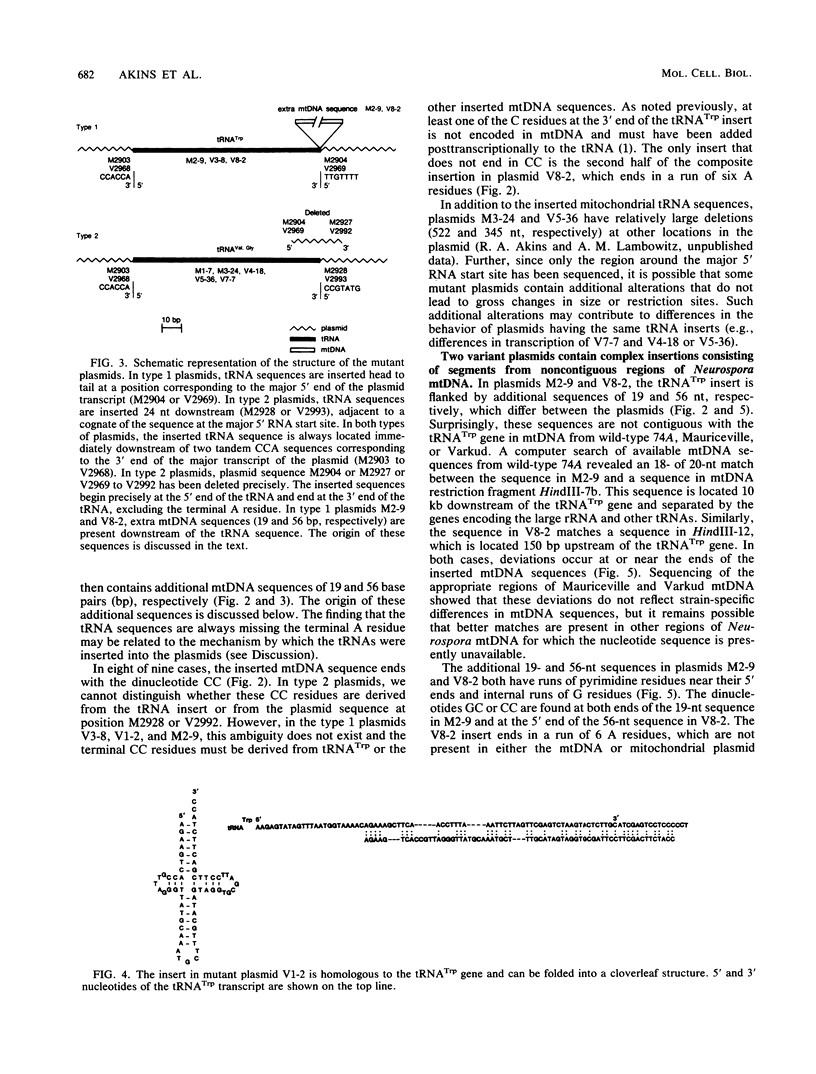
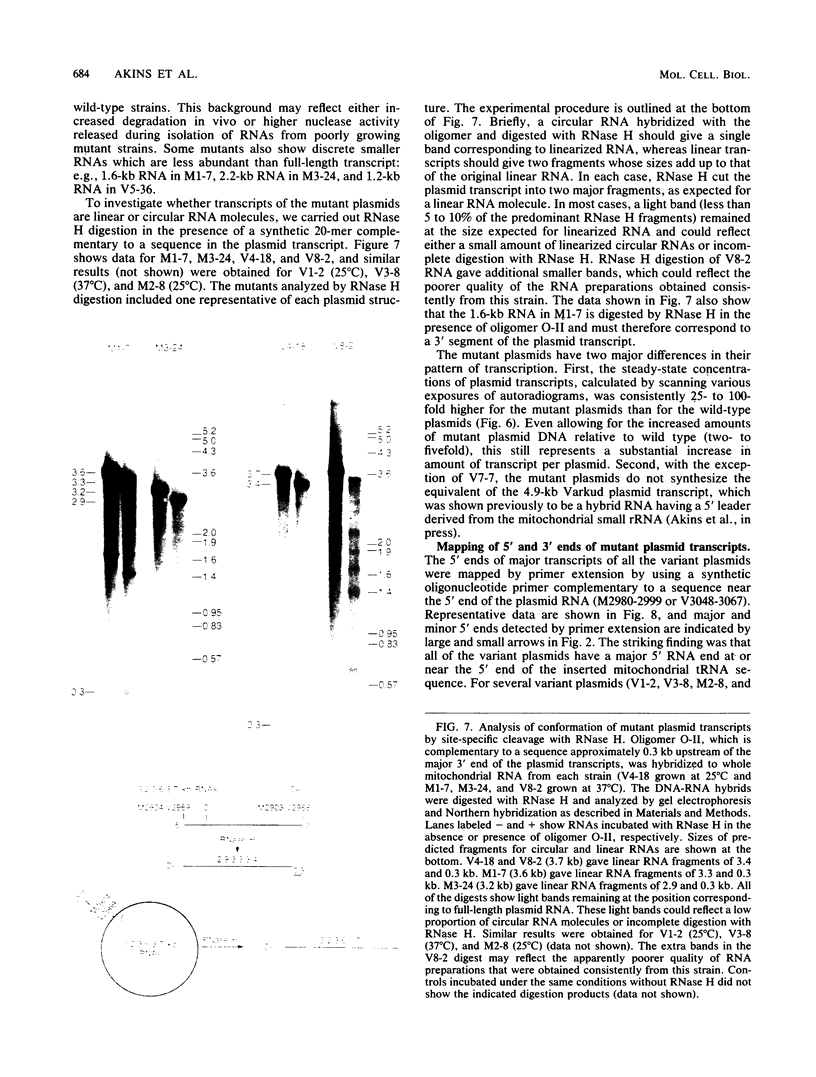
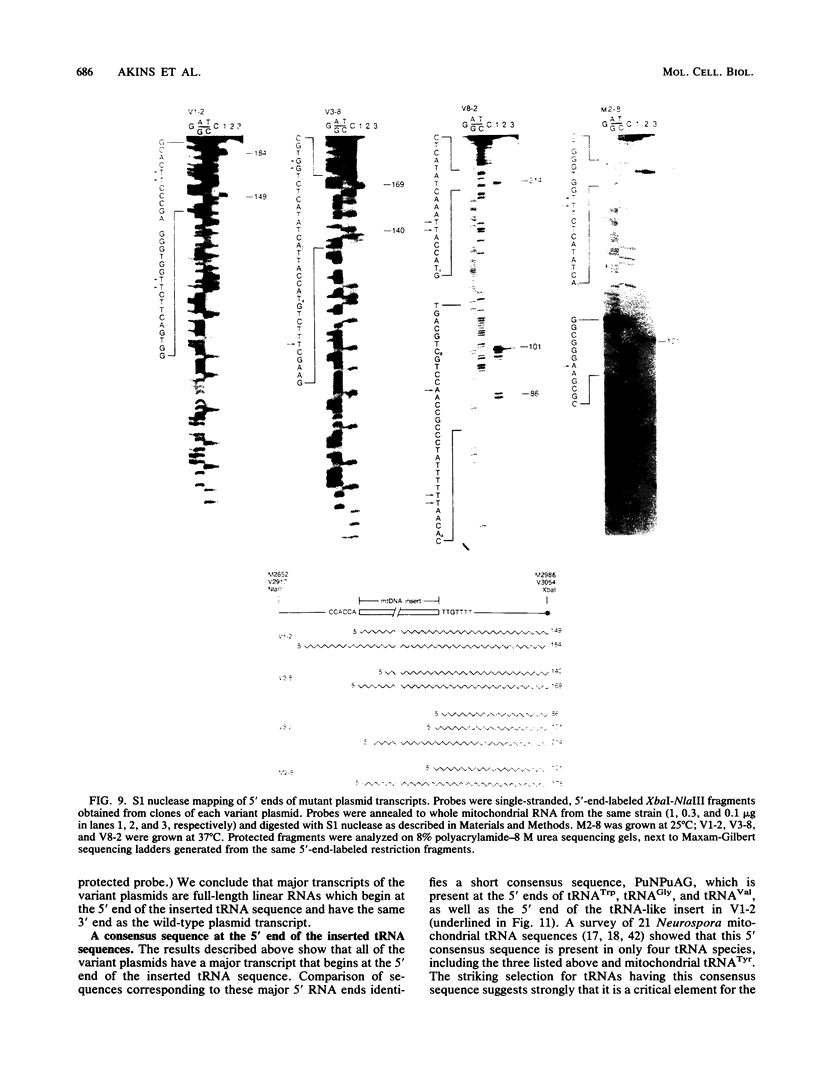
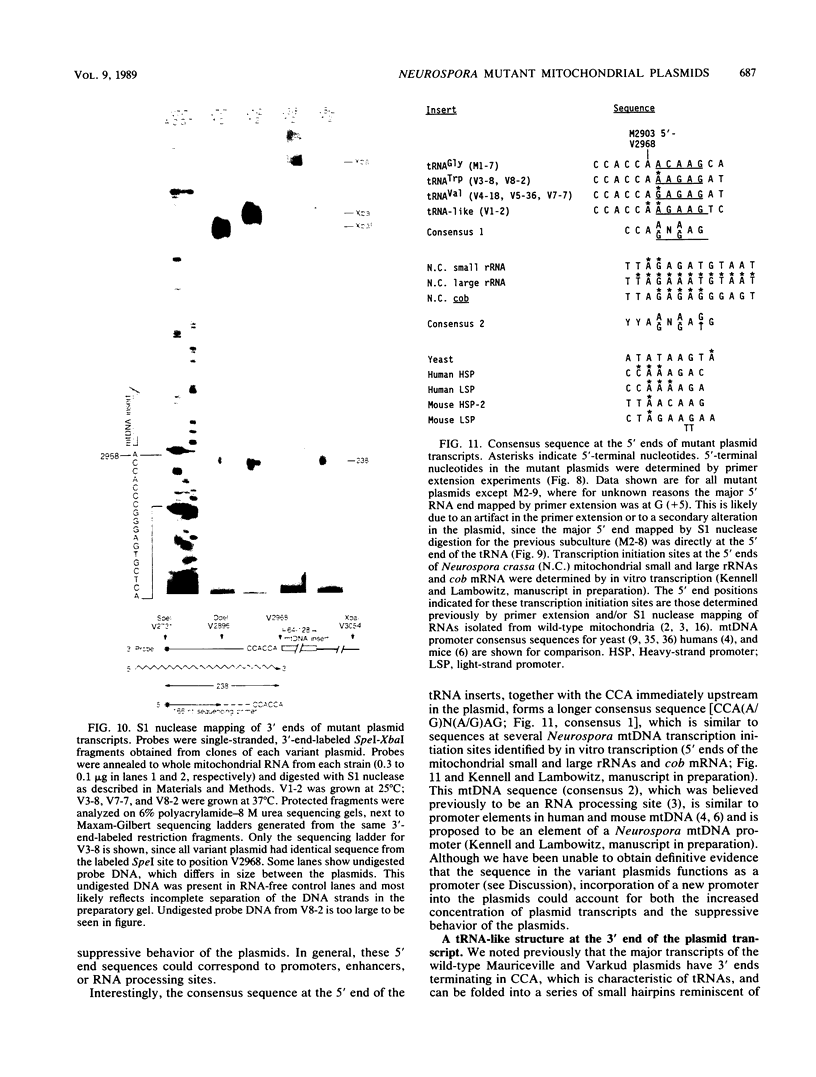
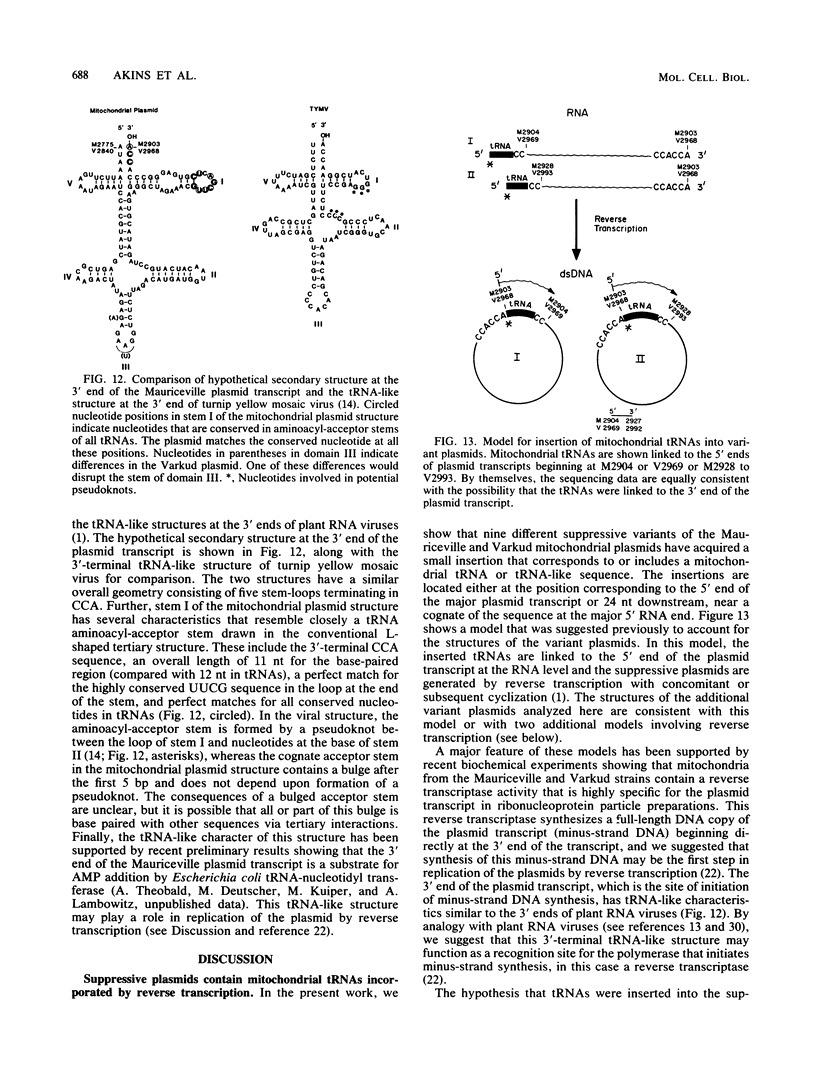
The Mauriceville and Varkud mitochondrial plasmids of Neurospora spp. are closely related, closed-circular DNAs (3.6 and 3.7 kilobases, respectively) whose nucleotide sequences and genetic organization suggest relationships to mitochondrial introns and retroelements. We have characterized nine suppressive mutants of these plasmids that outcompete mitochondrial DNA and lead to impaired growth. All nine suppressive plasmids contain small insertions, corresponding to or including a mitochondrial tRNA (tRNATrp, tRNAGly, or tRNAVal) or a tRNA-like sequence. The insertions are located at the position corresponding to the 5' end of the major plasmid transcript or 24 nucleotides downstream near a cognate of the sequence at the major 5' RNA end. The structure of the suppressive plasmids suggests that the tRNAs were inserted via an RNA intermediate. The 3' end of the wild-type plasmid transcript can itself be folded into a secondary structure which has tRNA-like characteristics, similar to the tRNA-like structures at the 3' ends of plant viral RNAs. This structure may play a role in replication of the plasmids by reverse transcription. Major transcripts of the suppressive plasmids begin at the 5' end of the inserted mitochondrial tRNA sequence and are present in 25- to 100-fold-higher concentrations than are transcripts of wild-type plasmids. Mapping of 5' RNA ends within the inserted mtDNA sequences identifies a short consensus sequence (PuNPuAG) which is present at the 5' ends of a subset of mitochondrial tRNA genes. This sequence, together with sequences immediately upstream in the plasmids, forms a longer consensus sequence, which is similar to sequences at transcription initiation sites in Neurospora mitochondrial DNA. The suppressive behavior of the plasmids is likely to be directly related to the insertion of tRNAs leading to overproduction of plasmid transcripts.