Introduction
Transmission of viral infections through transfusion of blood products and plasma derivatives has been known for decades but was considered as an unavoidable consequence of a life-saving treatment. Any viral infection has the potential for transmission by transfusion if it has a prolonged or even short asymptomatic blood-borne phase and if the infectious virus has the ability to survive/persist in collected blood products and to cause infection by the intravenous route. Since the introduction of laboratory testing for hepatitis B virus (HBV) surface antigen (HBsAg) in the early 70s, the risk of viral transfusion-transmitted infections (TTI) has steadily declined over the past four decades1. Alanine aminotransferase and antibody to HBV core antigen (anti-HBc) testing began in the 80s to reduce transmission of non-A, non-B viral hepatitis. Then, the advent of the human immunodeficiency virus (HIV) epidemic and the rapidly following identification of hepatitis C virus (HCV) triggered unprecedented efforts to avoid the transmission of these new blood-borne viruses, and the implementation of antibody assays for both of these agents. To exclude individuals at risk for transmissible viral infection, donor screening process has been developed and implemented that includes the medical history of the candidate donor, a brief physical examination, and serologic testing. The next critical step in reducing viral TTI was achieved with the implementation of NAT technology2.
Rapid progress and constant improvements in nucleic acid amplification technologies resulted in an unprecedented technological turning point in the molecular diagnostic and characterisation of viral infections. NAT combines the advantages of direct and highly sequence-specific detection of viral genomes (DNA or RNA) with an analytic sensitivity that is several orders of magnitude greater than that of antigen detection or virus isolation methods. Adequate specificity and high sensitivity combined with high throughput and the increasing availability of commercial assays, made NAT particularly suitable to screen blood donations for the presence of viral genomes. However, the considerable cost of NAT and challenges in automation led the users to develop strategies based on pooling of multiple donor samples2. The downside of this economic approach was reduced assay sensitivity and cases of transmission that would have been avoided by NAT applied to single unit testing. This situation led to decrease progressively the pool size from 512 to 96, 36, 24, 16, 8 and finally 6 while many blood centers opted for individual donation screening. Implementation of NAT screening also stimulated the development and the increasing use of molecular methods in transfusion laboratories in order to confirm nucleic acid testing results and to characterise viruses infecting both donors and recipients. Many blood centers used the services of external reference laboratories for confirmation of NAT yield samples or laboratories internal to large blood services dedicated to diagnostics and confirmation.
Viral nucleic acid screening in blood donations
NAT was initially introduced in transfusion settings to test whole blood and platelet apheresis donations in the late 1990s2. Then, its implementation for routine blood donation screening was successfully extended over the following years in terms of number of blood bank users worldwide and in terms of technological advances. Blood banks around the world established first minipool NAT for HCV rapidly followed by duplex testing of HIV-1 on a routine basis in 1999, followed by the introduction of parvovirus B19 (B19V) DNA testing in the plasma manufacturing sector. In 1999, reported cases of humans infected with West Nile virus (WNV) in New York City marked the onset of an epidemic that spread across the American continent3. Evidence of WNV transfusion-transmitted cases triggered the rapid development of molecular assays for blood donation screening, and by 2003, all donations were tested for WNV RNA in the USA and Canada4. Despite HBV being known as a major transfusion-transmitted virus, HBV DNA testing started to be globally implemented only after 2004 when multiplexed commercial NAT assays that included simultaneous detection of HBV, HCV and HIV-1 nucleic acids became available2. HIV-2 RNA detection has been recently included in some of these multiplex assays. Investigational NAT assays have also been developed to test donors during geographically limited outbreaks of dengue and chikungunya viruses that appear to present a low risk of transfusion transmission even in endemic areas5. In addition, a similar NAT screening approach is seriously considered by regulatory authorities with (re)-emerging viruses including hepatitis A virus (HAV), hepatitis E virus (HEV), human T-cell lymphotropic virus (HTLV-1/2) and human herpesvirus-8 (HHV-8)3. However, NAT for HTLV and HHV-8 poses additional difficulties due to these viruses being essentially cell-associated and consequently displaying barely no plasma viremia.
Over the last decade, wide-scale implementation of NAT has been supported by significant technology improvements generating high-throughput fully automated commercial platforms and assays that require less technical expertise and manpower. However, NAT implementation may be limited by the considerable cost of these new technologies, to a large extent related to patent charges, especially in resource-limited settings where it is needed most. These resource-limited settings mainly include countries of Africa, Asia, and Latin America with high prevalence of blood-borne virus infections. In addition to a high investment cost for instruments and a high running cost for reagents, a lack of maintenance support and the need for cold-chain transport and storage of reagents contribute to render NAT unaffordable in resource-limited areas. In contrast, in developed countries with usually low viral infection prevalence, NAT shows limited yield resulting in a clinical risk reduction benefit associated to an extremely low cost-effectiveness6. Consequently, economic pressure is added on blood banks regarding further implementation of any additional expensive molecular test to screen blood donations for new (re)-emerging viruses. Low cost-effectiveness instigated the development of several options intended to reduce the cost of NAT. Two non-mutually exclusive approaches have been mainly adopted. First, testing for viral genomes in plasma pools of various sizes (6 to 96 plasmas) rather than in individual donations but with the disadvantage of reducing sensitivity. An extensive head-to head comparison study evaluating the performance of the Procleix Ultrio Tigris (Chiron/Gen-Probe, Emeryville, CA, USA) and the Cobas s 201 (Roche Molecular Systems, Indianapolis, IN, USA) systems showed that individual donation (ID) testing was significantly more sensitive than testing in minipools of six donations (MP-6) in detecting HCV and HIV RNAs, but the difference in sensitivity was limited for HBV DNA7. In addition, several cases of infectious donations have been reported not detected by MP testing but reactive when tested with ID NAT8,9. The risk of false-negative result can be partially reduced in some cases by introducing additional procedures to concentrate viral particles in samples (eg. ultracentrifugation of pooled plasmas prior to nucleic acid purification, increased sample volume)10. Nevertheless, there has been a constant progression towards screening smaller pools of 6–8 plasma samples and to individual testing2. The second approach has been to successfully develop multiplex assays able to simultaneously detect, and eventually directly identify, three or more nucleic acid targets in a single reaction. Multiplexing reduces the reagent costs, the volume of sample to process, and the time required to obtain results, but at the same time considerably complicate the already multi-step and delicate methods developed for single virus nucleic acid testing11. Although single or multiplex assays initially developed in-house have been generally replaced by fully automated and relatively expensive commercial platforms/assays, they might still constitute a reliable and affordable alternative. Several proficiency testing studies, set out to examine the ability of public quality control laboratories, blood banks and plasma fractionation organizations to detect different viruses using NAT assays, reported similar performance of in-house assays compared to commercial assays12. Until recently, in-house NAT assays were still in use in Germany, Austria, and Scotland2,13,14. In Brazil, NAT testing is developing with the use of both commercial and in-house assays2. In Ghana, a new screening strategy associating pre-donation testing with serologic rapid test and post-donation in-house NAT on pools of 10 plasma samples improved transfusion safety and limited the cost of blood15. In-house assays can be more easily implemented at a fraction of the cost offered by commercial companies. However, assay quality depends on long-term procurement and supply of individual reagents (e.g. DNA polymerases, reverse transcriptases) that are not originally produced for diagnostic purposes. Any discontinuity by the manufacturer of one reagent will necessitate its replacement and the complete re-evaluation of the testing performance of the original assay, and eventually a costly and time-consuming re-optimization of the entire assay. Another limitation of the currently used NAT procedures is a lack of efficiency in detecting cell-associated transfusion-transmitted viruses like herpesviruses due to technical limitations regarding large-scale viral nucleic acid purification from whole blood.
NAT testing for the three major transfusion-transmitted viruses HBV, HCV, and HIV-1 significantly reduced the risk of transfusion-transmitted infections by reducing the diagnostic pre-seroconversion window period (WP) and by detecting immunovariant viruses. Indeed, HBV DNA testing uncovered infected, apparently healthy blood donors undetected by HBsAg testing. This condition known as occult HBV infection/carriage (OBI) is defined as a lack of reactivity with the most sensitive HBsAg assays after the WP of primary infection and the presence of very low levels of HBV DNA in circulation16. Antibodies against the HBV core (anti-HBc) are detectable in the majority of donors with OBI. The risk of transfusion-transmitted HBV infection has been widely estimated by using the incidence-window period model based on the incidence of new infection detected in repeat donors17. The introduction of a calculation method based on NAT-yield donations incidence allowed the determination of the incidence rate of infection in all donations, and it included both WP and occult carriage in the calculation of the residual risk17,18. Transfusion-transmission risk has been also calculated from clinical cases in Japan19. The residual risk of transfusion-transmitted infections with NAT in place along with serologic testing has been estimated to be below 1 per million donations for HCV and HIV-1 in most developed countries, and 0.69–8.69, 7.5–15.8, and 30.6–200 per million donations for HBV in areas of low, moderate, and high endemicity, respectively20,21. However, the recent introduction of the Ultrio Plus NAT assay (Novartis Diagnostics, Emeryville, CA, USA) with an increased 95% limit of detection of 3 IU/mL doubled the yield of both WP and OBI detection22,23.
