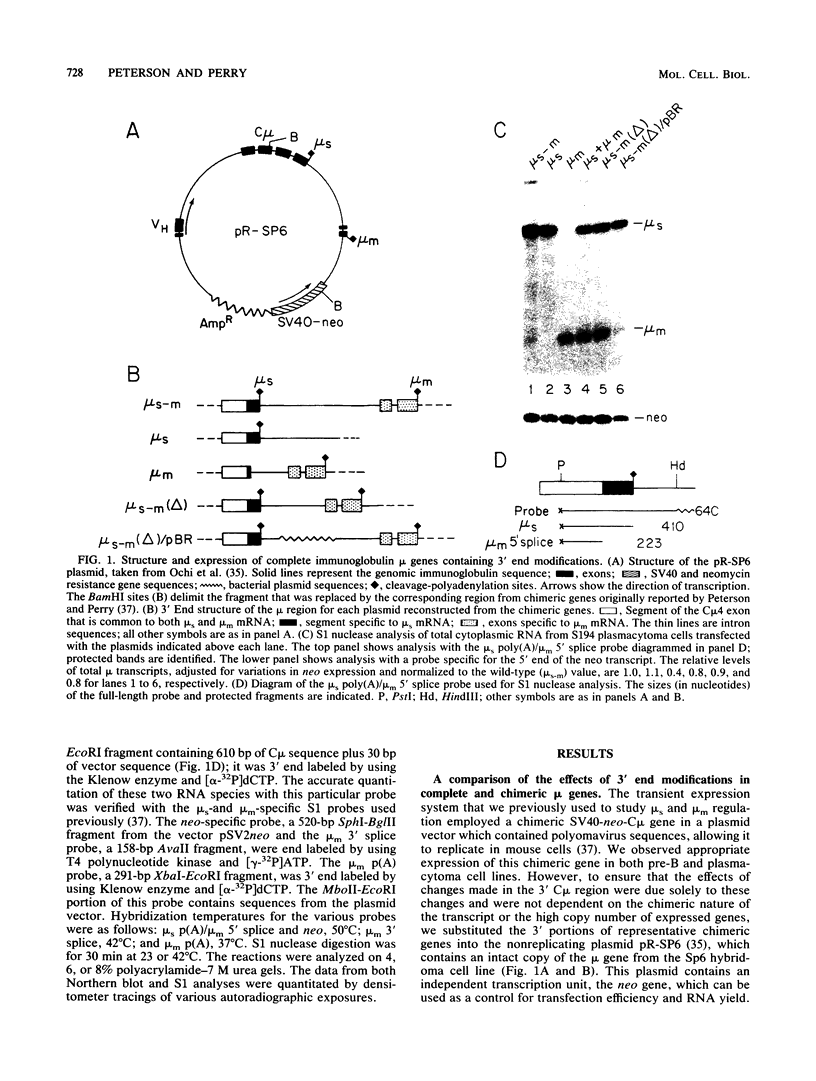
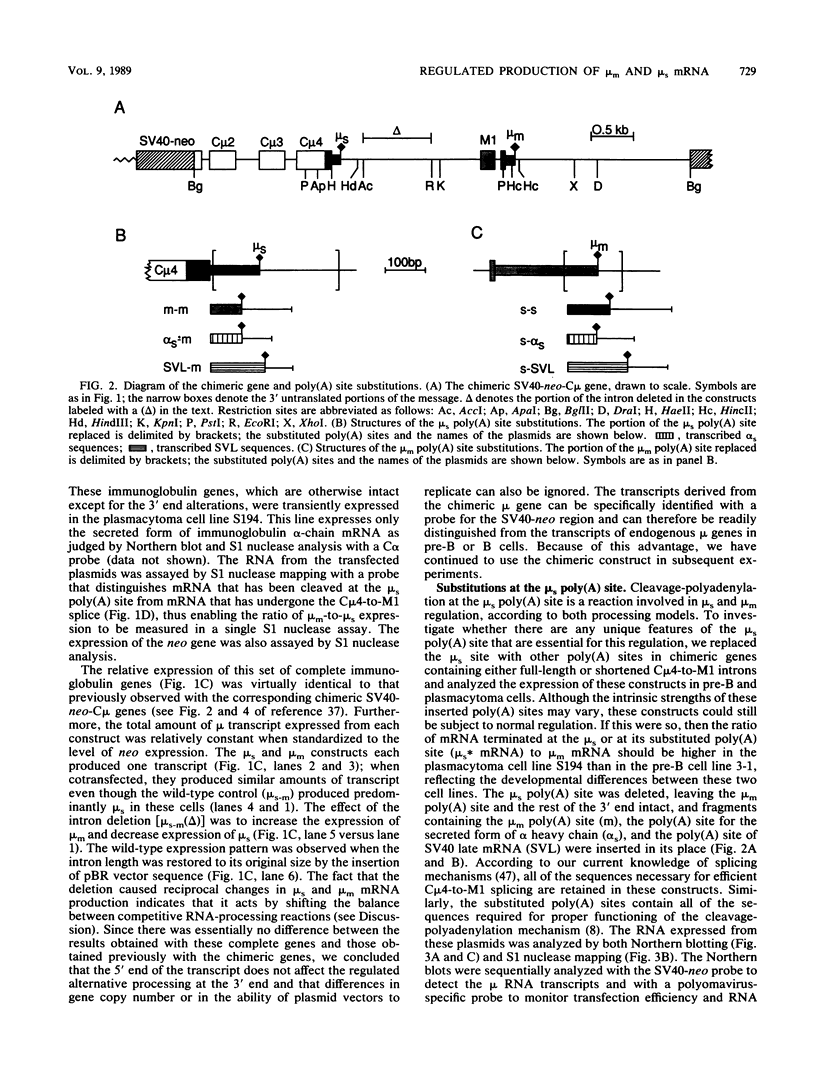
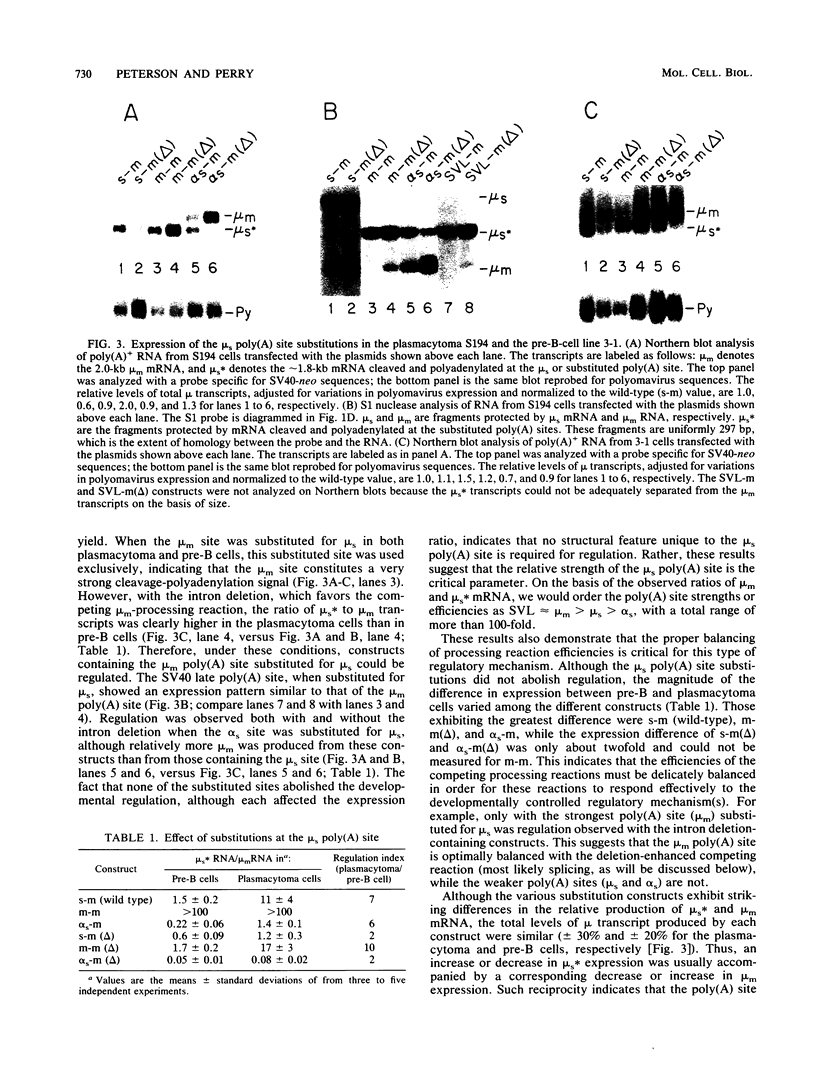
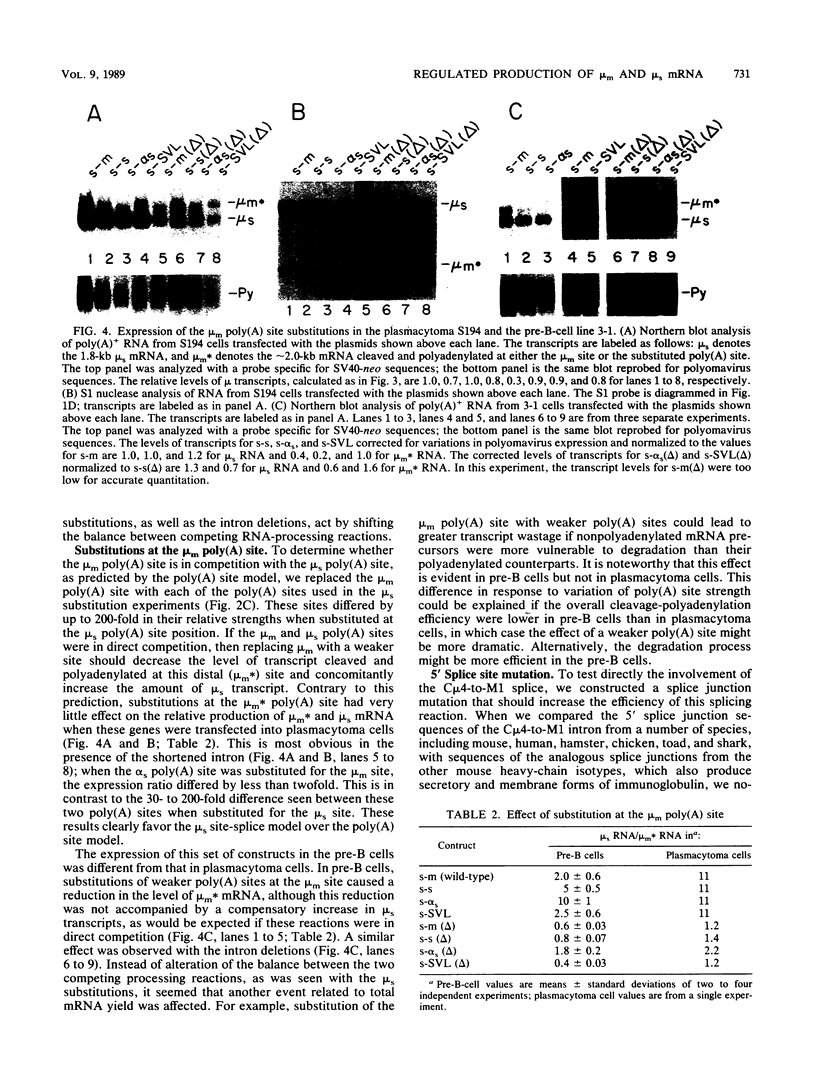
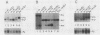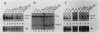Abstract
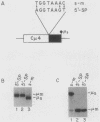
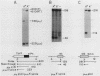
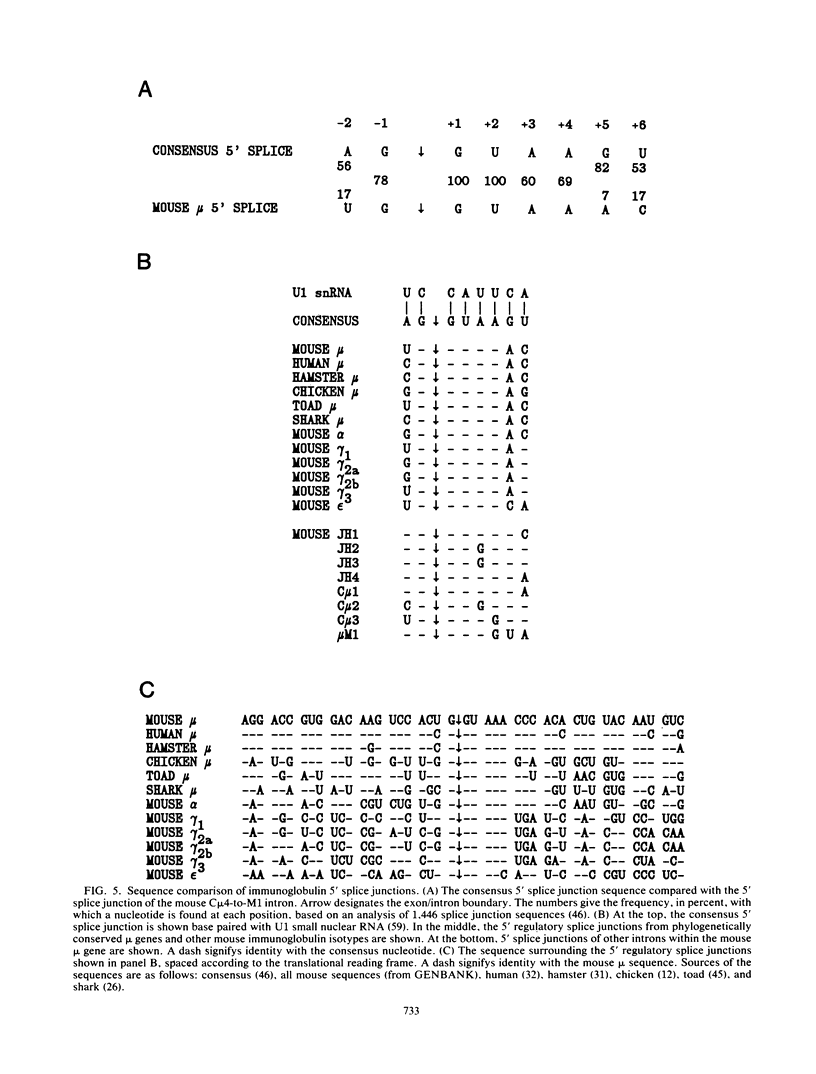
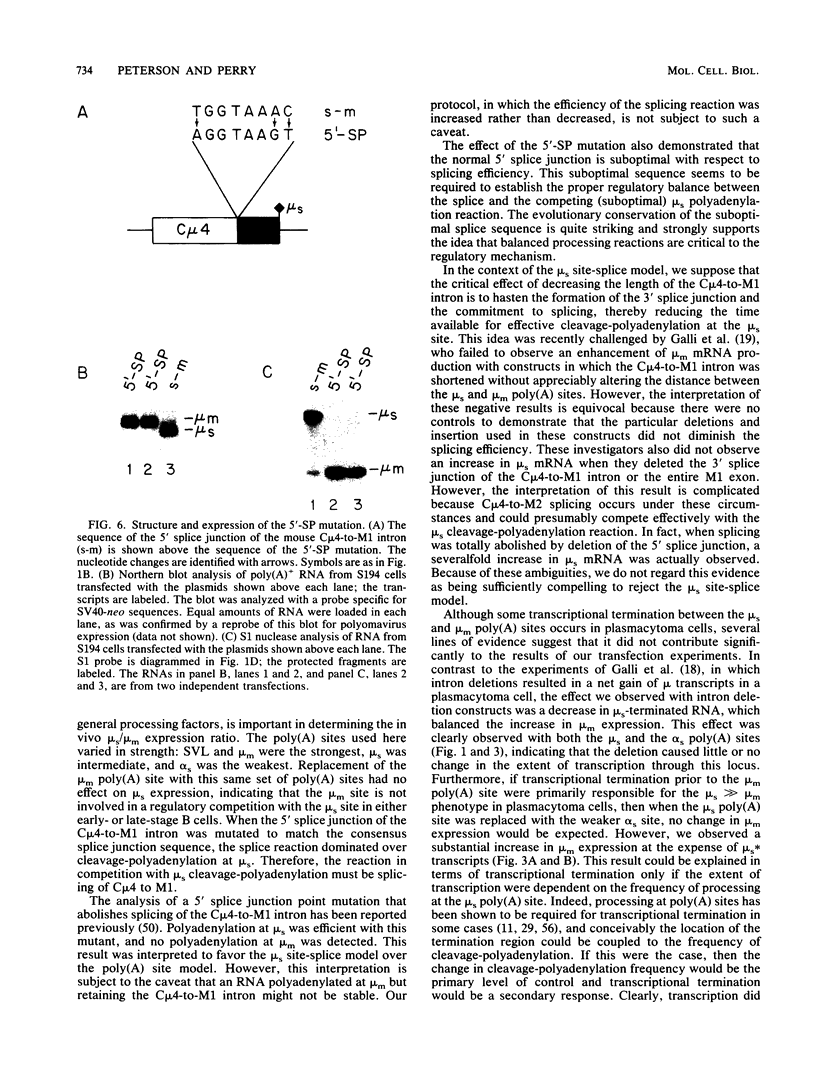
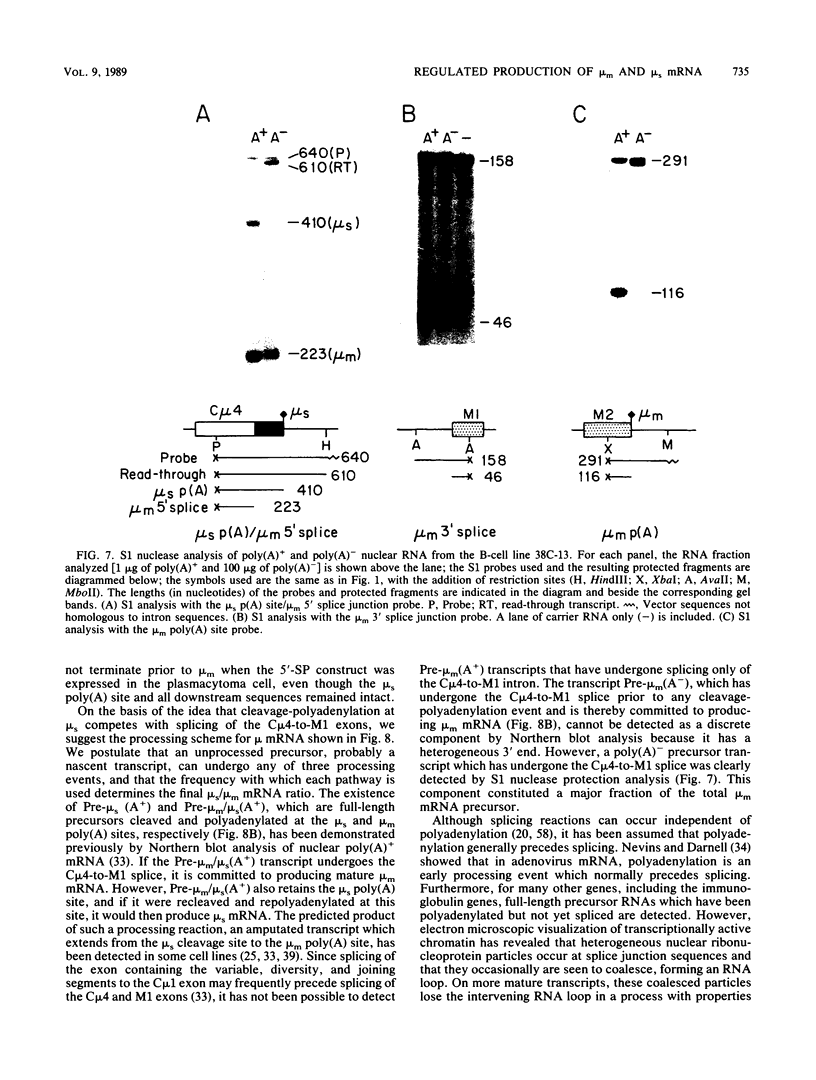
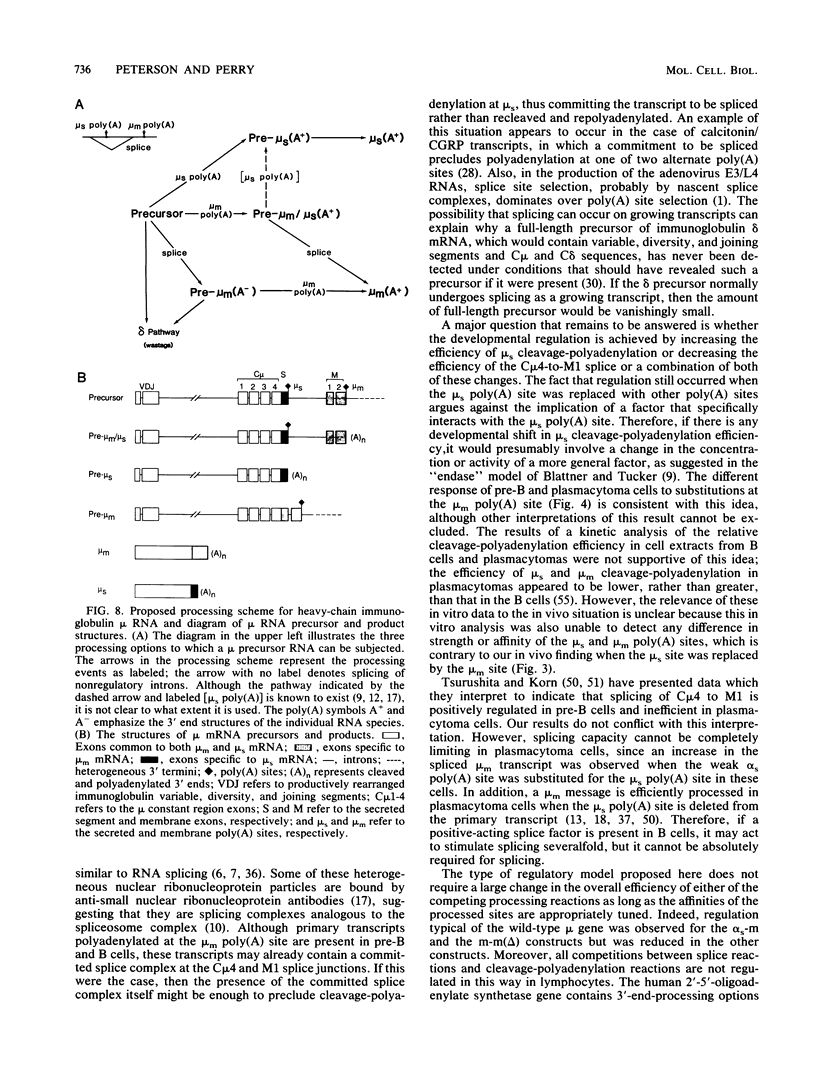
The relative abundance of the mRNAs encoding the membrane (mu m) and secreted (mu s) forms of immunoglobulin mu heavy chain is regulated during B-cell maturation by a change in the mode of RNA processing. Current models to explain this regulation involve either competition between cleavage-polyadenylation at the proximal (mu s) poly(A) site and cleavage-polyadenylation at the distal (mu m) poly(A) site [poly(A) site model] or competition between cleavage-polyadenylation at the mu s poly(A) site and splicing of the C mu 4 and M1 exons, which eliminates the mu s site (mu s site-splice model). To test certain predictions of these models and to determine whether there is a unique structural feature of the mu s poly(A) site that is essential for regulation, we constructed modified mu genes in which the mu s or mu m poly(A) site was replaced by other poly(A) sites and then studied the transient expression of these genes in cells representative of both early- and late-stage lymphocytes. Substitutions at the mu s site dramatically altered the relative usage of this site and caused corresponding reciprocal changes in the usage of the mu m site. Despite these changes, use of the proximal site was still usually higher in plasmacytomas than in pre-B cells, indicating that regulation does not depend on a unique feature of the mu s poly(A) site. Replacement of the distal (mu m) site had no detectable effect on the usage of the mu s site in either plasmacytomas or pre-B cells. These findings are inconsistent with the poly(A) site model. In addition, we noted that in a wide variety of organisms, the sequence at the 5' splice junction of the C mu 4-to-M1 intron is significantly different from the consensus 5' splice