Abstract
Purpose.
MicroRNAs (miRNAs) are known to participate in post-transcriptional regulation of gene expression and are involved in multiple pathogenic processes. Here, we identified miRNA expression changes in the retinas of Akita mice, a genetic model of type 1 diabetes, and investigated the potential role of miRNA in diabetic retinopathy.
Methods.
Visual function of Akita and control mice was evaluated by electroretinography. MiRNA expression changes in the retinas of Akita mice were identified by miRNA-specific microarray and confirmed by quantitative RT-PCR (qRT-PCR). The potential downstream targets of identified miRNAs were predicted by bioinformatic analysis using web-based applications and confirmed by dual luciferase assay. The mRNA and protein changes of identified downstream targets were examined by qRT-PCR and Western blot analysis.
Results.
MiRNA-specific microarray and qRT-PCR showed that miR-200b was upregulated significantly in the Akita mouse retina. Sequence analysis and luciferase assay identified oxidation resistance 1 (Oxr1) as a downstream target gene regulated by miR-200b. In a human Müller cell line, MIO-M1, transfection of a miR-200b mimic downregulated Oxr1 expression. Conversely, transfection of MIO-M1 with a miR-200b inhibitor resulted in upregulated Oxr1. Furthermore, overexpression of recombinant Oxr1 attenuated oxidative stress marker, nitration of cellular proteins, and ameliorated apoptosis induced by 4-hydroxynonenal (4-HNE), an oxidative stressor. Similarly, transfection of a miR-200b inhibitor decreased, whereas transfection of miR-200b mimic increased the number of apoptotic cells following 4-HNE treatment.
Conclusions.
These results suggested that miR-200b–regulated Oxr1 potentially has a protective role in diabetic retinopathy.
This study demonstrated that miR-200b was upregulated in the Akita retina, and miR-200b downregulates the expression of Oxr1, which potentially has a protective role in diabetic retinopathy.
Introduction
MicroRNAs (miRNAs) are evolutionally conserved, single-stranded, small (19–24 nucleotides) noncoding RNA molecules that have an inhibitory role in gene expression at the post-transcriptional level by binding to partially complementary sequences in the 3′ un-translated region (3′UTR) of target messenger RNAs (mRNAs).1–4 It is known that miRNAs are present widely in the body, and regulate a variety of developmental, physiologic, and pathologic processes.2 For example, it has been reported that some miRNAs change their expression during the development and progression of various diseases, such as cancer and diabetes.5,6 Recent studies have revealed that several miRNAs are involved in the pathogenesis of diabetic complications.3,6–8 However, an miRNA profile of the retina from a genetic model of diabetes has not been examined to our knowledge.
Diabetic retinopathy (DR) is a common microvascular complication of diabetes and a leading cause of blindness among the working age population in the United States.9 Accumulating evidence suggests that chronic inflammation and oxidative stress in the retina have important pathogenic roles in DR.1,10,11 Oxidative stress (i.e., the over-production of reactive oxygen species [ROS]) has been found in various tissues, including the retina, under diabetic conditions.12–14 Subsequently, ROS damages key cellular components, such as lipids, proteins, and DNA,12–14 as well as activates numerous cell signaling pathways involving stress responses and apoptosis.12–17
To protect against ROS and oxidative stress, cells use several mechanisms through oxygen detoxification enzymes, such as superoxide dismutase 1 (SOD1), SOD2, glutathione peroxidase (Gpx), and catalase.18,19 In addition, it was reported recently that oxidation resistance 1 (Oxr1) may have a protective role against oxidative stress.20 Oxr1 is encoded by an evolutionally conserved gene (Oxr1) and is found in several organisms, including plants and most eukaryotes (from yeast to human).21 The carboxyl-terminal TLDc domain of Oxr1, in particular, is highly conserved.22 The Oxr1 gene can produce multiple tissue-specific protein-encoding transcripts by alternative splicing; most transcripts share the region that encodes the TLDc domain.21,23 The Oxr1 protein is localized in the mitochondria and nuclei in human, rat, and mouse cells.21 In conditions of oxidative stress, Oxr1 confers a protective effect against oxidative DNA damages in Escherichia coli,24 and mutations of Oxr1 in yeast decreased survival.21,24 In the mouse retina25 and cultured cells,21 Oxr1 expression is upregulated transiently at early time points after oxidative stress. Furthermore, it has been shown recently that Oxr1 has a crucial role in the protection of neuron cells against oxidative stress by H2O2.20 It is speculated that Oxr1 regulates the expression of other ROS detoxification enzymes, such as SODs, catalase, and Gpx, possibly through the c-Jun NH2-terminal kinase (JNK) pathway.23
In our study, we identified miRNAs differentially regulated in the retinas of Akita mice, a genetic model of type 1 diabetes and age-matched nondiabetic mice using microarray analysis. Among them, we demonstrated that miR-200b was upregulated in the Akita retina, and miR-200b downregulates Oxr1 under diabetic conditions.
Materials and Methods
Animals
C57BL/6J-Ins2Akita (Akita) and C57BL/6J mice (Jackson Laboratories, Bar Harbor, ME) were housed and raised in 12-hour light and 12-hour dark cycles. All of the experiments involving mice were approved by the local Institutional Animal Care and Use Committees (IACUC) at the University of Oklahoma Health Sciences Center, and performed following the guidelines of the Association for Research in Vision and Ophthalmology (ARVO) Statement for the Use of Animals in Ophthalmic and Vision Research.
Electroretinography
Electroretinograms (ERGs) were recorded in the darkroom as described previously.26
RNA Extraction
Total RNA, including miRNA, was extracted from the retinas or cultured cells using a miRNeasy Mini kit (or RNeasy Mini kit for mRNA analysis) and treated with RNase-free DNase-I in the purification columns to eliminate genomic DNA contamination following the manufacturer's instructions (Qiagen, Valencia, CA).
MicroRNA Microarray Analysis
RNA (5 μg) was pooled from 3 nine-month-old heterozygous Akita male and 3 age-matched control (wild-type) mice. A miRNA-specific microarray was performed by LC Sciences (Houston, TX) using murine-specific miRNA chips that contained sequences from Sanger miRBase version 14.0.