The high analytical sensivity of NAT assays is a major factor in their ability to improve blood safety. Analytical sensivities (95% limits of detection) reported for ID testing ranged from 4.1 to 10.9 IU/mL for HBV DNA, 2.0 to 9.4 IU/mL for HCV RNA, 18.9 to 42.2 IU/mL for HIV-1 RNA, and 6.4 to 125 copies/mL for WNV RNA7,17,24. However, a recent international survey on HBV/HCV/HIV-1 NAT testing of blood donations showed that ~21% of HBsAg-confirmed positive donors depending on HBV genotypes, ~30% of anti-HCV positive donors, and ~2% of anti-HIV positive donors were tested NAT negative indicating that serological screening should be maintained even with the most sensitive NAT assay performed on individual donations2. These data, higher than those generally reported in individual studies, indicate that in the current level of sensitivity of both serological and genomic screening assays, both remain necessary to insure viral blood safety.
Confirmation of NAT screening
Discrepant results between molecular and serological testing made final interpretation difficult and confirmation process of NAT results needed. The two widely used commercial HBV/HCV/HIV-1 multiplex NAT systems (Procleix Ultrio, TIGRIS platform [Novartis Diagnostics], and cobas TaqScreen MPX, cobas s201 platform [Roche Molecular Systems]) indicate the presence of viral genomes in a sample with a single consensual signal that does not discriminate between the three viruses. Therefore, a secondary diagnostic step using three separate virus-specific amplification discriminatory assays is necessary to identify the origin of the initial test signal. These additional discriminatory assays do not qualify as confirmation assays since they are using the same methodology and the same reagents as the initial screening assays24. The two-step screening systems currently in large-scale use have introduced the difficult problem of samples reactive with the screening assay but non-reactive with the discriminatory assay. Some in-house real-time RT-qPCR-based assays have been specifically designed to provide direct, single step, identification of amplified viral genome by using specific probes labeled with distinct dyes25,26. However, nonreproducible reactivity (NRR) has been reported irrespective of the type of assay used. Different users have established different algorithms to solve this problem: repeat testing, improved assay sensitivity by increasing the nucleic acid extraction volume, and confirmation by alternative assays27. Using the cobas MPX or in-house real-time RT-qPCR assays, NRR is defined as an initial-reactive individual test that failed to react on repeat testing with the same assay. On the Procleix Ultrio system, NRR is usually defined as initially reactive, discriminatory assay-nonreactive, and repeat Ultrio-nonreactive. NRR rates of 0.1% and 0.29% have been reported for the Ultrio assay (individual donation testing) and the cobas MPX test (testing in minipools of 6 donations), respectively24.
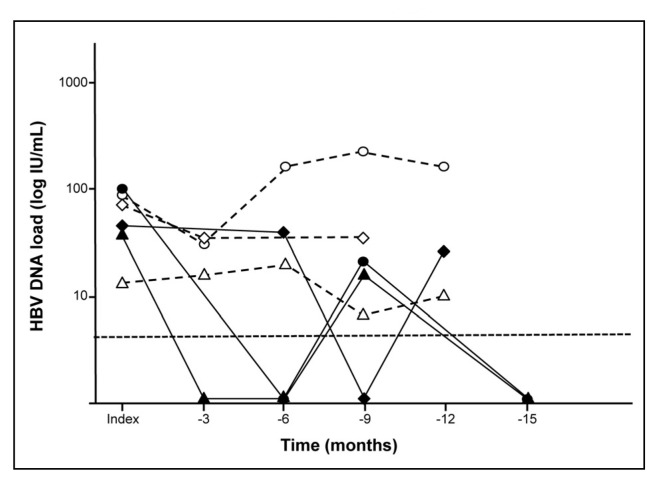
In the absence of serological evidence of infection, NRR most probably relates to the high sensitivity of NAT that increases the risk of false-positive results. Sample cross-contamination can be resolved by retesting a clean sample from the initial plasma bag. Alternatively, donor follow-up may conclusively rule out false positive NAT results, particularly for window period donors who lacked other marker of infection28. In the presence of detectable specific antibodies, lack of consistency of NAT results and false-negative results should be considered and investigated further. For instance, after introduction of individual-donation NAT screening for HBV/HCV/HIV-1, the South African National Blood Service reported the case of two anti-HIV positive donors who tested Ultrio non-reactive and Ultrio NRR, respectively29. Multiple repeats of the screening test confirmed HIV-1 infection with the first donor and the second donor being reactive in 20% (6/30 tests reactive) and 30% (9/30 reactive tests) replicates, respectively. This lack of consistency of NAT results suggested HIV-1 infection at an extremely low viral load in agreement with the Poisson distribution principle by which the chances of detecting a rare event (presence of viral genome) increases with repeating the assay. Similarly, a French study reported that MP-8 NAT and ID NAT failed to reliably detect 65% and 18% of HIV-positive individuals with HIV RNA levels persistently lower than 50 copies per mL8. Low viral loads near the detection limit of the assays may also explain the lack of repeatable HBV NAT reactivity observed in donors with occult HBV infection30. Amalgamation of published31,32 and unpublished data showed that HBV DNA presence was confirmed in the plasmas of 232 OBI donors infected with HBV genotypes A–D after viral particle concentration by ultracentrifugation. Initial HBV DNA quantification in plasma showed that 41 donors (18%) had HBV DNA below the 5 IU/mL detection limit of the quantitative PCR assay used, and 52 donor samples (22%) for whom HBV DNA was detected but viral load could not be determined precisely (<10 IU/mL). Viral load was quantified in 139 donors (60%) and ranged from 10 to 5,640 IU/mL (median: 39 IU/mL) with no significant difference between genotypes. Fifty-four donor samples had viral load <30 IU/mL. Consequently, 6-MP screening even with the most sensitive assay available may not detect ~63% of these occult HBV carriers. In addition, viral load may fluctuate over time in donors with occult infection31. As shown in Figure 1, in the first group of anti-HBc-positive OBI donors, viral DNA is detected in all samples and fluctuated between >10 IU/mL and 219 IU/mL, whereas in the second group HBV DNA was not detected in some of the samples but was successfully quantified in others (<10–99 IU/mL). Such intermittent detectability raises issues regarding blood safety as, in the absence of anti-HBc testing, a HBV-infected donor might test DNA negative in one donation and positive in the next, both being potentially infectious. Such low level HBV DNA samples needs to be tested extensively to correctly diagnose the donor status. In the case of HBV infection, it is difficult to confirm the NAT result solely on the serology results of the index donation due to the background prevalence of anti-HBc, especially in areas of high endemicity. Therefore, alternative investigational NAT approaches using real-time amplification and standard nested (RT)-PCR methods targeting different regions of the viral genome have been successfully developed as previously reviewed27.
Figure 1.
HBV DNA load detection over time in six anti-HBc-positive donors with occult HBV infection (Candotti et al. Gut 2012)31. Detection threshold is indicated by an horizontal dashed line.
Real-time (RT)-PCR assays have the advantage of showing an analytical sensitivity relatively close to that of the screening assays due in part to this method amplifying very small regions of the viral genome (60–120 nucleotides). These assays can also be quantitative when the tested sample is assessed against a standard curve obtained by testing in parallel dilutions of a calibrated reference sample. Direct amplification with nested (RT)-PCR methods can generate amplicons long enough to be sequenced. The nucleotide sequence obtained provides a definitive confirmation and identification of the presence of a viral pathogen. However, these amplification methods may lack the sensitivity needed to confirm the results of highly sensitive screening assays. The efficacy of the nested PCR assays can be increased by limiting the size of the amplified region but with the counter effect of reducing the informative value of sequencing27. Another approach is to increase the amount of nucleic acid template in the amplification reaction. This can be done by increasing the volume of plasma in the nucleic acid extraction procedure or by concentrating the amount of virus in the sample before extraction. Viral particles can be efficiently concentrated by high-speed centrifugation, affinity chromatography on heparin columns, or capture with magnetic particles33–35. However, the yield of viral particle recovery being variable, these methods are not suitable for viral load quantification.
Viral nucleic acid quantification
Determination of viral nucleic acid load in peripheral blood has become widely used in the clinical management of viral infections and to study the efficacy of antiviral therapies or the emergence of drug-resistant variants36. Consecutive assessment of the virus load is essential for both diagnosis and prognosis in patients with acute infection or with viral reactivation. Threshold levels have been defined as parameters for the initiation of antiviral therapy, for determining the efficacy and eventually the required duration of treatment. The most common genomic amplification methods used to quantify viral DNA or RNA include real-time (RT)-PCR, isothermal nucleic acid sequence-based amplification (NASBA), and branched DNA. It also provides new insights into the early dynamics of viral replication in blood donor during acute or persistent phases of infection.