junction sequence and is therefore suboptimal with respect to its complementary base pairing with U1 small nuclear RNA. When we mutated this suboptimal sequence into the consensus sequence, the mu mRNA production in plasmacytoma cells was shifted from predominantly mu s to exclusively mu m. This result unequivocally demonstrated that splicing of the C mu 4-to-M1 exon is in competition with usage of the mu s poly(A) site. A key feature of this regulatory phenomenon appears to be the appropriately balanced efficiencies of these two processing reactions. Consistent with predictions of the mu s site-splice model, B cells were found to contain mu m precursor RNA that had undergone the C mu 4-to-M1 splice but had not yet been polyadenylated at the mu m site.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adami G., Nevins J. R. Splice site selection dominates over poly(A) site choice in RNA production from complex adenovirus transcription units. EMBO J. 1988 Jul;7(7):2107–2116. doi: 10.1002/j.1460-2075.1988.tb03050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt F. W., Bothwell A. L., Knapp M., Siden E., Mather E., Koshland M., Baltimore D. Synthesis of secreted and membrane-bound immunoglobulin mu heavy chains is directed by mRNAs that differ at their 3' ends. Cell. 1980 Jun;20(2):293–301. doi: 10.1016/0092-8674(80)90615-7. [DOI] [PubMed] [Google Scholar]
- Alt F., Rosenberg N., Lewis S., Thomas E., Baltimore D. Organization and reorganization of immunoglobulin genes in A-MULV-transformed cells: rearrangement of heavy but not light chain genes. Cell. 1981 Dec;27(2 Pt 1):381–390. doi: 10.1016/0092-8674(81)90421-9. [DOI] [PubMed] [Google Scholar]
- Atchison M. L., Perry R. P. Tandem kappa immunoglobulin promoters are equally active in the presence of the kappa enhancer: implications for models of enhancer function. Cell. 1986 Jul 18;46(2):253–262. doi: 10.1016/0092-8674(86)90742-7. [DOI] [PubMed] [Google Scholar]
- Benech P., Merlin G., Revel M., Chebath J. 3' end structure of the human (2'-5') oligo A synthetase gene: prediction of two distinct proteins with cell type-specific expression. Nucleic Acids Res. 1985 Feb 25;13(4):1267–1281. doi: 10.1093/nar/13.4.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer A. L., Bouton A. H., Miller O. L., Jr Correlation of hnRNP structure and nascent transcript cleavage. Cell. 1981 Oct;26(2 Pt 2):155–165. doi: 10.1016/0092-8674(81)90299-3. [DOI] [PubMed] [Google Scholar]