Quantitative Real-Time RT-PCR (qRT-PCR) for MicroRNA and mRNA Expression Analysis
cDNA synthesis was conducted using 50 ng of extracted total RNA, including microRNAs and specific stem-loop primers for miR-200b or small nuclear RNA U6 (an endogenous small nuclear RNA, snRNA) using TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems, Carlsbad, CA). The expression of miR-200b and U6 was detected with TaqMan MicroRNA Assay (Applied Biosystems). The qRT-PCR was conducted using a CFX96 qRT-PCR instrument (BioRad, Hercules, CA) with 45 cycles of 94°C for 15 seconds (denature) and 60°C for 60 seconds (annealing and extension), using sequence specific primers and probes from TaqMan MicroRNA Assay and iQ Supermix (BioRad). MiRNA expression was normalized to that of U6 and expressed as fold of control group. Data were analyzed according to the comparative Ct method.27,28
For quantification of mRNAs in mouse tissues and cultured human cells, the synthesized cDNA, iQ SYBR Green Supermix (BioRad) and set of primers (hOxr1-F, TGTCCGGAGTTTGAGGTCTT; hOxr1-R, GATCTCCATCAAGCCAAAGC; mOxr1-F, TTCTGCCCAGAATTTGAGGT; mOxr1-R, TCTCCATCAAGCCAAAGAGC) were used in qRT-PCR with 40 cycles of 94°C for 15 seconds and 60°C for 30 seconds followed by melt-curve analysis. The primers for Oxr1 mRNA detection were designed to amplify regions that encode the respective TDLc domains to cover all splicing variants. The Oxr1 mRNA level was normalized to that of endogenous control, hypoxanthine phosphoribosyltransferase 1, (Hprt1; hHprt1-F, GACCAGTCAACAGGGGACAT; hHprt1-R, CCTGACCAAGGAAAGCAAAG; mHprt1-F, CAGGCCAGACTTTGTTGGAT; mHprt1-R, TTGCGCTCATCTTAGGCTTT), and expressed as fold of control. Data were analyzed according to the comparative Ct method.27,28 The primers were designed using the Primer3Plus application.29
Bioinformatic Analysis for Possible MicroRNA Target Genes
To predict the target genes of the miRNA(s) of interest, bioinformatic analyses via internet databases (e.g., microRNA.org, available in the public domain at www.microrna.org30), TargetScan Release 6.2 (available in the public domain at http://targetscan.org31), and MicroCosm targets version 5 (available in the public domain at http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/) were used. The 3′-UTR sequences of potential target genes were examined to determine whether selected microRNAs showed “seed” matches (i.e., complementary at positions 2–7) and the minimum free energy was lower than −20 kcal/mol by using RNAhybrid application (available in the public domain at http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/submission.html32). Further, the potential targets were narrowed down by examination of genes that are known to be involved in DR.
Validations of Potential miRNA Targets
The potential 3′UTR binding site of miR-200b in the target gene, as predicted by the bioinformatic analysis, was cloned into the pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega Corporation, Madison, WI) for in vitro analysis following the manufacturer's protocol.33
For the luciferase assay, 50 nM of miR-200b mimic (Applied Biosystems) and the pmirGLO vectors containing the gene fragment of interest (0.2 μg) were cotransfected into ARPE-19 cells, a cell line derived from human retinal pigment epithelial cells,34 in 24-well plates using Lipofectamine 2000 reagent following manufacturer's instructions (Invitrogen, Carlsbad, CA). As negative controls, an empty vector (mock, 0.2 μg) and the same amount of vector containing a mismatched gene fragment were used for each gene assay. After 36 hours of incubation, the cells were harvested and dual luciferase assays were conducted using the Dual Luciferase Reporter Assay System (Promega Corporation) following the manufacturer's instructions.
Cell Culture
To identify the downstream target of miR-200b, ARPE-1934 and a human Müller cell line (MIO-M135) were selected to evaluate the effects of miR-200b in this study. These cell lines were cultured in low glucose (5.5 mM D-glucose) Dulbecco's modified Eagle's medium (DMEM; CellGro, Manassas, VA) supplemented with 10% fetal bovine serum (FBS). MIO-M1 cells were incubated with low glucose DMEM media containing 10 or 20 μM of 4-hydroxynonenal (4-HNE; EMD Chemicals, Billerica, MA), an oxidized lipid that induces oxidative stress, or the same amount of vehicle and cultured for another 16 hours for protein and RNA assays, unless specified.
To analyze the impacts of miRNA in the cultured cells, the mirVana miRNA mimic negative control (5 nM), miR-200b mimic (5 nM), mirVana miRNA inhibitor negative control (50 nM), or miR-200b inhibitor (50 nM; Ambion, Austin, TX) were transfected separately into MIO-M1 cells using siPORT NeoFX reagent (Ambion) following manufacture's instruction. At 48 hours post-transfection, the cells were harvested, and the protein and RNA levels of identified target genes were analyzed by Western blot analysis and qRT-PCR, respectively.
Western Blot Analysis
The treated MIO-M1 cells were lysed by sonication and total cellular protein concentrations were measured by Bradford assay.36 Equal amounts of protein (25 μg) were resolved by electrophoresis through 10% Tris-glycine SDS polyacrylamide gel and electrotransferred onto a nitrocellulose membrane. The membrane was blocked with 5% (wt/vol) nonfat dry milk in Tris-buffered saline with 0.1% Tween-20 (TBST) for 30 minutes and subsequently incubated overnight at 4°C with a 1:3000 dilution of an antihuman Oxr1 polyclonal antibody (BETHYL Laboratories, Montgomery, TX). After three washes with TBST, the membrane was incubated for 1.5 hours with a 1:5000 dilutions of an HRP-conjugated anti-rabbit IgG antibody (Vector Laboratories, Burlingame, CA) in TBST containing 1% nonfat dry milk. After four washes with TBST, the bands were detected using Super Signal West Dura Extended Duration Substrate (Pierce, Rockford, IL) or noncommercial ECL solution.37 As needed, the membrane was stripped in stripping buffer (Pierce) and reblotted with an antibody specific for β-actin (Sigma-Aldrich, St. Louis, MO) for loading control. The band intensities were semiquantified by densitometry using Fluorochem-Q software (AlphaInnotech, San Leandro, CA); values are an average of at least 3 independent experiments.
Cloning and Construction of a Mouse Oxr1 Adenovirus
Briefly, the full-length mouse Oxr1 short isoform, variant C (accession number NM_001130164) was amplified by PCR using the C57BL/6J brain cDNA (human Oxr1 is highly expressed in the brain21) and a set of gene-specific primers containing restriction enzyme sites (bold) and Kozak sequence (underline)38 in the forward primer (mOxr1Vc Fwd: GCGGCCGCCACCATGTCCCGTCTCTGGTATGGG) and the reverse primer (mOxr1Vc Rev: GATATCGTTCAAAAGCCCAGATTTCAATGTC). The amplified fragments were cloned into pGEM-T easy vector (Promega Corporation), and the insert was sequenced using the T7 primer to identify any mutations. The confirmed mOxr1Vc cDNA was cloned between NotI and EcoRV sites of a shuttle vector with a 3×FLAG-epitope, pShuttle-IRES-hrGFP-1 (Agilent Tech, Santa Clara, CA), for construction of the adenovirus (AD-Oxr1). Preparation, amplification and titration of the recombinant adenoviruses were performed as described previously.39,40
Statistical Analysis
To determine statistical significance, at least three individual measurements were conducted and data entered into Microsoft Excel (Microsoft, Redmond, WA) were subjected to Student's t-test. Significance was denoted when there was a P value < 0.05. All of the values were expressed as Mean ± SEM.