Quantitative analysis of pre-seroconversion replication rates can be used to estimate the time period between infectivity and the detectability of infection by molecular and serological viral assays37. When combined with incidence rate data, such estimates can be used for risk analysis modeling of the safety level associated with various viral serological and molecular screening strategies, particularly when debating the value of implementing individual donation or mini-pool NAT screening systems.
An understanding of the relationship between viral load and infectivity during the natural course of infection is also important for evaluating the risk of transfusion-transmission. For some viruses, the stage of infection may influence whether or not a blood component will transmit infection as evidenced by retrospective analysis of transfusion-transmission cases and studies in animal experimental models18. For example, inocula from the acute phase of HBV infection seem to carry a greater infectivity (about 100-fold) in a chimeric mouse model than inocula from late acute phase or chronic phase38. In addition, data indicate that HBV transmission by blood components collected from donors in the window period is greater than 10-fold higher than transmission by blood collected from donors with occult infection. This is likely due to the higher viral load detected during the WP compared to that during late chronic phase18,38,39. Similar observation was not reported for transfusion-transmitted HIV or HCV infections in the clinical setting. The nature and the amount of blood products transfused may also impact infectivity. For instance, fresh-frozen plasma and platelet concentrates from HBV-, HCV- or HIV-infected donors suspended in 50–200 mL of plasma were 3–20 fold more infectious than red cell concentrates containing approximately 5–50 mL of plasma40–42. Quantification of viral nucleic acids in circulation may provide valuable information to evaluate the infectivity associated with individual blood components (red cells, platelets, plasma) and to understand viral compartmentalization and persistence43.
Knowing the minimal infectious dose for a given virus is essential to estimate the efficacy of strategies implemented to reduce the residual risk of transmission. Experimental animal models have been used to determine infectious dose. These models accurately quantified viral DNA or RNA but inocula may significantly differ from transfused blood components in terms of storage or handling resulting in possible enhanced or decreased infectivity combined with species susceptibility differences. Another approach is to extrapolate the minimal infectious dose from clinical evidence by determining retrospectively the viral load in donations involved in documented transfusion-transmission cases. For instance, HBV transmission was reported with products from donors in the WP phase and OBI donors showing HBV DNA load transfused <20 IU/mL21. In contrast, a linked donor-recipient study showed that blood components with B19V DNA less than 106 IU/mL are unlikely to transmit B19V infection because most recipients have circulating IG to B19V44. However, a review of clinical cases indicates that transfusion-transmission occurrence varied across recipients exposed to a given viral nucleic acid dose. There are several reported cases of absence of infection in recipients of blood components containing a viral nucleic acid load that was equivalent or exceeded the minimal infectious dose either observed in other transfused patients or calculated from animal experimental systems19,42,45,46. The lack of clear relationship between infectivity and viral load in blood components may be related to the innate immunity of the recipients that may affect susceptibility to infection, the presence of immune-modulating factors like neutralising antibodies either in the recipient or the donor(s), viral genetic factors, inaccurate estimation of the viral nucleic acid concentration in the transfused blood products, detection of nonvirion-associated nucleic acids or replication-defective virions19,21,47,48.
Molecular analysis of nucleic acid testing failure
Extremely low viral nucleic acid levels in a blood product may not be the only cause of NAT failure. Recently, six cases of non-detection of HIV-1 RNA by NAT have been reported in Germany (n =5) and Italy (n =1)49,50. Two of them involving WP donors resulted in infection of two RBCs recipients50. HIV-1 RNA load measured in these donors ranged from 650 to 200,000 IU/mL, largely exceeding the sensitivity limit of four different NAT systems initially used for screening. Comparative testing with several CE-marked and in-house NAT systems confirmed the presence of viral RNA in all samples even if the detection efficiencies varied according to assays. As viral genetic diversity may theoretically reduce or abolish primers/probes hybridization and thus affects the performance of molecular detection and/or quantification methods, the underlying viral sequences (LTR and gag) targeted by the different assays were investigated. It revealed multiple nucleotide mismatches between HIV-1 variants and the primers and probes used by the screening assays leading to false-negative results. Similar genetic polymorphisms in the LTR region have been reported to affect the detection of anti-HIV positive donations with an in-house real-time PCR-based screening assay51. Mismatches located at the 3′-end of amplification primers were shown to strongly interfere with the efficiency of both qualitative and quantitative assays. Such interference was clearly demonstrated by recovering full amplification efficiency with primers and probes modified to match the index viral sequence49,51.
The effect of genetic variability on molecular detection and viral load quantification is usually unpredictable and it is not limited to viruses described as genetically hypervariable. For decades, the genetic diversity of B19V strains was reported to be very low until two new variants were characterised. Phylogenetic analysis of B19V sequences identified three genotypes: genotype 1 that includes the original prototype B19V and two new genotypes (2 and 3) showing >9% nucleotide divergence over the whole genome52. Two subtypes (3a and 3b) were further identified within B19V genotype 353. Commercial assays were initially developed to detect and quantify B19V genotype 1 DNA and did not take into account the two new genotypes. When the performance of two commercially available NAT assays for the detection of all genotypes was evaluated, B19V genotype 1 DNA was readily detected and quantified using both assays but, one assay failed to detect any of the genotype 2 or 3 viruses, while the other detected the new genotypes although with marked difference in sensitivity according to genotype54. Sequence variations in the probe region were documented and in-house novel assays were developed to improve viral variants detection in plasma pool screening55,56.
From the reported examples of HIV-1 variants escaping detection, it appears that the risk of false-negative test result is significantly lower when more than one amplification target region is included in the screening NAT assay. All reported false-negative results were obtained with monotarget NAT assays targeting different regions of the HIV-1 genome. Detection failure has not been documented for all monotarget assays used for blood screening, but in the long term these assays might be affected by HIV-1 genetic evolution. It has been shown that the performance of dual-target assays may be also compromised in the case of failure of one of the targets50. However, it is expected that a second amplification system would be able to compensate for the failure of the other component. Therefore, the general use of dual-target NAT assays to screen blood donations for hypervariable viruses such as HIV-1 should be considered as suggested50. Theoretical rules have been established for the design of primers and probes predicting tolerance to mismatches, but assay performance should be (re)-assessed experimentally and regularly as knowledge of viral genetic drift increases.
Molecular epidemiology
Emergence of variants is particularly expected for retroviruses (HIV-1/2, HTLV), flaviviruses (HCV, WNV, Dengue), and hepdnaviruses (HBV) that are characterised by a high genetic variability resulting from the combination of a high mutation rate of the viral reverse transcriptases or the RNA-dependent RNA polymerases they used to replicate, a potential high rate of genomic recombination between viral strains, and generally a high replication rate. In addition, passive immunization and antiviral treatments contribute to selecting escape mutants. This genetic diversity results in the classification of these viruses in multiple genotypes and subtypes (or subgenotypes). Different genotypes and subtypes are prevalent in different areas of the world. Moreover, these viruses are circulating in infected individuals as a population of viral variants referred to as quasispecies.
Viral diversity can dramatically affect the performance of both serological and nucleic acid detection assays and viral load quantification as described above. The performance optimization and standard calibration of both serologic and molecular assays are mainly based on standards derived from viral genotypes that are prevalent in North America and Western Europe. However, an extensive head-to-head comparison study of two commercial NAT screening systems clearly demonstrated the existence of significant differences between NAT assays in detecting limiting dilutions of HBV, HCV, and HIV-1 strains of different genotypes7. Several studies reported false-negative detection results and underestimation of viral load using various commercial assays in individuals infected with HIV-1 non-subtype B strains, mainly subtypes A, E and G57,58.