- Beyer A. L., Osheim Y. N. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988 Jun;2(6):754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Blattner F. R., Tucker P. W. The molecular biology of immunoglobulin D. Nature. 1984 Feb 2;307(5950):417–422. doi: 10.1038/307417a0. [DOI] [PubMed] [Google Scholar]
- Brody E., Abelson J. The "spliceosome": yeast pre-messenger RNA associates with a 40S complex in a splicing-dependent reaction. Science. 1985 May 24;228(4702):963–967. doi: 10.1126/science.3890181. [DOI] [PubMed] [Google Scholar]
- Connelly S., Manley J. L. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev. 1988 Apr;2(4):440–452. doi: 10.1101/gad.2.4.440. [DOI] [PubMed] [Google Scholar]
- Dahan A., Reynaud C. A., Weill J. C. Nucleotide sequence of the constant region of a chicken mu heavy chain immunoglobulin mRNA. Nucleic Acids Res. 1983 Aug 25;11(16):5381–5389. doi: 10.1093/nar/11.16.5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danner D., Leder P. Role of an RNA cleavage/poly(A) addition site in the production of membrane-bound and secreted IgM mRNA. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8658–8662. doi: 10.1073/pnas.82.24.8658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeChiara T. M., Erlitz F., Tarnowski S. J. Procedures for in vitro DNA mutagenesis of human leukocyte interferon sequences. Methods Enzymol. 1986;119:403–415. doi: 10.1016/0076-6879(86)19060-4. [DOI] [PubMed] [Google Scholar]
- Early P., Rogers J., Davis M., Calame K., Bond M., Wall R., Hood L. Two mRNAs can be produced from a single immunoglobulin mu gene by alternative RNA processing pathways. Cell. 1980 Jun;20(2):313–319. doi: 10.1016/0092-8674(80)90617-0. [DOI] [PubMed] [Google Scholar]
- Eperon L. P., Estibeiro J. P., Eperon I. C. The role of nucleotide sequences in splice site selection in eukaryotic pre-messenger RNA. Nature. 1986 Nov 20;324(6094):280–282. doi: 10.1038/324280a0. [DOI] [PubMed] [Google Scholar]