Results
Progressive Loss of Visual Function in Akita Mice
The visual function of Akita mice was evaluated by ERG recording at several time points (1, 2, 3, 4, 5, and 8 months of age). ERG results showed that there were significant reductions in the scotopic a-wave amplitude at 5 and 8 months of age (Fig. 1A), suggesting impaired rod function. In addition, the reduction of second order neuronal function (scotopic b-wave) was observed at 4, 5, and 8 months of age (Fig. 1B). Moreover, photopic b-wave amplitude was significantly reduced at 8 months of age (Fig. 1C), suggesting an impaired cone function in this diabetic model.
Figure 1.
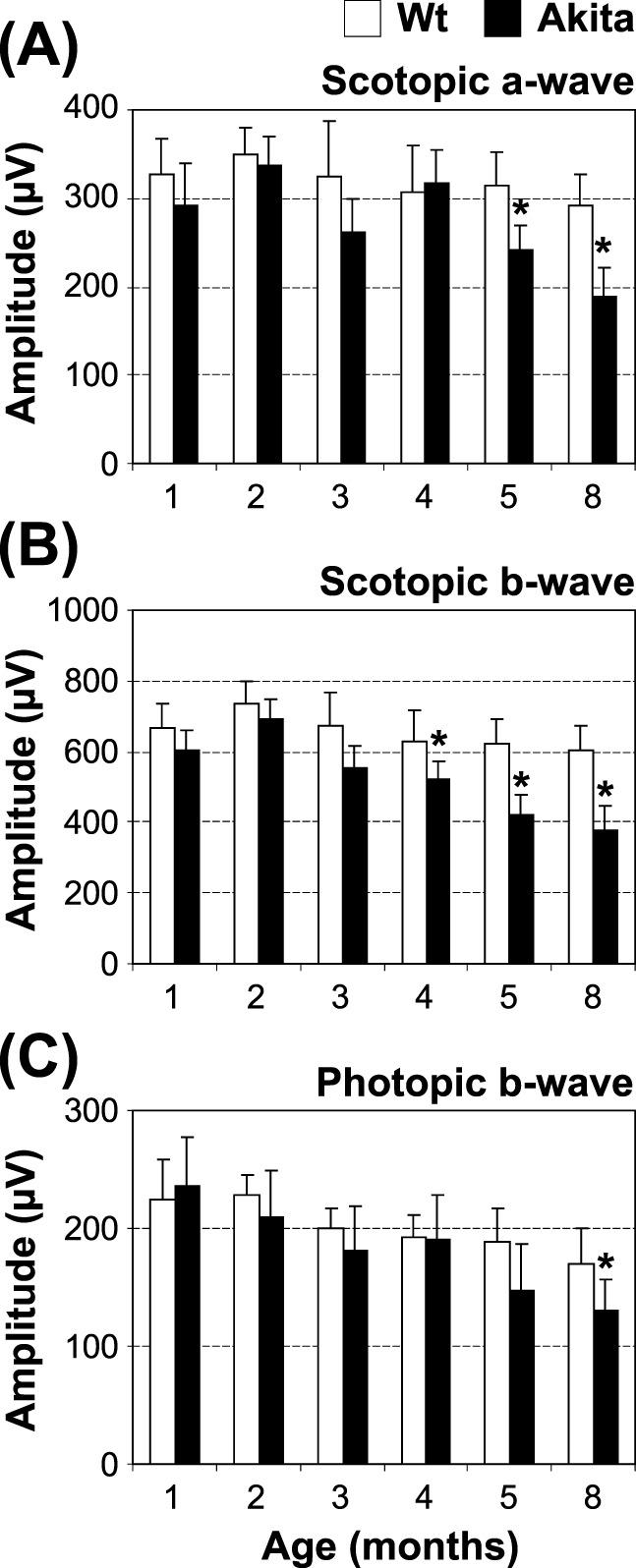
Functional analysis of the Akita mouse retina. Retinal function of Akita mice and age-matched wild-type controls was evaluated by ERG recording at the indicated age: scoptopic a-wave (A), scotopic b-wave (B), and photopic b-wave (C) are shown (mean ± SEM, n = 10, *P < 0.05).
Differentially Expressed miRNas in the Akita Mouse Retina
Akita mice have shown many DR-related pathologic changes, such as retinal oxidative stress, inflammation, neuron degeneration, and vascular leakage.41 Although altered transcription in a number of genes has been identified, which contributes to these pathologic changes of DR, the changes in miRNA expression or DNA methylation status that contribute to the pathogenesis of DR in the Akita model have not been well understood. Here, we identified changes in miRNA expression that may be associated with DR in Akita mice. The miRNA expression profile was examined using an miRNA-specific microarray, and the microarray identified a number of miRNAs that were upregulated and downregulated (Fig. 2A) in the Akita mouse retina compared to the nondiabetic control. Among them, miR-200b was confirmed to be upregulated by 4-fold in the Akita retina using qRT-PCR assay specific for miR-200b (Fig. 2B). Other miRNA-200 family members also tended to be upregulated in the Akita retinas, although the changes were not statistically significant (see Supplementary Material and Supplemental Fig. S1, http://www.iovs.org/content/54/3/1689/suppl/DC1).
Figure 2.