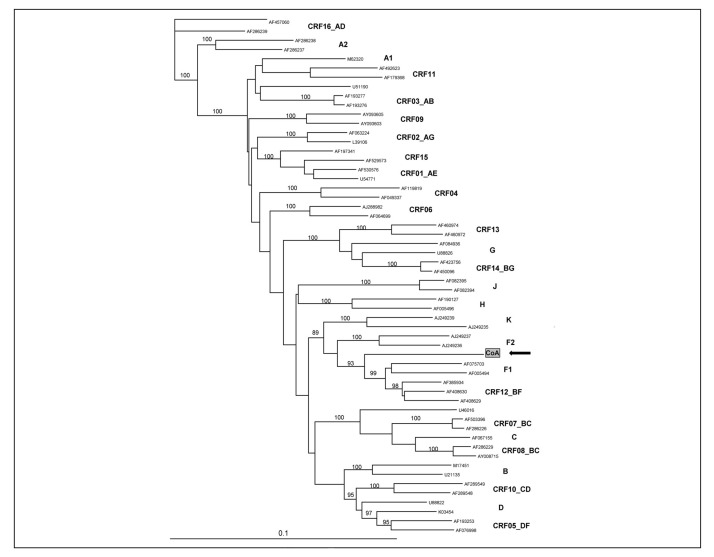
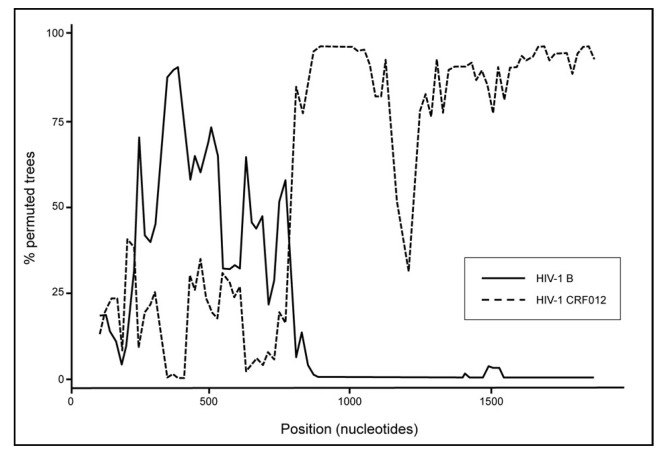
In a recent study from Italy describing a HIV-1 variant escaping NAT detection, the phylogenetic analysis of 1,700 bp spanning the gag-pol region and 670 bp in the env region (C2V4) identified a novel complex HIV-1 recombinant B/F related to the South-American circulating recombinant form CRF012 (Figure 2)49. This was further confirmed by identifying two B/F breakpoints within the gag-pol region using the SimPlot software (Figure 3). The identification of this complex HIV-1 variant in a blood donor triggered extended genetic investigation in HIV-infected patients. Subsequently, similar B/F recombinant strains were identified in 11 out of 19 HIV-1-infected patients with evidence of underestimated viral load. These results indicated that a cluster of unrelated individuals was infected with unusual B/F recombinant strains circulating in Lombardia and entering the blood system. In addition, two of eight additional patients were infected with HIV-1 recombinant forms related to CRF02_AG and CRF14_BG, and five were infected with subtype B strains. Interestingly, in the similar NAT false-negative cases reported from Germany, the limited HIV-1 sequences (242 bp) obtained from cases 4 and 5 might suggest infection with a B/F recombinant strain50. Further analysis of longer nucleotide sequences is needed to clarify this point. Nevertheless, these data obtained from blood donors confirmed the rapidly increasing prevalence of non-B subtype strains observed not only in blood donors but also in the general or exposed populations.
Figure 2.
Phylogenetic analysis of a HIV-1 strain infecting a NAT-negative blood donor (Foglieni et al. Transfusion 2011)49. Neighbour-joining tree analysis of a 1,700 bp in the gag-pol region, the donor sequence (IC) is identified with a black arrow, bootstrap values of 1,000 replicates are indicated, reference sequences from GenBank are identified by accession number, HIV-1 subtypes and circulating recombinant forms are indicated.
Figure 3.
SIMPLOT analysis of the gag-pol region of the HIV-1 strain infecting a NAT-negative blood donor (Foglieni et al. Transfusion 2011)49. The black line and the dashed line correspond to HIV-1 subtype B and circulating recombinant form CRF012, respectively.
Genotyping of blood-borne viruses detected through large-scale routine NAT and serological screening of blood donors provide insight into the origins and the dynamics of the infections. Changes in the distribution of HBV, HCV and HIV-1 genotypes in blood donor populations have been reported over the past decade. Genotypes of these viruses common in the developing world are becoming increasingly frequent in Western Europe and North America in agreement with human population migration patterns59–61. A recent survey of the characteristics of HBV infected blood donors from United Kingdom, Channel Islands and Republic of Ireland showed that 75% of new infections were associated with an endemic country (NHSBT/HPA Colindale Epidemiology Unit; www.hpa.org.uk/webc/HPAwebFile/HPAweb_C/1317130910492). Similarly, a study based on the phylogenetic analysis of B19V sequences collected from 11 countries worldwide suggested the spread of the West African genotype 3b62. Co-existence of different viral genotypes within a given population also increases the risk of inter-genotype recombination, and the emergence of recombinant forms establishing new variants should be expected63. Such new circulating recombinant forms should be incorporated into the global dimension of the viral epidemics and carefully monitored as their prevalence may increase at the expense of the parental strains as previously described for HIV-1 B/F recombinants64.
It is essential to monitor the constant genetic evolution of viruses in terms of changes in genotype distribution and emergence of variants escaping screening by using molecular epidemiologic techniques. Surveillance and/or haemovigilance programs have been implemented in most developed countries but routine analysis of viral genetic variability in blood donors remains limited5. It is also important to introduce epidemiologic observatories in developing countries where high levels of viral diversity are generally observed. In addition, molecular characterisation data obtained in blood donors showed general concordance with those in the general population or in highly exposed populations. Therefore, the monitoring of blood donors can be used as a generally valid indicator of the evolution of blood-borne virus epidemiology. Sequence data generated by molecular surveillance studies can assist in evaluating transmission risk and can be important to select screening assays or to adjust primers/probes used in NAT assays, as well as the specificity of antibodies and antigens used in serological assays.
Determining the origin of infection can be of value for infected donor management and counseling. In a recent study, transient occult HBV infection was characterised in six vaccinated blood donors. Transmission through sexual exposure was evidenced in four of these donors by phylogenetic analysis of the viral sequences65. Molecular analysis also showed that the pre-dominant genotypes in five of these donors were different from genotype A2 that is the parent genotype of the HBV vaccine. Dual infection with genotype A2 and genotype D strains was documented in two donors. Dual infection was first suggested by the presence of overlapping fluorescent base-calling peaks in the sequencing chromatogram when HBV-specific PCR products were directly sequenced. Further cloning of the PCR products and sequencing of multiple clones confirmed the concomitant presence of genotype A2 and genotype D variants in the infected donors. In addition, the analysis of the deduced amino acid sequences indicated that one donor was infected with a genotype A2 strain that was carrying a substitution (Glycine → Arginine; position 145) previously associated with immune escape. This variant was present as a minor strain among the quasispecies distribution observed in the transmitting partner. Hence, molecular characterisation of viruses infecting blood donors can provide data about the prevalence of variants carrying mutations responsible for immune escape or antiviral treatment resistance. Such information can be useful to adapt antiviral management in infected individuals and prophylactic strategies in the populations. As counterpoint, in patient care settings, the therapeutic management of viral infection is often based on sequence information of the genes coding for viral proteins targeted by drugs or passive immunization. However, the use of these sequence data for NAT or serological assays design or improvement remains limited. For instance, it has been shown that lamivudine treatment may select mutations in the HBV pol gene that also affect the overlapping S gene resulting in amino acid changes in the HBsAg and detection failure with screening assays. Collaborative effort to create and share databases of newly identified virus sequences might be of mutual benefit for blood product suppliers, patient care and public health institutions, and assay manufacturers.
Identification of transfusion-transmitted viral infections
Monitoring and characterising transfusion-transmitted infection (TTI) is essential to evaluate (1) the safety of the blood supply, (2) the impact of new testing strategies and their associated residual risk, and (3) transmissibility by transfusion of (re)-emerging viruses and associated threat to blood safety. Suspected cases of transfusion transmission are usually reported to the blood supplier and trace-back procedures are initiated to identify the implicated donor. In addition, identification of a recent infection in a repeat donor can initiate look-back investigations in recipients of potentially infectious previous donations from the implicated donor. The main limitations of assessing TTI are the difficulties in identifying infection in recipients post-transfusion and to confirm the source of infection because a pre-transfusion sample is usually unavailable.
Acute viral infection in recipients may not be recognised due to a lack of clinical evidence and proper diagnosis66,67. Diagnosis might be missed due to (1) acute infection being often asymptomatic (HBV, HCV) or associated with mild unspecific symptom (HIV) irrespective of the amount of virus transfused, (2) unusual prolonged serologic window period in recipients with an impaired immune system or receiving passive or active immunotherapy, and (3) the ~50% mortality rate within 6–12 months transfusion reported in recipients68,69. Trace-back and look-back procedures also require archived donation samples that can be paired with recipient follow-up samples. However, repository of frozen samples from both donors and recipients are rare, sample volume is usually limited, and donors or recipients cannot be traced and/or may not be willing to provide samples.
In the absence of recipient pre-transfusion sample free of viral markers, definitive evidence of transfusion transmission can only be obtained by the genomic analysis of the viral strain(s) present in both donor and recipient. In several studies, transfusion transmission was proven by sequence data showing high genetic similarity between viral sequences obtained in both donor and recipient. Transmission is proven when donor and recipient sequences are identical or show >99% homology as previously reported. HIV-1 transmission by transfusion of red blood cells was verified by molecular analysis showing that viral sequences obtained for donor and recipient were identical over 1,188 nucleotides in the pol region and showed only one mismatch (99.7% identity) over 493 nucleotides in the hypervariable env region10. Similarly, identical partial sequences of two distinct regions (core and HVR1) of the HCV genome and nearly identical HBV whole genome sequences (2 mismatches over 3,200 nucleotides) for donor and recipient supported transfusion transmission of HCV and HBV infections, respectively70,71. Transfusion-transmitted HEV infection has also been proven on the basis of a strict sequence homology between donor and recipient strains72,73. Efficiency of viral genome sequencing in identifying transfusion transmission has been further evidenced in a recent study reporting HBV infection in two transfused newborns74. Symptomatic acute HBV infection in a newborn recipient of a quarter of a regular red cell concentrate and his mother triggered look-back investigations that identified a second unrelated newborn recipient of the same RCC to be asymptomatically HBV infected. Serologic and molecular investigations in archived and follow-up samples suggested that the donor was in the window period at the time of the index donation with low level of HBV DNA. Unfortunately, no viral DNA retrieved from the donor was available for sequencing. However, full-length genome sequence identity between HBV strains from the two unrelated newborn recipients and the mother strongly supported transfusion transmission and an unusual reverse vertical transmission from newborn to mother.