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akins R. A., Kelley R. L., Lambowitz A. M. Mitochondrial plasmids of Neurospora: integration into mitochondrial DNA and evidence for reverse transcription in mitochondria. Cell. 1986 Nov 21;47(4):505–516. doi: 10.1016/0092-8674(86)90615-x. [DOI] [PubMed] [Google Scholar]
- Akins R. A., Lambowitz A. M. The [poky] mutant of Neurospora contains a 4-base-pair deletion at the 5' end of the mitochondrial small rRNA. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3791–3795. doi: 10.1073/pnas.81.12.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G., Helmer Citterich M., Nelson M. A., Werner S., Macino G. RNA processing in Neurospora crassa mitochondria: transfer RNAs punctuate a large precursor transcript. EMBO J. 1985 Jan;4(1):197–204. doi: 10.1002/j.1460-2075.1985.tb02336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D. D., Clayton D. A. Precise assignment of the light-strand promoter of mouse mitochondrial DNA: a functional promoter consists of multiple upstream domains. Mol Cell Biol. 1986 Sep;6(9):3253–3261. doi: 10.1128/mcb.6.9.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D. D., Clayton D. A. Precise identification of individual promoters for transcription of each strand of human mitochondrial DNA. Cell. 1984 Mar;36(3):635–643. doi: 10.1016/0092-8674(84)90343-x. [DOI] [PubMed] [Google Scholar]