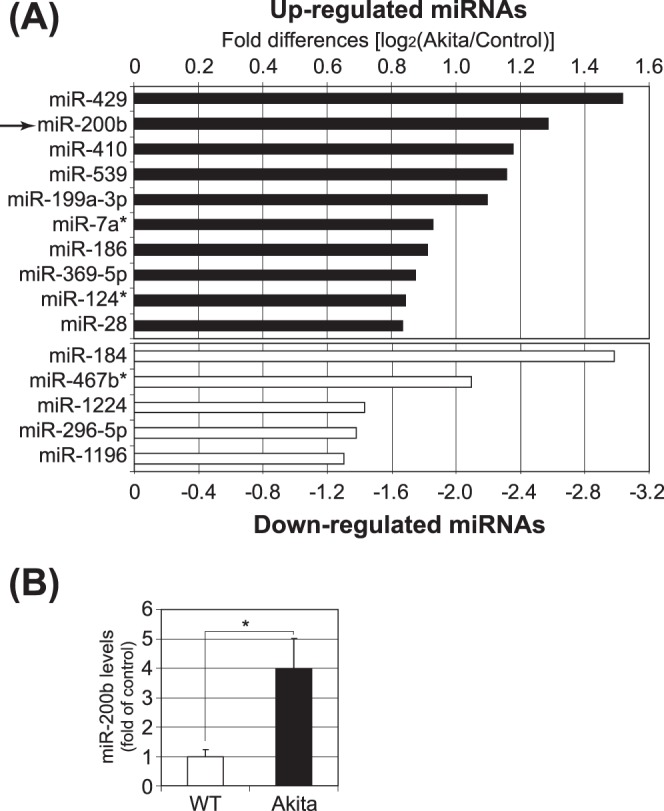
MicroRNA expression profile in the Akita mouse retina. (A) Retinal RNA was isolated from three Akita mice and three controls at age nine months: the RNA from each group was pooled and the miRNA-specific microarray was conducted by LC Sciences. The log2 of microarray signal intensities is shown in bar graph for top 10 upregulated (black bars) and top 5 downregulated (white bars) miRNAs in the Akita retinas. (B) The retinal levels of miR-200b in the Akita mice were analyzed by qRT-PCR and expressed as the fold change over that in the age-matched nondiabetic control (mean ± SEM, n = 8 wild type and n = 11 Akita, *P < 0.05).
Identification of Possible miR-200b Target Genes Involved in DR
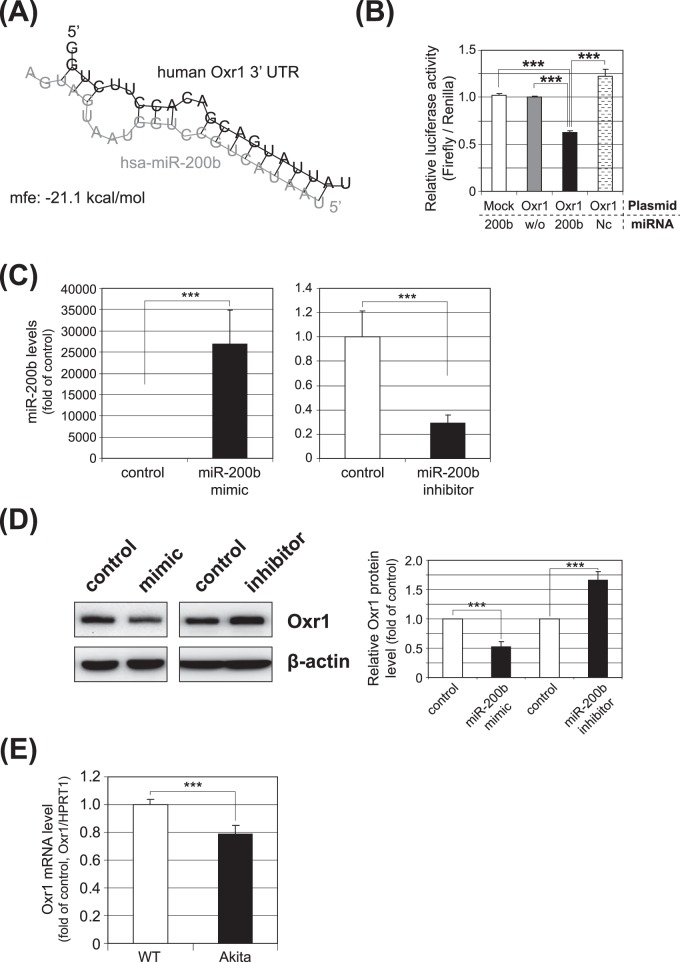
To evaluate the functional significance of the elevated expression of miR-200b in the Akita retina, the potential downstream miR-200b target genes were predicted and examined. Bioinformatic analysis using the internet databases microRNA.org (available in the public domain at www.microrna.org) and TargetScan (available in the public domain at www.targetscan.org) identified a number of putative target genes for miR-200b. Among the potential targets, we focused on those involved potentially in DR-associated oxidative stress and inflammation. Among them, Oxr1 was identified as a putative target gene of miR-200b, as microRNA.org and Targetscan.org identified the miR-200b target sequence in the 3′UTR of Oxr1. An RNAhybrid application showed a minimum free energy of −21 kcal/mol on miR-200b at the potential binding site in 3′UTR of Oxr1 (Fig. 3A), suggesting that miRNA-200b is likely to bind to the predicted target site in the 3′UTR of Oxr1. We chose Oxr1 for this study, since it recently was suggested that Oxr1 has a protective role against oxidative stress.20,23
Figure 3.
Validation of Oxr1 as a target gene of miRNA-200b. (A) The predicted secondary structure of the miR-200b binding site in the 3′UTR of human Oxr1 is shown; minimum free energy (mfe) is calculated and shown. (B) Luciferase reporter vectors containing the miR-200b target sequence of Oxr1 (Oxr1), or the same vector without the target sequence (Mock) were cotransfected with miR-200b mimic (200b), without miRNA (w/o), or with mismatched oligos (Nc, negative control) into ARPE-19 cells for 36 hours, and a dual luciferase assay subsequently was conducted. The luciferase activity was decreased significantly by cotransfection with the miR-200b mimic (***P < 0.001). (C) qRT-PCR analysis of miR-200b expression following transfection of the miR-200b mimic (left) or the miR-200b inhibitor (right) in MIO-M1 cells. The results are presented as fold change over the control (mean ± SEM, n = 6; ***P < 0.001). (D) Regulation of Oxr1 expression by miR-200b mimic and its inhibitor. Western blot analyses of Oxr1 were conducted 48 hours after transfection of miR-200b mimic or its inhibitor (left). Oxr1 protein levels were semiquantified by densitometry (right, mean ± SEM, n = 3, ***P < 0.001). (E) Oxr1 mRNA levels in the retina of Akita mice and Wt mice were analyzed by qRT-PCR (mean ± SEM, n = 7, ***P < 0.001).