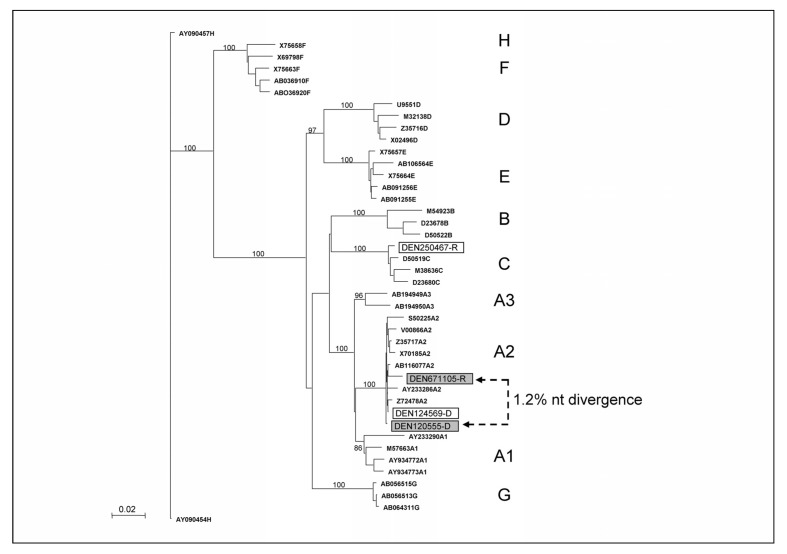
Complete sequence identity between viral strains infecting donor and recipient is not always observed. Due to the generally long delay between the transfusion and the sample testing at recall, viral genome or quasispecies might evolve independently over time under selective pressure in both donor and recipient75. The possibility of errors made by polymerases/reverse transcriptases during the genomic amplification and sequencing procedures may also be considered as suggested in a recent study assessing a B19V transmission76. Despite the endogenous genetic variability of some viruses, evolutionary relationship can be inferred among various viral species infecting different individuals by molecular phylogenetic analysis. However, molecular phylogenies depend on the assumptions and models utilised. Consequently, strikingly different results may be obtained by applying different models to the same dataset77. Nevertheless, phylogenetic comparison of donor and recipient sequences is extensively used to document transfusion transmission. Phylogenetic trees showing sequences from both donor and recipient clustering together within the same genotype or subgenotype clade indicate transfusion transmission10,71,74,78. Transmission is further supported by the intra-group nucleotide divergence of donor and recipient not differing significantly from each other or from the donor-recipient intergroup divergence, and donor-recipient intra-group divergence being significantly lower than intra-group divergence for unrelated control sequences. Defining a significant level of nucleotide divergence can be challenging, especially for viruses showing limited if any inter-strain sequence variability (e.g. B19V, herpesviruses). For instance, transfusion of a HBV window period donation triggered look-back investigations eight months later that identified a living recipient with HBV infection. Molecular analysis showed that both donor and recipient were infected with HBV genotype A2 but the viral sequences clustered separately within clade A2 and showed ~1.2% nucleotide divergence (Figure 4). In the absence of a recipient pre-transfusion sample, it was not possible to determine whether donor and recipient were infected either with unrelated HBV genotype A2 strains, or with the same strain that evolved separately in the two hosts over an eight-month period, and transfusion transmission remained uncertain (unpublished data).
Figure 4.
Neighbour-joining tree analysis of HBV sequences (1,600 nucleotides) of HBV strains infecting two donor-recipient pairs (DEN124569-D/DEN250467-R and DEN120555-D/DEN671105-R). Bootstrap values of 1,000 replicates are indicated, reference HBV sequences from GenBank are identified by accession number, HBV genotypes are indicated.
Post-transfusion infection is not necessarily transfusion-transmitted and iatrogenic sources of infection should be systematically investigated before concluding that infected blood donors are involved in viral transmission especially in high endemicity areas21. In a recent look-back procedure, HBV infection was detected in a patient who received blood components from a donor with occult HBV infection. As shown in Figure 4, the phylogenetic analysis showed that donor and recipient were infected with HBV strains of different genotypes (A2 and C), therefore excluding transfusion as the source of infection in the recipient (Allain et al. submitted). In addition, approximately 50% of recipients of blood components in Western Europe present some degree of immunodeficiency79. Among them are patients receiving highly immunosuppressive chemotherapy for haematological malignancies and who often need multiple transfusions and may be exposed to a large number of donors over the course of their treatment. Consequently, TTI is commonly suspected in patients developing symptoms of viral infections. However, reactivation of persistent viral infections has been largely documented in immunocompromised individuals80. For example, phylogenetic analysis was successfully used to characterise HBV reactivation in an HBsAg negative multi-transfused patient treated with Fludarabine who had recovered from acute infection over 30 years earlier75. Such case suggests that molecular analysis might provide a differential diagnosis between transfusion transmission and reactivation that is critical for medical and legal reasons. Patients should be screened for viral markers in advance of instituting immunosuppressive therapy and an archive sample should be retained.
Prospective testing of donor-recipient pairs remains extremely limited since it requires a large number of patients, allocation of considerable resources, and it might be considered unethical. However, such approach might be of interest to estimate the risk of transfusion-transmitted infection related to (re)-emerging viruses in order to preserve the confidence of both clinicians and patients in the safety of blood transfusion, and to provide regulators with clinical evidence to determine whether new costly testing measures and exclusion of positive donations should be implemented. For instance, there is an ongoing debate about the status of HHV-8. While HHV-8 transmission through exposure to blood and plasma-derived products is biologically plausible and has been convincingly demonstrated under epidemiologic and transfusion conditions present in Africa (absence of leukodepletion), the risk in low-prevalence Western countries appears significantly lower81–83. HHV-8 transfusion transmission has been mainly based on seroconversion in recipients transfused with seropositive products with or without detectable HHV-8 DNA. However, due to the absence of viral DNA in recipients, no direct link could be established with the donor strain, and acquisition of HHV-8 from other background sources should be considered especially in high-prevalence areas. In a large study in the US, a transfusion recipient seroconverted despite having not received any HHV-8-seropositive blood units, suggesting that the infection was not related to transfusion81. Recently, Qu and colleagues failed to find HHV-8 DNA with a sensitive PCR in a donor population with a 7.3% seroprevalence, suggesting that infected cells may be infrequent in healthy contemporary seropositive donors84. These data suggest that, even if units collected from donors with unusually high viraemia were capable of transmitting HHV-8 to transfusion recipients, such a transmission risk would represent a rare event that may not warrant implementation of safety measures at least in low-prevalence areas. In recent years, studies have found asymptomatic HEV viremia in donor blood suggesting asymptomatic infection and a potential risk for transmission of HEV through blood products85. Transfusion transmission of HEV from donor to recipient has been documented by serology and molecular methods73,86. However, the infection has been asymptomatic or presented as a mild self-limiting illness. At present, this absence of significant morbidity in most recipients does not support universal screening for this virus. However, HEV NAT was implemented in 2006 in Hokkaido, Japan, an endemic area for HEV infection87. In endemic areas, screening for immunocompromised recipients and recipients at risk of hepatic failure or chronicity might be considered86. Similarly, although transfusion-transmission of the recently discovered parvovirus 4 (PARV4) has been reported with pooled plasma derivatives, no clear pathogenicity or particular susceptible population has been identified to support large-scale blood donation testing88.
Conclusions
Over the past decades, molecular virology methods have been increasingly introduced in blood services resulting in a significant improvement of blood safety. The introduction of nucleic acid testing-based laboratory methods to screen blood donations dramatically reduced the risk of the most common transfusion-transmitted infections HIV-1, HCV, and to a lesser extent HBV. Molecular assays can be rapidly developed in case of emerging epidemic crisis involving new pathogens considered potential threat for blood safety. Due to its high sensitivity and specificity, NAT efficiently complements serologic testing in detecting pre-seroconversion window period infections, infections with immunovariant viruses, and occult/persistent viral carriage. However, the existence of seropositive but NAT-negative donations indicates that serological testing should be maintained even with the most sensitive NAT assay performed on individual donation. Confirmation of the initial screening result requires the development of alternative molecular detection assays with sensitivity equal or higher than the screening assay. In addition, viral sequence characterisation is increasingly used for definitive confirmation.