- Chang D. D., Clayton D. A. Priming of human mitochondrial DNA replication occurs at the light-strand promoter. Proc Natl Acad Sci U S A. 1985 Jan;82(2):351–355. doi: 10.1073/pnas.82.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D. D., Hauswirth W. W., Clayton D. A. Replication priming and transcription initiate from precisely the same site in mouse mitochondrial DNA. EMBO J. 1985 Jun;4(6):1559–1567. doi: 10.1002/j.1460-2075.1985.tb03817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Christianson T., Rabinowitz M. Identification of multiple transcriptional initiation sites on the yeast mitochondrial genome by in vitro capping with guanylyltransferase. J Biol Chem. 1983 Nov 25;258(22):14025–14033. [PubMed] [Google Scholar]
- Collins R. A., Stohl L. L., Cole M. D., Lambowitz A. M. Characterization of a novel plasmid DNA found in mitochondria of N. crassa. Cell. 1981 May;24(2):443–452. doi: 10.1016/0092-8674(81)90335-4. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Dreher T. W., Hall T. C. Mutational analysis of the sequence and structural requirements in brome mosaic virus RNA for minus strand promoter activity. J Mol Biol. 1988 May 5;201(1):31–40. doi: 10.1016/0022-2836(88)90436-6. [DOI] [PubMed] [Google Scholar]
- Ganem D., Varmus H. E. The molecular biology of the hepatitis B viruses. Annu Rev Biochem. 1987;56:651–693. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- Garriga G., Bertrand H., Lambowitz A. M. RNA splicing in Neurospora mitochondria: nuclear mutants defective in both splicing and 3' end synthesis of the large rRNA. Cell. 1984 Mar;36(3):623–634. doi: 10.1016/0092-8674(84)90342-8. [DOI] [PubMed] [Google Scholar]
- Heckman J. E., Alzner-Deweerd B., RajBhandary U. L. Interesting and unusual features in the sequence of Neurospora crassa mitochondrial tyrosine transfer RNA. Proc Natl Acad Sci U S A. 1979 Feb;76(2):717–721. doi: 10.1073/pnas.76.2.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman J. E., Sarnoff J., Alzner-DeWeerd B., Yin S., RajBhandary U. L. Novel features in the genetic code and codon reading patterns in Neurospora crassa mitochondria based on sequences of six mitochondrial tRNAs. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3159–3163. doi: 10.1073/pnas.77.6.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A. M., McCahon D., Slade W. R., Newman J. W. Recombination in RNA. Cell. 1982 Jul;29(3):921–928. doi: 10.1016/0092-8674(82)90454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard K., Baltimore D. The mechanism of RNA recombination in poliovirus. Cell. 1986 Nov 7;47(3):433–443. doi: 10.1016/0092-8674(86)90600-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug R. M. The role of RNA priming in viral and trypanosomal mRNA synthesis. Cell. 1985 Jul;41(3):651–652. doi: 10.1016/S0092-8674(85)80041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper M. T., Lambowitz A. M. A novel reverse transcriptase activity associated with mitochondrial plasmids of Neurospora. Cell. 1988 Nov 18;55(4):693–704. doi: 10.1016/0092-8674(88)90228-0. [DOI] [PubMed] [Google Scholar]
- Lambowitz A. M., LaPolla R. J., Collins R. A. Mitochondrial ribosome assembly in Neurospora. Two-dimensional gel electrophoretic analysis of mitochondrial ribosomal proteins. J Cell Biol. 1979 Jul;82(1):17–31. doi: 10.1083/jcb.82.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambowitz A. M. Preparation and analysis of mitochondrial ribosomes. Methods Enzymol. 1979;59:421–433. doi: 10.1016/0076-6879(79)59103-4. [DOI] [PubMed] [Google Scholar]
- Lazzarini R. A., Keene J. D., Schubert M. The origins of defective interfering particles of the negative-strand RNA viruses. Cell. 1981 Oct;26(2 Pt 2):145–154. doi: 10.1016/0092-8674(81)90298-1. [DOI] [PubMed] [Google Scholar]