To confirm that Oxr1 is a target gene of miR-200b, the miR-200b binding sequence in the 3′UTR of Oxr1 (Fig. 3A) was cloned into a miRNA target assay plasmid (pmirGLO; Promega) and examined for its potential regulation by miR-200b using dual luciferase assays. To validate the downstream target of miR-200b, ARPE-19 cells were selected as a transfection host. A miR-200b mimic and the miRNA target assay plasmids containing potential target sequence of 3′UTR in Oxr1 were cotransfected into ARPE-19 cells. Dual luciferase assay 36 hours post-transfection revealed that the luciferase reporter activity was reduced significantly by miR-200b, indicating that Oxr1 is regulated by miR-200b (Fig. 3B). Furthermore, to analyze the potential role of miR-200b and Oxr1 in Müller cells, transfection of a miR-200b mimic into the human retinal Müller cell line, MIO-M1, resulted in a remarkable increase in miR-200b levels, while transfection of a miR-200b inhibitor caused a significant reduction in the level of miR-200b (Fig. 3C). Western blot analysis showed that Oxr1 protein levels were downregulated by the miR-200b mimic, while upregulated by the miR-200b inhibitor (Fig. 3D). Taken together, these results showed clearly that Oxr1 is a downstream target of miR-200b. Examination of Oxr1 mRNA levels in the Akita retina revealed that the levels of Oxr1 mRNA were decreased significantly compared to the age-matched nondiabetic control (Fig. 3E), indicating a negative correlation between Oxr1 expression and miR-200b levels in the retina of this DR model.
Differential Regulation of miR-200b and Oxr1 by Oxidative Stress in Retinal Müller Cells
To examine if changes in miR-200b are related to increased oxidative stress, MIO-M1 cells were treated for 16 hours with 10 μM and 20 μM of 4-HNE with vehicle (ethanol), a commonly used diabetic stressor, or the vehicle alone as control. Examination of miR-200b levels by qRT-PCR revealed that miR-200b was upregulated in the cells that were exposed to 10 μM or 20 μM of 4-HNE when compared to cells exposed to vehicle alone (Fig. 4A). Furthermore, transcript levels of Oxr1 were downregulated after exposure to 4-HNE in 10 and 20 μM (Fig. 4B). Western blot analysis showed that the Oxr1 protein levels in MIO-M1 cells were decreased upon exposure to 4-HNE (Figs. 4C, 4D), further supporting that overexpression of miR-200b may be responsible for the downregulation of Oxr1 under diabetic conditions.
Figure 4.
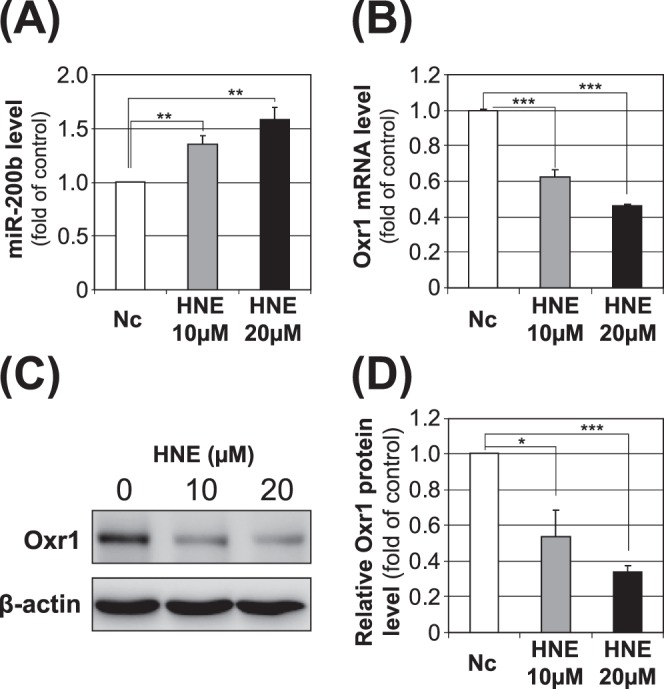
Impacts of 4-HNE on miRNA-200b and Oxr1 expression in retinal Müller cells. Human Müller (MIO-M1) cells were treated with 10 and 20 μM of 4-HNE, or the same volume of vehicle (ethanol) for 16 hours. The cells subsequently were harvested, and miR-200b (A), Oxr1 mRNA (B), and Oxr1 protein (C, D) levels were measured by qRT-PCR and Western blot analysis (mean ± SEM, [A, B] n = 4, [C, D] n = 3, *P < 0.05, **P < 0.01; ***P < 0.001).
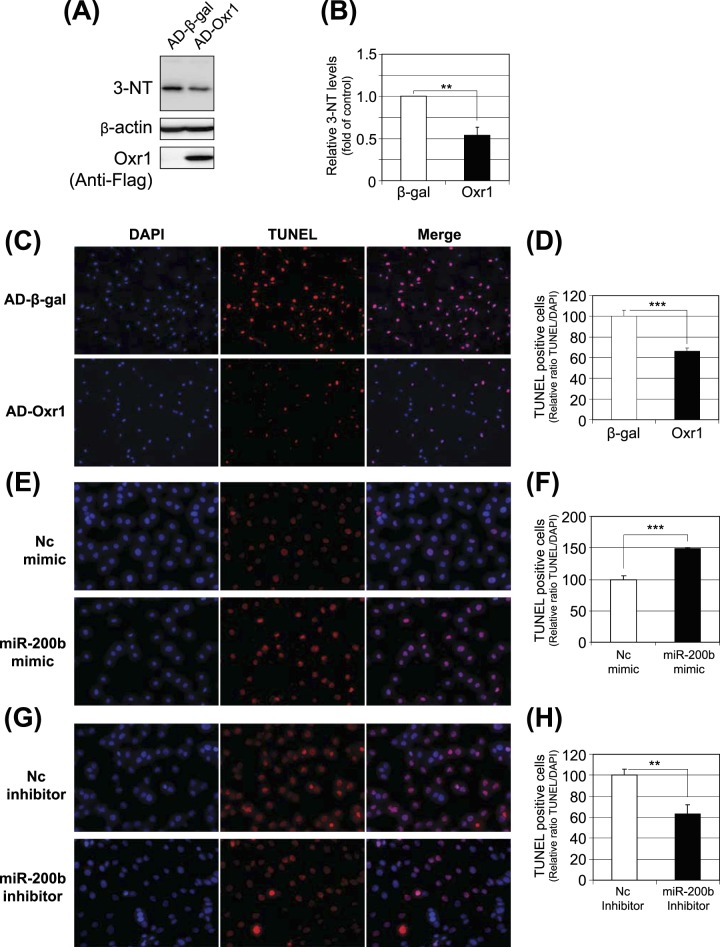
Protective Effect of Oxr1 against Oxidative Stress and Apoptosis under Diabetic Conditions
To evaluate the role of Oxr1 in DR, we examined whether overexpression of Oxr1 has a protective effect against oxidative stress. Overexpression of Oxr1 in MIO-M1 cells attenuated an increase of 3-nitrotyrosine (3-NT), an oxidative stress marker, induced by 4-HNE (Figs. 5A, 5B). TUNEL analysis showed that overexpression of Oxr1 decreased the number of apoptotic cells in MIO-M1 cells exposed to 4-HNE (Figs. 5C, 5D), suggesting that downregulation of Oxr1 induced by miR-200b contributed to oxidative stress and retinal cell death in diabetes. Furthermore, MIO-M1 cells were exposed to 4-HNE following the transfection of the miR-200b mimic or the miR-200b inhibitor. TUNEL staining showed that miR-200b mimic transfection significantly increased the number of apoptotic cells compared to the cells transfected with negative control miRNA (Figs. 5E, 5F), whereas miR-200b inhibitor transfection significantly reduced the number of TUNEL-positive cells (Figs. 5G, 5H).