Furthermore, viral sequence characterisation is increasingly used in the transfusion setting not only to provide confirmation of NAT screening results, but also for epidemiologic surveillance and monitoring of viral genetic diversity in blood donors. Surveillance of the evolution of viral diversity in the blood donor population is essential to evaluate the future geographical changes in viral variant prevalence and to continuously assess and improve the performance of the screening assays to ensure blood safety. Surveillance of transfusion-transmissible infections in blood donors as a sentinel group can provide large databases for the active surveillance of viral infections and the evaluation of risk factors for infections in a healthy low-risk population that may benefit health of the general community. The ability to rapidly test large numbers of blood samples for viral nucleic acids in individuals of different age groups and geographic origins may be particularly useful to identify and evaluate the potential threat related to emerging blood-borne pathogens.
Despite the decrease of residual risk provided by NAT testing, several cases of transfusion-transmitted infections have been reported. Identifying clearly the source of infection in the infected recipient is essential to estimate blood safety and for medical and legal reasons. Infection in transfused patients is not necessarily transfusion-transmitted. Iatrogenic sources of infection and viral reactivation in patients previously exposed should be systematically investigated before concluding that infected blood products are responsible for viral transmission. Genomic analysis of the viral strains found in both donor and recipient is required to differentiate between transfusion-transmission and viral reactivation or external infection.
Facing the continuing (re)-emergence of new viruses and variants of known viruses, transfusion safety remains a major driving force for the development of technically innovative and challenging new molecular technologies applicable not only to blood screening but also to diagnosis of transmissible viruses.
Footnotes
Presented in part at the 40° Convegno Nazionale di Studi di Medicina Trasfusionale (Rimini, Italy, 23–26 May 2012).
The Authors declare no conflicts of interest.
References
- 1.Comanor L, Holland P. Hepatitis B virus blood screening: unfinished agendas. Vox Sang. 2006;91:1–12. doi: 10.1111/j.1423-0410.2006.00773.x. [DOI] [PubMed] [Google Scholar]
- 2.Roth WK, Busch MP, Schuller A, et al. International survey on NAT testing of blood donations: expanding implementation and yield from 1999 to 2009. Vox Sang. 2012;102:82–90. doi: 10.1111/j.1423-0410.2011.01506.x. [DOI] [PubMed] [Google Scholar]
- 3.Stramer SL, Hollinger FB, Katz LM, et al. Emerging infectious disease agents and their potential threat to transfusion safety. Transfusion. 2009;49(Suppl 2):1S–29S. doi: 10.1111/j.1537-2995.2009.02279.x. [DOI] [PubMed] [Google Scholar]
- 4.Busch MP, Tobler LH, Saldanha J, et al. Analytical and clinical sensitivity of West Nile virus RNA screening and supplemental assays available in 2003. Transfusion. 2005;45:492–9. doi: 10.1111/j.0041-1132.2005.04382.x. [DOI] [PubMed] [Google Scholar]
- 5.O’Brien SF, Zou S, Laperche S, et al. Surveillance of transfusion-transmissible infections: comparison of systems in five developed countries. Transfus Med Rev. 2012;26:38–57. doi: 10.1016/j.tmrv.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Busch M, Walderhaug M, Custer B, et al. Risk assessment and cost-effectiveness/utility analysis. Biologicals. 2009;37:78–87. doi: 10.1016/j.biologicals.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 7.Assal A, Barlet V, Deschaseaux M, et al. Comparison of the analytical and operational performance of two viral nucleic acid test blood screening systems: Procleix Tigris and cobas s 201. Transfusion. 2009;49:289–300. doi: 10.1111/j.1537-2995.2008.01965.x. [DOI] [PubMed] [Google Scholar]
- 8.Laperche S, Morel P, Deschaseaux M, et al. HIV antibody screening remains indispensable for ensuring viral safety of blood components despite NAT implementation. Transfusion. 2003;43:1428–32. doi: 10.1046/j.1537-2995.2003.00541.x. [DOI] [PubMed] [Google Scholar]
- 9.Nübling CM, Heiden M, Chudy M, et al. Experience of mandatory nucleic acid testing (NAT) screening across all blood organizations in Germany: NAT yield versus breakthrough transmissions. Transfusion. 2009;49:1850–8. doi: 10.1111/j.1537-2995.2009.02212.x. [DOI] [PubMed] [Google Scholar]
- 10.Schmidt M, Korn K, Nübling CM, et al. First transmission of human immunodeficiency virus type 1 by a cellular blood product after mandatory nucleic acid screening in Germany. Transfusion. 2009;49:1836–44. doi: 10.1111/j.1537-2995.2009.02203.x. [DOI] [PubMed] [Google Scholar]
- 11.Candotti D, Allain JP. The utility of multiplex NAT in blood screening. Dev Biol (Basel) 2007;127:71–86. [PubMed] [Google Scholar]
- 12.Fryer JF, Baylis SA, Gottlieb AL, et al. Development of working reference materials for clinical virology. J Clin Virol. 2008;43:367–71. doi: 10.1016/j.jcv.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 13.Roth WK, Seifried E. The German experience with NAT. Transfus Med. 2002;12:255–8. doi: 10.1046/j.1365-3148.2002.00383.x. [DOI] [PubMed] [Google Scholar]
- 14.Jarvis LM, Simmonds P. Scottish experience with NAT. Transfus Med. 2002;12:259–64. doi: 10.1046/j.1365-3148.2002.00384.x. [DOI] [PubMed] [Google Scholar]
- 15.Owusu-Ofori S, Temple J, Sarkodie F, et al. Predonation screening of blood donors with rapid tests: implementation and efficacy of a novel approach to blood safety in resource-poor settings. Transfusion. 2005;45:133–40. doi: 10.1111/j.1537-2995.2004.04279.x. [DOI] [PubMed] [Google Scholar]
- 16.Raimondo G, Allain JP, Brunetto MR, et al. Statements from the Taormina expert meeting on occult hepatitis B virus infection. J Hepatol. 2008;49:652–7. doi: 10.1016/j.jhep.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 17.Busch MP, Glynn SA, Stramer SL, et al. A new strategy for estimating risks of transfusion-transmitted viral infections based on rates of detection of recently infected donors. Transfusion. 2005;45:254–64. doi: 10.1111/j.1537-2995.2004.04215.x. [DOI] [PubMed] [Google Scholar]
- 18.Kleinman SH, Lelie N, Busch MP. Infectivity of human immunodeficiency virus-1, hepatitis C virus, and hepatitis B virus and risk of transmission by transfusion. Transfusion. 2009;49:2454–89. doi: 10.1111/j.1537-2995.2009.02322.x. [DOI] [PubMed] [Google Scholar]
- 19.Satake M, Taira R, Yugi H, et al. Infectivity of blood components with low hepatitis B virus DNA levels identified in a lookback program. Transfusion. 2007;47:1197–205. doi: 10.1111/j.1537-2995.2007.01276.x. [DOI] [PubMed] [Google Scholar]
- 20.Zou S, Dorsey KA, Notari EP, et al. Prevalence, incidence, and residual risk of human immunodeficiency virus and hepatitis C virus infections among United States blood donors since the introduction of nucleic acid testing. Transfusion. 2010;50:1495–504. doi: 10.1111/j.1537-2995.2010.02622.x. [DOI] [PubMed] [Google Scholar]
- 21.Candotti D, Allain JP. Transfusion-transmitted hepatitis B virus infection. J Hepatol. 2009;51:798–809. doi: 10.1016/j.jhep.2009.05.020. [DOI] [PubMed] [Google Scholar]
- 22.Tsoi WC, Lelie N, Lin CK. Enhanced HBV detection in Hong Kong blood donors with the second generation triplex transcription mediated amplification assay. Vox sang. 2012;103(Suppl 1):P-296. [Google Scholar]
- 23.Vermeulen M, Coleman C, Lelie N, et al. Ehancement of HBV NAT yield rate after introduction of the Ultrio Plus assay. Vox Sang. 2012;103(Suppl 1):P-295. [Google Scholar]
- 24.Margaritis AR, Brown SM, Seed CR, et al. Comparison of two automated nucleic acid testing systems for simultaneous detection of human immunodeficiency virus and hepatitis C virus RNA and hepatitis B virus DNA. Transfusion. 2007;47:1783–93. doi: 10.1111/j.1537-2995.2007.01343.x. [DOI] [PubMed] [Google Scholar]