- Levens D., Ticho B., Ackerman E., Rabinowitz M. Transcriptional initiation and 5' termini of yeast mitochondrial RNA. J Biol Chem. 1981 May 25;256(10):5226–5232. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Lang B. F. Mitochondrial class II introns encode proteins related to the reverse transcriptases of retroviruses. Nature. 1985 Aug 15;316(6029):641–643. doi: 10.1038/316641a0. [DOI] [PubMed] [Google Scholar]
- Miller W. A., Bujarski J. J., Dreher T. W., Hall T. C. Minus-strand initiation by brome mosaic virus replicase within the 3' tRNA-like structure of native and modified RNA templates. J Mol Biol. 1986 Feb 20;187(4):537–546. doi: 10.1016/0022-2836(86)90332-3. [DOI] [PubMed] [Google Scholar]
- Murphy W. J., Watkins K. P., Agabian N. Identification of a novel Y branch structure as an intermediate in trypanosome mRNA processing: evidence for trans splicing. Cell. 1986 Nov 21;47(4):517–525. doi: 10.1016/0092-8674(86)90616-1. [DOI] [PubMed] [Google Scholar]
- Nargang F. E., Bell J. B., Stohl L. L., Lambowitz A. M. The DNA sequence and genetic organization of a Neurospora mitochondrial plasmid suggest a relationship to introns and mobile elements. Cell. 1984 Sep;38(2):441–453. doi: 10.1016/0092-8674(84)90499-9. [DOI] [PubMed] [Google Scholar]
- Nargang F. E. Conservation of a long open reading frame in two Neurospora mitochondrial plasmids. Mol Biol Evol. 1986 Jan;3(1):19–28. doi: 10.1093/oxfordjournals.molbev.a040375. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Osinga K. A., De Haan M., Christianson T., Tabak H. F. A nonanucleotide sequence involved in promotion of ribosomal RNA synthesis and RNA priming of DNA replication in yeast mitochondria. Nucleic Acids Res. 1982 Dec 20;10(24):7993–8006. doi: 10.1093/nar/10.24.7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osinga K. A., Tabak H. F. Initiation of transcription of genes for mitochondrial ribosomal RNA in yeast: comparison of the nucleotide sequence around the 5'-ends of both genes reveals a homologous stretch of 17 nucleotides. Nucleic Acids Res. 1982 Jun 25;10(12):3617–3626. doi: 10.1093/nar/10.12.