- Fakan S., Leser G., Martin T. E. Immunoelectron microscope visualization of nuclear ribonucleoprotein antigens within spread transcription complexes. J Cell Biol. 1986 Oct;103(4):1153–1157. doi: 10.1083/jcb.103.4.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli G., Guise J. W., McDevitt M. A., Tucker P. W., Nevins J. R. Relative position and strengths of poly(A) sites as well as transcription termination are critical to membrane versus secreted mu-chain expression during B-cell development. Genes Dev. 1987 Jul;1(5):471–481. doi: 10.1101/gad.1.5.471. [DOI] [PubMed] [Google Scholar]
- Galli G., Guise J., Tucker P. W., Nevins J. R. Poly(A) site choice rather than splice site choice governs the regulated production of IgM heavy-chain RNAs. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2439–2443. doi: 10.1073/pnas.85.8.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. R., Maniatis T., Melton D. A. Human beta-globin pre-mRNA synthesized in vitro is accurately spliced in Xenopus oocyte nuclei. Cell. 1983 Mar;32(3):681–694. doi: 10.1016/0092-8674(83)90054-5. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Baltimore D. Cell-type specificity of immunoglobulin gene expression is regulated by at least three DNA sequence elements. Cell. 1985 Jul;41(3):885–897. doi: 10.1016/s0092-8674(85)80069-6. [DOI] [PubMed] [Google Scholar]
- Guise J. W., Lim P. L., Yuan D., Tucker P. W. Alternative expression of secreted and membrane forms of immunoglobulin mu-chain is regulated by transcriptional termination in stable plasmacytoma transfectants. J Immunol. 1988 Jun 1;140(11):3988–3994. [PubMed] [Google Scholar]
- Kelley D. E., Perry R. P. Transcriptional and posttranscriptional control of immunoglobulin mRNA production during B lymphocyte development. Nucleic Acids Res. 1986 Jul 11;14(13):5431–5447. doi: 10.1093/nar/14.13.5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley D. E., Pollok B. A., Atchison M. L., Perry R. P. The coupling between enhancer activity and hypomethylation of kappa immunoglobulin genes is developmentally regulated. Mol Cell Biol. 1988 Feb;8(2):930–937. doi: 10.1128/mcb.8.2.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp D. J., Morahan G., Cowman A. F., Harris A. W. Production of RNA for secreted immunoglobulin mu chains does not require transcriptional termination 5' to the microM exons. Nature. 1983 Jan 6;301(5895):84–86. doi: 10.1038/301084a0. [DOI] [PubMed] [Google Scholar]