Figure 5.
Roles of Oxr1 and miR-200b in oxidative stress and apoptosis in retinal Müller cells. MIO-M1 cells were infected with either adenovirus expressing β-gal (AD-β-gal) or Oxr1 (AD-Oxr1) at a multiplicity of infection at 20 for 24 hours and then treated with 20 μM of 4-HNE for another 16 hours. (A, B) 3-NT, Oxr1, and β-actin were measured by Western blot analysis (A) and semiquantified by densitometry (B, mean ± SEM, n = 3, **P < 0.01). (C) Apoptotic cells were detected by TUNEL staining (red) using the in situ cell death detection kit (Roche, Indianapolis, IN). The nuclei were counterstained with DAPI (blue). (D) TUNEL-positive cells were counted and presented as a relative ratio of TUNEL-positive cells to total DAPI stained cells (mean ± SEM, n = 3, ***P < 0.001). (E–H) MIO-M1 cells were transfected separately with the miR-200b mimic or its negative control and the miR-200b inhibitor or its negative control, respectively. The cells were exposed to 20 μM of 4-HNE for 1 hour at 48 hours post-transfection. Apoptotic cells were detected by TUNEL staining (red) using the in situ cell death detection kit and the nuclei were counterstained with DAPI (blue; E, G). TUNEL-positive cells were counted and presented as a relative ratio of TUNEL-positive cells to DAPI-stained total cells. (E, F) miRNA mimic. (G, H) miRNA inhibitor. Mean ± SEM, n = 3, ***P < 0.001, **P < 0.01).
Discussion
Akita mice are a commonly used type 1 diabetes model for DR research.41 Earlier studies showed that Akita mice develop progressively increased vascular permeability and leukocytes infiltration in the retina.41 Furthermore, there is documented evidence of retinal neuron degeneration (i.e., increased apoptotic cells and decreased retinal thickness) in Akita mice.41,42 To our knowledge, our study is the first to document a progressive reduction in ERG responses in Akita mice. This finding is consistent with a recent study showing that Akita mice exhibited significant reductions in optomotor behavior.43 Taken together, these observations suggested that Akita mice develop slow and progressive diabetes-induced retinal degeneration, likely due to oxidative stress and ROS over-production in diabetes.13
To investigate the impact of miRNAs on diabetes-induced oxidative stress and ROS production in the retina, we examined the differential expression of miRNAs in the Akita mouse retina. A microRNA-specific microarray identified a number of miRNAs that were regulated differentially in the diabetic retina. In our study, we focused on miR-200b, since it was reported recently that the expression of miR-200b is regulated by hypoxia and modulates angiogenesis, a process prevalent in DR.44,45 Our qRT-PCR analysis confirmed the results of the microarray and demonstrated a 4-fold increase in miR-200b levels in the Akita retina. Although miR-200b in the Akita mouse retina has never been measured to our knowledge, some groups have examined miR-200b expression in the retina of the streptozotocin (STZ)-induced diabetic model.46,47 However, there are contradictory findings regarding miR-200b in DR. Kovas et al. reported that miR-200b was upregulated in the retina and in retinal endothelial cells of STZ-induced diabetic rats 3 months after the onset of diabetes.47 In contrast, McArthur et al. reported that miR-200b was downregulated in STZ-induced diabetic rat retinas 1 month after the onset of diabetes.46 As VEGF is one of the downstream targets of miR-200b, the downregulation of miR-200b in the STZ-induced diabetic retina was proposed to be responsible for the elevation of VEGF expression in DR.46 The disparities in miR-200b levels in the retina of diabetic models may be ascribed to different durations of diabetes or perhaps different models (genetic model of diabetes versus STZ-induced diabetes). Indeed, we analyzed the miR-200b levels in the retina of Akita and nondiabetic control mice at 3 months of age in addition to 9 months. The levels of miR-200b did not show statistically significant changes in 3-month-old Akita mouse retinas (see Supplementary Material and Supplemental Fig. S2, http://www.iovs.org/content/54/3/1689/suppl/DC1). This result suggests that miR-200b expression changes may be dependent on diabetes durations. It also is possible that the chemical (STZ) in the STZ-induced diabetic animal model may lead to changes in mRNA and miRNA expression due to possible toxic effects of STZ, as an earlier study demonstrated toxic effects in the STZ model.48,49
Previous studies of miR-200b in the retina also were performed in retinal endothelial cells (RECs).46,47 One of the studies reported that the expression level of miR-200b in RECs was substantially lower than that in total retinas,47 suggesting that other retinal cell types may be the major sources of miR-200b. Another study showed that miR-200b is expressed in retinal capillaries, ganglion cells, and a type of cell whose cell body is located in the inner nuclear layers (likely Müller cells).46 Müller cell is the principal glial cell in the vertebrate retina, maintaining the retinal neurons by exchanging molecules,50 and also acting as the modulators of immune and inflammatory responses by releasing pro-inflammatory cytokines under pathologic conditions. Therefore, in this study, we selected the human Müller cell (MIO-M1) as the cell model.
Although bioinformatic analyses identified a number of potential target genes of miR-200b, we chose to verify those involved in known pathologic changes of DR: oxidative stress, angiogenesis, and hypoxia.51–54 As a result, only Oxr1 among the candidates analyzed was confirmed to be regulated by mR-200b by luciferase assay. The regulation of Oxr1 by miR-200b also was confirmed at protein levels using transfection of the miR-200b mimic and inhibitor. Documented studies suggest that Oxr1 may have a role in cell apoptosis. It has been reported that Oxr1 is expressed in a biphasic fashion under ischemic conditions: showing an initial upregulation and then, upon the initiation of cell death, its expression is reduced in the hyperoxic mouse retina.25 Its implication in DR, however, has not been reported previously to our knowledge. Our study demonstrated that Oxr1 is, indeed, downregulated in the retina of Akita mice, negatively correlating with an increase in miR-200b. Furthermore, 4-HNE, a commonly used diabetic stressor, also significantly upregulated the expression of miR-200b, while downregulating Oxr1 in cultured retinal cells. Taken together, these results suggested that oxidative stress contributes to the upregulation of miR-200b and subsequent downregulation of Oxr1 in the Akita retina.