- 25.Candotti D, Temple J, Owusu-Ofori s, Allain JP. Multiplex real-time quantitative RT-PCR assay for hepatitis B virus, hepatitis C virus, and human immunodeficiency virus type 1. J Virol Methods. 2004;118:39–47. doi: 10.1016/j.jviromet.2004.01.017. [DOI] [PubMed] [Google Scholar]
- 26.Mine H, Emura H, Miyamoto M, et al. High throughput screening of 16 million serologically negative blood donors for hepatitis B virus, hepatitis C virus and human immunodeficiency virus type-1 by nucleic acid amplification testing with specific and sensitive multiplex reagent in Japan. J Virol Methods. 2003;112:145–51. doi: 10.1016/s0166-0934(03)00215-5. [DOI] [PubMed] [Google Scholar]
- 27.Allain JP, Candotti D. Diagnostic algorithm for HBV safe transfusion. Blood Trans. 2009;7:174–82. doi: 10.2450/2008.0062-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L, Chen PJ, Chen MH, et al. A pilot study for dcreening blood donors in Taiwan by nucleic acid amplification technology: detecting occult hepatitis B virus infections and closing the serologic window period for hepatitis C virus. Transfusion. 2008;48:1198–1206. doi: 10.1111/j.1537-2995.2008.01672.x. [DOI] [PubMed] [Google Scholar]
- 29.Vermeulen M, Lelie N, Sykes W, et al. Impact of individual-donation nucleic acid testing on risk of human immunodeficiency virus, hepatitis B virus, and hepatitis C virus transmission by blood transfusion in South Africa. Transfusion. 2009;49:1115–25. doi: 10.1111/j.1537-2995.2009.02110.x. [DOI] [PubMed] [Google Scholar]
- 30.Stramer SL, Krysztof DE, Brodsky JP, et al. Sensitivity comparison of two Food and Drug Administration-licensed, triplex nucleic acid test automated assays for hepatitis B virus DNA detection and associated projections of United States yield. Transfusion. 2011;51:2012–22. doi: 10.1111/j.1537-2995.2011.03140.x. [DOI] [PubMed] [Google Scholar]
- 31.Candotti D, Lin CK, Belkhiri D, et al. Occult hepatitis B infection in blood donors from South East Asia: molecular characterization and potential mechanisms of occurrence. Gut. 2012 doi: 10.1136/gutjnl-2011-301281. gutjnl-2011-301281. [DOI] [PubMed] [Google Scholar]
- 32.Candotti D, Grabarczyk P, Ghiazza P, et al. Characterization of occult hepatitis B virus from blood donors carrying genotype A2 or genotype D strains. J Hepatol. 2008;49:537–47. doi: 10.1016/j.jhep.2008.04.017. [DOI] [PubMed] [Google Scholar]
- 33.Roth WK, Weber M, Seifried E. Feasibility and efficacy of routine PCR screening of blood donations for hepatitis C virus, hepatitis B virus, and HIV-1 in a blood-bank setting. Lancet. 1999;353:359–63. doi: 10.1016/S0140-6736(98)06318-1. [DOI] [PubMed] [Google Scholar]
- 34.Zahn A, Allain JP. Hepatitis C virus and hepatitis B virus bind to heparin: purification of largely IgG-free virions from infected plasma by heparin chromatography. J Gen Virol. 2005;86:677–85. doi: 10.1099/vir.0.80614-0. [DOI] [PubMed] [Google Scholar]
- 35.Satoh K, Iwata-Takakura A, Yoshikawa A, et al. A new method of concentrating hepatitis B virus (HBV) DNA and HBV surface antigen: an application of the method to the detection of occult HBV infection. Vox sang. 2008;95:174–80. doi: 10.1111/j.1423-0410.2008.01091.x. [DOI] [PubMed] [Google Scholar]
- 36.Berger A, Preiser W. Viral genome quantification as a tool for improving patient management: the example of HIV, HBV, HCV and CMV. J Antimicrob Chemother. 2002;49:713–21. doi: 10.1093/jac/dkf050. [DOI] [PubMed] [Google Scholar]
- 37.Fiebig EW, Wright DJ, Rawaal BD, et al. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS. 2003;17:1871–79. doi: 10.1097/00002030-200309050-00005. [DOI] [PubMed] [Google Scholar]
- 38.Tabuchi A, Tanaka J, Katayama K, et al. Titration of hepatitis B virus infectivity in the sera of pre-acute and late acute phases of HBV infection: transmission experiments to chimeric mice with human liver repopulated hepatocytes. J Med Virol. 2008;80:2064–8. doi: 10.1002/jmv.21320. [DOI] [PubMed] [Google Scholar]
- 39.Yoshikawa A, Gotanda Y, Minegishi K, et al. Lengths of hepatitis B viremia and antigenemia in blood donors: preliminary evidence of occult (hepatitis b surface antigen-negative) infection in the acute phase. Transfusion. 2007;47:1162–71. doi: 10.1111/j.1537-2995.2007.01234.x. [DOI] [PubMed] [Google Scholar]
- 40.Schüttler CG, Caspari G, Jursch CA, et al. Hepatitis C transmission by a blood donation negative in nucleic acid amplification tests for viral RNA. Lancet. 2000;355:41–2. doi: 10.1016/S0140-6736(99)04719-4. [DOI] [PubMed] [Google Scholar]
- 41.Busch MP, Tobler LH, Gerlich WH, et al. Very low level viremia in HCV infectious unit missed by NAT. Transfusion. 2003;43:1173–4. doi: 10.1046/j.1537-2995.2003.00524.x. [DOI] [PubMed] [Google Scholar]
- 42.Ferreira MC, Nel TJ. Differential transmission of human immunodeficiency virus (HIV) via blood components from an HIV-infected donor. Transfusion. 2006;46:156–7. doi: 10.1111/j.1537-2995.2006.00688.x. [DOI] [PubMed] [Google Scholar]
- 43.Lee TH, Kleinman SH, Wen L, et al. Distribution of parvovirus B19 DNA in blood compartments and persistence of virus in blood donors. Transfusion. 2011;51:1896–1908. doi: 10.1111/j.1537-2995.2010.03035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kleinman SH, Glynn SA, Lee TH, et al. A linked donor-recipient study to evaluate parvovirus B19 transmission by blood component transfusion. Blood. 2009;114:3677–83. doi: 10.1182/blood-2009-06-225706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsumoto C, Tadokoro K, Fujimura K, et al. Analysis of HBV infection after blood transfusion in Japan through investigation of a comprehensive donor specimen repository. Transfusion. 2001;41:878–84. doi: 10.1046/j.1537-2995.2001.41070878.x. [DOI] [PubMed] [Google Scholar]
- 46.Wang JT, Lee CZ, Chen PJ, et al. Transfusion-transmitted HBV infection in an endemic area: the necessity of more sensitive screening for HBV carriers. Transfusion. 2002;42:1592–7. doi: 10.1046/j.1537-2995.2002.00274.x. [DOI] [PubMed] [Google Scholar]
- 47.Levicnik-Stezinar S, Rahne-Potokar U, Candotti D, et al. Anti-HBs positive occult hepatitis B virus carrier blood infectious in two transfusion recipients. J Hepatol. 2008;48:1022–5. doi: 10.1016/j.jhep.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 48.El Chaar M, El Jisr T, Allain JP. Hepatitis B virus DNA splicing in lebanese blood donors and genotype a to e strains: implications for hepatitis B virus DNA quantification and infectivity. J Clin Microbiol. 2012;50:3159–67. doi: 10.1128/JCM.01251-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foglieni B, Candotti D, Guarnori I, et al. A cluster of human immunodeficiency virus type 1 recombinant form escaping detection by commercial genomic amplification assays. Transfusion. 2011;51:719–30. doi: 10.1111/j.1537-2995.2010.02942.x. [DOI] [PubMed] [Google Scholar]
- 50.Chudy M, Weber-Schehl M, Pichl L, et al. Blood screening nucleic acid amplification tests for human immunodeficiency virus type 1 may require two different amplification targets. Transfusion. 2012;52:431–9. doi: 10.1111/j.1537-2995.2011.03281.x. [DOI] [PubMed] [Google Scholar]
- 51.Edelmann A, Kalus U, Oltmann A, et al. Improvement of an ultrasensitive human immunodeficiency virus type 1 real-time reverse transcriptase-polymerase chain reaction targeting the long terminal repeat region. Transfusion. 2010;50:685–92. doi: 10.1111/j.1537-2995.2009.02477.x. [DOI] [PubMed] [Google Scholar]
- 52.Servant A, Laperche S, Lallemand F, et al. Genetic diversity within human erythroviruses: identification of three genotypes. J Virol. 2002;76:9124–34. doi: 10.1128/JVI.76.18.9124-9134.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parsyan A, Szmaragd C, Allain JP, Candotti D. Identification and genetic diversity of two human parvovirus B19 genotype 3 subtypes. J Gen Virol. 2007;88:428–31. doi: 10.1099/vir.0.82496-0. [DOI] [PubMed] [Google Scholar]