- Kokubu F., Hinds K., Litman R., Shamblott M. J., Litman G. W. Complete structure and organization of immunoglobulin heavy chain constant region genes in a phylogenetically primitive vertebrate. EMBO J. 1988 Jul;7(7):1979–1988. doi: 10.1002/j.1460-2075.1988.tb03036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law R., Kuwabara M. D., Briskin M., Fasel N., Hermanson G., Sigman D. S., Wall R. Protein-binding site at the immunoglobulin mu membrane polyadenylylation signal: possible role in transcription termination. Proc Natl Acad Sci U S A. 1987 Dec;84(24):9160–9164. doi: 10.1073/pnas.84.24.9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leff S. E., Evans R. M., Rosenfeld M. G. Splice commitment dictates neuron-specific alternative RNA processing in calcitonin/CGRP gene expression. Cell. 1987 Feb 13;48(3):517–524. doi: 10.1016/0092-8674(87)90202-9. [DOI] [PubMed] [Google Scholar]
- Logan J., Falck-Pedersen E., Darnell J. E., Jr, Shenk T. A poly(A) addition site and a downstream termination region are required for efficient cessation of transcription by RNA polymerase II in the mouse beta maj-globin gene. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8306–8310. doi: 10.1073/pnas.84.23.8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather E. L., Nelson K. J., Haimovich J., Perry R. P. Mode of regulation of immunoglobulin mu- and delta-chain expression varies during B-lymphocyte maturation. Cell. 1984 Feb;36(2):329–338. doi: 10.1016/0092-8674(84)90226-5. [DOI] [PubMed] [Google Scholar]
- McGuire K. L., Duncan W. R., Tucker P. W. Phylogenetic conservation of immunoglobulin heavy chains: direct comparison of hamster and mouse Cmu genes. Nucleic Acids Res. 1985 Aug 12;13(15):5611–5628. doi: 10.1093/nar/13.15.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milstein C. P., Deverson E. V., Rabbitts T. H. The sequence of the human immunoglobulin mu-delta intron reveals possible vestigial switch segments. Nucleic Acids Res. 1984 Aug 24;12(16):6523–6535. doi: 10.1093/nar/12.16.