To investigate the role of Oxr1 downregulation in DR, we have overexpressed Oxr1 in cultured cells. Overexpression of Oxr1 decreased 3-NT levels induced by a diabetic stressor (4-HNE), suggesting an antioxidant role for Oxr1. Furthermore, overexpression of Oxr1 also protected retinal cells against apoptosis under diabetic conditions. This is consistent with the finding by Oliver et al. showing that the recombinant Oxr1 significantly reduces DNA damage and the number of apoptotic brain granule cells caused by H2O2-induced oxidative stress.20 Furthermore, overexpression and knockdown of miR-200b in the MIO-M1 cells by transfection of the miR-200b mimic and inhibitor, respectively, significantly altered the number of apoptotic cells. Taken together, these observations suggested that the diabetes-induced increase of miR-200b and subsequent downregulation of Oxr1 in the retina contribute to oxidative stress and retinal degeneration in Akita mice.
Although miR-200b was the focus of our study, investigating the other miRNAs that are significantly changed in the diabetic retina will be an interesting avenue to pursue in future studies. It is plausible that the upregulation and downregulation of specific miRNAs in the diabetic retina could contribute to the visual loss in DR. Future studies also will focus on defining the target genes of other differentially regulated (both up- and downregulated) miRNAs identified in our study. Finally, it should be noted that physiologic response to the stresses or disease conditions is unlikely ascribed to single miRNA changes. It was well accepted that one target gene can be regulated by multiple microRNAs,55 as well as other regulation mechanisms. The complexity of miRNA-related regulations must be studied systemically.
Supplementary Material
Footnotes
Supported by NIH Grants EY018659, EY012231, and EY019309; Grant P20GM104934 from the National Center for Research Resources; and Grant HR12-103 from Oklahoma Center for the Advancement of Science & Technology (OCAST).
Disclosure: A.R. Murray, None; Q. Chen, None; Y. Takahashi, None; K.K. Zhou, None; K. Park, None; J.-X. Ma, None
References
- 1. Xu S. MicroRNA expression in the eyes and their significance in relation to functions. Prog Retin Eye Res. 2009; 28: 87–116 [DOI] [PubMed] [Google Scholar]
- 2. Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol. 2010; 11: 252–263 [DOI] [PubMed] [Google Scholar]
- 3. Kolfschoten IG, Roggli E, Nesca V, Regazzi R. Role and therapeutic potential of microRNAs in diabetes. Diabetes Obes Metab. 2009; 11: 118–129 [DOI] [PubMed] [Google Scholar]
- 4. van den Berg A, Mols J, Han J. RISC-target interaction: cleavage and translational suppression. Biochim Biophys Acta. 2008; 1779: 668–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bartels CL, Tsongalis GJ. MicroRNAs: novel biomarkers for human cancer. Clin Chem. 2009; 55: 623–631 [DOI] [PubMed] [Google Scholar]
- 6. Pandey AK, Agarwal P, Kaur K, Datta M. MicroRNAs in diabetes: tiny players in big disease. Cell Physiol Biochem. 2009; 23: 221–232 [DOI] [PubMed] [Google Scholar]
- 7. Hezova R, Slaby O, Faltejskova P, et al. MicroRNA-342, microRNA-191 and microRNA-510 are differentially expressed in T regulatory cells of type 1 diabetic patients. Cell Immunol. 2010; 260: 70–74 [DOI] [PubMed] [Google Scholar]
- 8. Ferland-McCollough D, Ozanne SE, Siddle K, Willis AE, Bushell M. The involvement of microRNAs in Type 2 diabetes. Biochem Soc Trans. 2010; 38: 1565–1570 [DOI] [PubMed] [Google Scholar]
- 9. Centers for Disease Control and Prevention General information and national estimates on diabetes in the United States. National Diabetes Fact Sheet. 2007. Available at: http://www.cdc.gov/diabetes/pubs/pdf/ndfs_2007.pdf [Google Scholar]
- 10. Cheung N, Mitchell P, Wong TY. Diabetic retinopathy. Lancet. 2010; 376: 124–136 [DOI] [PubMed] [Google Scholar]
- 11. Antonetti DA, Barber AJ, Bronson SK, et al. Diabetic retinopathy: seeing beyond glucose-induced microvascular disease. Diabetes. 2006; 55: 2401–2411 [DOI] [PubMed] [Google Scholar]
- 12. Kaneto H, Matsuoka TA, Katakami N, et al. Oxidative stress and the JNK pathway are involved in the development of type 1 and type 2 diabetes. Curr Mol Med. 2007; 7: 674–686 [DOI] [PubMed] [Google Scholar]
- 13. Kowluru RA, Chan PS. Oxidative stress and diabetic retinopathy. Exp Diabetes Res. 2007; 2007: 43603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Al-Shabrawey M, Smith S. Prediction of diabetic retinopathy: role of oxidative stress and relevance of apoptotic biomarkers. EPMA Journal. 2010; 1: 56–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang Y, Hayden MR, Sowers S, Bagree SV, Sowers JR. Retinal redox stress and remodeling in cardiometabolic syndrome and diabetes. Oxid Med Cell Longev. 2010; 3: 392–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. El-Remessy AB, Tang Y, Zhu G, et al. Neuroprotective effects of cannabidiol in endotoxin-induced uveitis: critical role of p38 MAPK activation. Mol Vis. 2008; 14: 2190–2203 [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17. Lamb RE, Goldstein BJ. Modulating an oxidative-inflammatory cascade: potential new treatment strategy for improving glucose metabolism, insulin resistance, and vascular function. Int J Clin Pract. 2008; 62: 1087–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fukai T, Ushio-Fukai M. Superoxide dismutases: role in redox signaling, vascular function, and diseases. Antioxid Redox Signal. 2011; 15: 1583–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lubos E, Loscalzo J, Handy DE. Glutathione peroxidase-1 in health and disease: from molecular mechanisms to therapeutic opportunities. Antioxid Redox Signal. 2011; 15: 1957–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Oliver PL, Finelli MJ, Edwards B, et al. Oxr1 is essential for protection against oxidative stress-induced neurodegeneration. PLoS Genet. 2011; 7: e1002338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Elliott NA, Volkert MR. Stress induction and mitochondrial localization of Oxr1 proteins in yeast and humans. Mol Cell Biol. 2004; 24: 3180–3187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Durand M, Kolpak A, Farrell T, et al. The OXR domain defines a conserved family of eukaryotic oxidation resistance proteins. BMC Cell Biol. 2007; 8: 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jaramillo-Gutierrez G, Molina-Cruz A, Kumar S, Barillas-Mury C. The Anopheles gambiae oxidation resistance 1 (OXR1) gene regulates expression of enzymes that detoxify reactive oxygen species. PLoS One. 2010; 5: e11168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Volkert MR, Elliott NA, Housman DE. Functional genomics reveals a family of eukaryotic oxidation protection genes. Proc Natl Acad Sci U S A. 2000; 97: 14530–14535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Natoli R, Provis J, Valter K, Stone J. Expression and role of the early-response gene Oxr1 in the hyperoxia-challenged mouse retina. Invest Ophthalmol Vis Sci. 2008; 49: 4561–4567 [DOI] [PubMed] [Google Scholar]