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer P., Hohn T. Involvement of reverse transcription in the replication of cauliflower mosaic virus: a detailed model and test of some aspects. Cell. 1983 Jul;33(3):781–789. doi: 10.1016/0092-8674(83)90020-x. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Hong G. F., Hill D. F., Petersen G. B. Nucleotide sequence of bacteriophage lambda DNA. J Mol Biol. 1982 Dec 25;162(4):729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. E., Summers M. D. The bidirectional transfer of DNA and RNA to nitrocellulose or diazobenzyloxymethyl-paper. Anal Biochem. 1980 Nov 15;109(1):123–129. doi: 10.1016/0003-2697(80)90019-6. [DOI] [PubMed] [Google Scholar]
- Sprinzl M., Gauss D. H. Compilation of tRNA sequences. Nucleic Acids Res. 1984;12 (Suppl):r1–57. [PMC free article] [PubMed] [Google Scholar]
- Summers J., Mason W. S. Replication of the genome of a hepatitis B--like virus by reverse transcription of an RNA intermediate. Cell. 1982 Jun;29(2):403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- Sutton R. E., Boothroyd J. C. Evidence for trans splicing in trypanosomes. Cell. 1986 Nov 21;47(4):527–535. doi: 10.1016/0092-8674(86)90617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmus H. RNA viruses. Reverse transcription in plants? Nature. 1983 Jul 14;304(5922):116–117. doi: 10.1038/304116a0. [DOI] [PubMed] [Google Scholar]
- Weiner A. M., Deininger P. L., Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- Yin S., Heckman J., RajBhandary U. L. Highly conserved GC-rich palindromic DNA sequences flank tRNA genes in Neurospora crassa mitochondria. Cell. 1981 Nov;26(3 Pt 1):325–332. doi: 10.1016/0092-8674(81)90201-4. [DOI] [PubMed] [Google Scholar]