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson K. J., Haimovich J., Perry R. P. Characterization of productive and sterile transcripts from the immunoglobulin heavy-chain locus: processing of micron and muS mRNA. Mol Cell Biol. 1983 Jul;3(7):1317–1332. doi: 10.1128/mcb.3.7.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins J. R., Darnell J. E., Jr Steps in the processing of Ad2 mRNA: poly(A)+ nuclear sequences are conserved and poly(A) addition precedes splicing. Cell. 1978 Dec;15(4):1477–1493. doi: 10.1016/0092-8674(78)90071-5. [DOI] [PubMed] [Google Scholar]
- Ochi A., Hawley R. G., Hawley T., Shulman M. J., Traunecker A., Köhler G., Hozumi N. Functional immunoglobulin M production after transfection of cloned immunoglobulin heavy and light chain genes into lymphoid cells. Proc Natl Acad Sci U S A. 1983 Oct;80(20):6351–6355. doi: 10.1073/pnas.80.20.6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osheim Y. N., Miller O. L., Jr, Beyer A. L. RNP particles at splice junction sequences on Drosophila chorion transcripts. Cell. 1985 Nov;43(1):143–151. doi: 10.1016/0092-8674(85)90019-4. [DOI] [PubMed] [Google Scholar]
- Peterson M. L., Perry R. P. Regulated production of mu m and mu s mRNA requires linkage of the poly(A) addition sites and is dependent on the length of the mu s-mu m intron. Proc Natl Acad Sci U S A. 1986 Dec;83(23):8883–8887. doi: 10.1073/pnas.83.23.8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy V. B., Thimmappaya B., Dhar R., Subramanian K. N., Zain B. S., Pan J., Ghosh P. K., Celma M. L., Weissman S. M. The genome of simian virus 40. Science. 1978 May 5;200(4341):494–502. doi: 10.1126/science.205947. [DOI] [PubMed] [Google Scholar]
- Rogers J., Fasel N., Wall R. A novel RNA in which the 5' end is generated by cleavage at the poly(A) site of immunoglobulin heavy-chain secreted mRNA. Mol Cell Biol. 1986 Dec;6(12):4749–4752. doi: 10.1128/mcb.6.12.4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruether J. E., Maderious A., Lavery D., Logan J., Fu S. M., Chen-Kiang S. Cell-type-specific synthesis of murine immunoglobulin mu RNA from an adenovirus vector. Mol Cell Biol. 1986 Jan;6(1):123–133. doi: 10.1128/mcb.6.