- 54.Baylis SA, Shah N, Minor PD. Evaluation of different assays for the detection of parvovirus B19 DNA in human plasma. J Virol Methods. 2004;121:7–16. doi: 10.1016/j.jviromet.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 55.Baylis SA, Fryer JF, Grabarczyk P. Effects of probe binding mutations in an assay designed to detect parvovirus B19: implications for the quantitation of different virus genotypes. J Virol Methods. 2007;139:97–9. doi: 10.1016/j.jviromet.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 56.Koppelman MH, Rood IG, Fryer JF, et al. Parvovirus B19 genotypes 1 and 2 detection with real-time polymerase chain reaction assays. Vox Sang. 2007;93:208–15. doi: 10.1111/j.1423-0410.2007.00957.x. [DOI] [PubMed] [Google Scholar]
- 57.Cunningham P, Marriott D, Harris C, et al. False negative HIV-1 proviral DNA polymerase chain reaction in a patient with primary infection acquired in Thailand. J Clin Virol. 2003;26:163–9. doi: 10.1016/s1386-6532(02)00115-4. [DOI] [PubMed] [Google Scholar]
- 58.Wirden M, Tubiana R, Marguet F, et al. Impact of discrepancies between the Abbott realtime and cobas TaqMan assays for quantification of human immunodeficiency virus type 1 group M non-B subtypes. J Clin Microbiol. 2009;47:1543–5. doi: 10.1128/JCM.02134-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koppelman MH, Zaaijer HL. Diversity and origin of hepatitis B virus in Dutch blood donors. J Med Virol. 2004;73:29–32. doi: 10.1002/jmv.20057. [DOI] [PubMed] [Google Scholar]
- 60.Davidson F, Lycett C, Sablon E, et al. Hepatitis B virus genotypes and precore mutations in Scottish blood donors. Vox Sang. 2005;88:87–92. doi: 10.1111/j.1423-0410.2005.00597.x. [DOI] [PubMed] [Google Scholar]
- 61.Delwart E, Slikas E, Stramer SL, et al. Genetic diversity of recently acquired and prevalent HIV, hepatitis B virus, and hepatitis C virus infections in US blood donors. J Infect Dis. 2012;205:875–85. doi: 10.1093/infdis/jir862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hübschen JM, Mihneva Z, Mentis AF, et al. Phylogenetic analysis of human parvovirus b19 sequences from eleven different countries confirms the predominance of genotype 1 and suggests the spread of genotype 3b. J Clin Microbiol. 2009;47:3735–8. doi: 10.1128/JCM.01201-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mahgoub S, Candotti D, El Ekiaby M, Allain JP. Hepatitis B virus (HBV) infection and recombination between HBV genotypes D and E in asymptomatic blood donors from Khartoum, Sudan. J Clin Microbiol. 2011;49:298–306. doi: 10.1128/JCM.00867-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hemelaar J, Gouws E, Ghys PD, Osmanov S. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS. 2006;20:W13–23. doi: 10.1097/01.aids.0000247564.73009.bc. [DOI] [PubMed] [Google Scholar]
- 65.Stramer SL, Wend U, Candotti D, et al. Nucleic acid testing to detect HBV infection in blood donors. N Engl J Med. 2011;364:236–47. doi: 10.1056/NEJMoa1007644. [DOI] [PubMed] [Google Scholar]
- 66.Soldan K, Barbara JAJ, Dow BC. Transfusion-transmitted hepatitis B virus infection in the UK: a small and moving target. Vox Sang. 2002;83:305–8. doi: 10.1046/j.1423-0410.2002.00228.x. [DOI] [PubMed] [Google Scholar]
- 67.Inaba S, Ito A, Miyata Y, et al. Individual nucleic amplification technology does not prevent all hepatitis B virus transmission by blood transfusion. Transfusion. 2006;46:2028–9. doi: 10.1111/j.1537-2995.2006.01011.x. [DOI] [PubMed] [Google Scholar]
- 68.Wendel S, Levi JE, Biagini S, et al. Aprobable case of hepatitis B virus transfusion transmission revealed after a 13-month-long window period. Transfusion. 2008;48:1602–8. doi: 10.1111/j.1537-2995.2008.01723.x. [DOI] [PubMed] [Google Scholar]
- 69.Hollinger FB, Dodd RY. Hepatitis B virus traceback and lookback: factors to consider. Transfusion. 2009;49:176–184. doi: 10.1111/j.1537-2995.2008.01961.x. [DOI] [PubMed] [Google Scholar]
- 70.Kretzschmar E, Chudy M, Nübling CM, et al. First case of hepatitis C virus transmission by a red blood cell concentrate after introduction of nucleic acid amplification technique screening in Germany: a comparative study with various assays. Vox Sang. 2007;92:297–301. doi: 10.1111/j.1423-0410.2007.00903.x. [DOI] [PubMed] [Google Scholar]
- 71.Niederhauser C, Weingand T, Candotti D, et al. Fatal outcome of a hepatitis B virus transfusion-transmitted infection. Vox Sang. 2010;98:504–7. doi: 10.1111/j.1423-0410.2009.01304.x. [DOI] [PubMed] [Google Scholar]
- 72.Tamura A, Shimizu YK, Tanaka T, et al. Persistent infection of hepatitis E virus transmitted by blood transfusion in a patient with T-cell lymphoma. Hepatol Res. 2007;37:113–20. doi: 10.1111/j.1872-034X.2007.00024.x. [DOI] [PubMed] [Google Scholar]
- 73.Haïm-Boukobza S, Ferey MP, Vetillard AL, et al. Transfusion-transmitted hepatitis E in a misleading context of autoimmunity and drug-induced toxicity. J Hepatol. 2012 doi: 10.1016/j.jhep.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 74.Niederhauser C, Candotti D, Weingand T, et al. Reverse vertical transmission of hepatitis B virus (HBV) infection from a transfusion-infected newborn to her mother. J Hepatol. 2012;56:734–7. doi: 10.1016/j.jhep.2011.07.034. [DOI] [PubMed] [Google Scholar]
- 75.Power JP, El Chaar M, Temple J, et al. HBV reactivation after fludarabine chemotherapy identified on investigation of suspected transfusion-transmitted hepatitis B virus. J Hepatol. 2010;53:780–7. doi: 10.1016/j.jhep.2010.04.034. [DOI] [PubMed] [Google Scholar]
- 76.Servant-Delmas A, Laperche S, Mercier M, et al. Limits of sequencing and phylogenetic analysis to assess B19V transmission by single-donor blood component. Vox Sang. 2011;100:254–5. doi: 10.1111/j.1423-0410.2010.01390.x. [DOI] [PubMed] [Google Scholar]
- 77.Philippe H, Brinkmann H, Lavrov DV, et al. Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biology. 2011 doi: 10.1371/journal.pbio.1000602.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vermeulen M, Dickens C, Lelie N, et al. Hepatitis B virus transmission by blood transfusion during 4 years of individual-donation nucleic acid testing in South Africa: estimated and observed window period risk. Transfusion. 2012;52:880–92. doi: 10.1111/j.1537-2995.2011.03355.x. [DOI] [PubMed] [Google Scholar]
- 79.Llewelyn CA, Wells AW, Amin M, et al. The EASTR study: a new approach to determine the reasons for transfusion in epidemiological studies. Transfus Med. 2009;19:89–98. doi: 10.1111/j.1365-3148.2009.00911.x. [DOI] [PubMed] [Google Scholar]
- 80.Liang R. How I treat and monitor viral hepatitis B infection in patients receiving intensive immunosuppressive therapies or undergoing hematopoietic stem cell transplantation. Blood. 2009;113:3147–53. doi: 10.1182/blood-2008-10-163493. [DOI] [PubMed] [Google Scholar]
- 81.Cannon MJ, Operskalski EA, Mosley JW, et al. J Infect Dis. 2009;199:1592–8. doi: 10.1086/598859. [DOI] [PubMed] [Google Scholar]
- 82.Vamvakas EC. Is human herpesvirus-8 transmitted by transfusion? Transfus Med Rev. 2010;24:1–14. doi: 10.1016/j.tmrv.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 83.Gobbini F, Owusu-Ofori S, Marcelin AG, et al. Human herpesvirus 8 transfusion transmission in Ghana, an endemic region of West Africa. Transfusion. 2012 doi: 10.1111/j.1537-2995.2012.03607.x.. [DOI] [PubMed] [Google Scholar]
- 84.Qu L, Triulzi DJ, Rowe DT, Jenkins FJ. Detection of HHV-8 (human herpesvirus-8) genomes in induced peripheral blood mononuclear cells (PBMCs) from US blood donors. Vox Sang. 2011;100:267–71. doi: 10.1111/j.1423-0410.2010.01404.x. [DOI] [PubMed] [Google Scholar]
- 85.Sakata H, Matsubayashi K, Takeda H, et al. A nationwide survey for hepatitis E virus prevalence in Japanese blood donors with elevated alanine aminotransferase. Transfusion. 2008;48:2568–76. doi: 10.1111/j.1537-2995.2008.01910.x. [DOI] [PubMed] [Google Scholar]
- 86.Bajpai M, Gupta E. Transfusion-transmitted hepatitis E: is screening warranted? Indian J Med Microbiol. 2011;29:353–8. doi: 10.4103/0255-0857.90158. [DOI] [PubMed] [Google Scholar]
- 87.Ikeda H, Matsubayashi K, Sakata H, et al. Prevalence of hepatitis E virus infection among Japanese blood donors. ISBT Science Series. 2009;4:299–301. [Google Scholar]
- 88.Delwart E. Human parvovirus 4 in the blood supply and transmission by pooled plasma-derived clotting factors: does it matter? Transfusion. 2012;52:1398–403. doi: 10.1111/j.1537-2995.2012.03721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]