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders M. E., Gewert D. R., Tugwell M. E., McMahon M., Williams B. R. Human 2-5A synthetase: characterization of a novel cDNA and corresponding gene structure. EMBO J. 1985 Jul;4(7):1761–1768. doi: 10.1002/j.1460-2075.1985.tb03848.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U., Marcu K. B., Perry R. P. The synthesis and processing of the messenger RNAs specifying heavy and light chain immunoglobulins in MPC-11 cells. Cell. 1978 Dec;15(4):1495–1509. doi: 10.1016/0092-8674(78)90072-7. [DOI] [PubMed] [Google Scholar]
- Schibler U., Wyler T., Hagenbüchle O. Changes in size and secondary structure of the ribosomal transcription unit during vertebrate evolution. J Mol Biol. 1975 May 25;94(3):503–517. doi: 10.1016/0022-2836(75)90217-x. [DOI] [PubMed] [Google Scholar]
- Schwager J., Mikoryak C. A., Steiner L. A. Amino acid sequence of heavy chain from Xenopus laevis IgM deduced from cDNA sequence: implications for evolution of immunoglobulin domains. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2245–2249. doi: 10.1073/pnas.85.7.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro M. B., Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987 Sep 11;15(17):7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A. Splicing of messenger RNA precursors. Science. 1987 Feb 13;235(4790):766–771. doi: 10.1126/science.3544217. [DOI] [PubMed] [Google Scholar]
- Solnick D. An adenovirus mutant defective in splicing RNA from early region 1A. Nature. 1981 Jun 11;291(5815):508–510. doi: 10.1038/291508a0. [DOI] [PubMed] [Google Scholar]
- Treisman R., Orkin S. H., Maniatis T. Specific transcription and RNA splicing defects in five cloned beta-thalassaemia genes. Nature. 1983 Apr 14;302(5909):591–596. doi: 10.1038/302591a0. [DOI] [PubMed] [Google Scholar]
- Tsurushita N., Avdalovic N. M., Korn L. J. Regulation of differential processing of mouse immunoglobulin mu heavy-chain mRNA. Nucleic Acids Res. 1987 Jun 11;15(11):4603–4615. doi: 10.1093/nar/15.11.4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurushita N., Ho L., Korn L. J. Nuclear factors in B lymphoma enhance splicing of mouse membrane-bound mu mRNA in Xenopus oocytes. Science. 1988 Jan 29;239(4839):494–497. doi: 10.1126/science.3124268. [DOI] [PubMed] [Google Scholar]
- Tsurushita N., Korn L. J. Effects of intron length on differential processing of mouse mu heavy-chain mRNA. Mol Cell Biol. 1987 Jul;7(7):2602–2605. doi: 10.1128/mcb.7.7.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker P. W., Slightom J. L., Blattner F. R. Mouse IgA heavy chain gene sequence: implications for evolution of immunoglobulin hinge axons. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7684–7688. doi: 10.1073/pnas.78.12.7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ness B. G., Weigert M., Coleclough C., Mather E. L., Kelley D. E., Perry R. P. Transcription of the unrearranged mouse C kappa locus: sequence of the initiation region and comparison of activity with a rearranged V kappa-C kappa gene. Cell. 1981 Dec;27(3 Pt 2):593–602. doi: 10.1016/0092-8674(81)90401-3. [DOI] [PubMed] [Google Scholar]
- Virtanen A., Sharp P. A. Processing at immunoglobulin polyadenylation sites in lymphoid cell extracts. EMBO J. 1988 May;7(5):1421–1429. doi: 10.1002/j.1460-2075.1988.tb02959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw E., Proudfoot N. Alpha-thalassaemia caused by a poly(A) site mutation reveals that transcriptional termination is linked to 3' end processing in the human alpha 2 globin gene. EMBO J. 1986 Nov;5(11):2915–2922. doi: 10.1002/j.1460-2075.1986.tb04587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan D., Tucker P. W. Transcriptional regulation of the mu-delta heavy chain locus in normal murine B lymphocytes. J Exp Med. 1984 Aug 1;160(2):564–583. doi: 10.1084/jem.160.2.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevi M., Nevins J. R., Darnell J. E., Jr Nuclear RNA is spliced in the absence of poly(A) addition. Cell. 1981 Oct;26(1 Pt 1):39–46. doi: 10.1016/0092-8674(81)90031-3. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Leung H., Weiner A. M. The natural 5' splice site of simian virus 40 large T antigen can be improved by increasing the base complementarity to U1 RNA. Mol Cell Biol. 1987 Aug;7(8):3018–3020. doi: 10.1128/mcb.7.8.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in U1 snRNA suppresses a 5' splice site mutation. Cell. 1986 Sep 12;46(6):827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]