- 26. Chen Y, Hu Y, Zhou T, et al. Activation of the Wnt pathway plays a pathogenic role in diabetic retinopathy in humans and animal models. Am J Pathol. 2009; 175: 2676–2685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001; 25: 402–408 [DOI] [PubMed] [Google Scholar]
- 28. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008; 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- 29. Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007; 35: W71–W74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008; 36: D149–D153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005; 120: 15–20 [DOI] [PubMed] [Google Scholar]
- 32. Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004; 10: 1507–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zeng Y, Cullen BR. Sequence requirements for micro RNA processing and function in human cells. RNA. 2003; 9: 112–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dunn KC, Aotaki-Keen AE, Putkey FR, Hjelmeland LM. ARPE-19, a human retinal pigment epithelial cell line with differentiated properties. Exp Eye Res. 1996; 62: 155–169 [DOI] [PubMed] [Google Scholar]
- 35. Limb GA, Salt TE, Munro PM, Moss SE, Khaw PT. In vitro characterization of a spontaneously immortalized human Muller cell line (MIO-M1). Invest Ophthalmol Vis Sci. 2002; 43: 864–869 [PubMed] [Google Scholar]
- 36. Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976; 72: 248–254 [DOI] [PubMed] [Google Scholar]
- 37. Haan C, Behrmann I. A cost effective non-commercial ECL-solution for Western blot detections yielding strong signals and low background. J Immunol Methods. 2007; 318: 11–19 [DOI] [PubMed] [Google Scholar]
- 38. Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987; 15: 8125–8148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Takahashi Y, Moiseyev G, Chen Y, Ma JX. Identification of conserved histidines and glutamic acid as key residues for isomerohydrolase activity of RPE65, an enzyme of the visual cycle in the retinal pigment epithelium. FEBS Lett. 2005; 579: 5414–5418 [DOI] [PubMed] [Google Scholar]
- 40. Takahashi Y, Moiseyev G, Chen Y, Ma JX. The roles of three palmitoylation sites of RPE65 in its membrane association and isomerohydrolase activity. Invest Ophthalmol Vis Sci. 2006; 47: 5191–5196 [DOI] [PubMed] [Google Scholar]
- 41. Barber AJ, Antonetti DA, Kern TS, et al. The Ins2Akita mouse as a model of early retinal complications in diabetes. Invest Ophthalmol Vis Sci. 2005; 46: 2210–2218 [DOI] [PubMed] [Google Scholar]
- 42. Smith SB, Duplantier J, Dun Y, et al. In vivo protection against retinal neurodegeneration by sigma receptor 1 ligand (+)-pentazocine. Invest Ophthalmol Vis Sci. 2008; 49: 4154–4161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Akimov NP, Renteria RC. Spatial frequency threshold and contrast sensitivity of an optomotor behavior are impaired in the Ins2Akita mouse model of diabetes. Behav Brain Res. 2012; 226: 601–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chan YC, Khanna S, Roy S, Sen CK. miR-200b targets Ets-1 and is downregulated by hypoxia to induce angiogenic response of endothelial cells. J Biol Chem. 2011; 286: 2047–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Magenta A, Cencioni C, Fasanaro P, et al. miR-200c is upregulated by oxidative stress and induces endothelial cell apoptosis and senescence via ZEB1 inhibition. Cell Death Differ. 2011; 18: 1628–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McArthur K, Feng B, Wu Y, Chen S, Chakrabarti S. MicroRNA-200b regulates vascular endothelial growth factor-mediated alterations in diabetic retinopathy. Diabetes. 2011; 60: 1314–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kovacs B, Lumayag S, Cowan C, Xu S. MicroRNAs in early diabetic retinopathy in streptozotocin-induced diabetic rats. Invest Ophthalmol Vis Sci. 2011; 52: 4402–4409 [DOI] [PubMed] [Google Scholar]
- 48. Bolzan AD, Bianchi MS. Genotoxicity of streptozotocin. Mutat Res. 2002; 512: 121–134 [DOI] [PubMed] [Google Scholar]
- 49. Freeman WM, Bixler GV, Brucklacher RM, et al. Transcriptomic comparison of the retina in two mouse models of diabetes. J Ocul Biol Dis Infor. 2009; 2: 202–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bringmann A, Pannicke T, Grosche J, et al. Muller cells in the healthy and diseased retina. Prog Retin Eye Res. 2006; 25: 397–424 [DOI] [PubMed] [Google Scholar]
- 51. Frank RN, Amin RH, Eliott D, Puklin JE, Abrams GW. Basic fibroblast growth factor and vascular endothelial growth factor are present in epiretinal and choroidal neovascular membranes. Am J Ophthalmol. 1996; 122: 393–403 [DOI] [PubMed] [Google Scholar]
- 52. Kwak N, Okamoto N, Wood JM, Campochiaro PA. VEGF is major stimulator in model of choroidal neovascularization. Invest Ophthalmol Vis Sci. 2000; 41: 3158–3164 [PubMed] [Google Scholar]
- 53. Miller JW, Adamis AP, Shima DT, et al. Vascular endothelial growth factor/vascular permeability factor is temporally and spatially correlated with ocular angiogenesis in a primate model. Am J Pathol. 1994; 145: 574–584 [PMC free article] [PubMed] [Google Scholar]
- 54. Vinores SA, Youssri AI, Luna JD, et al. Upregulation of vascular endothelial growth factor in ischemic and non-ischemic human and experimental retinal disease. Histol Histopathol. 1997; 12: 99–109 [PubMed] [Google Scholar]
- 55. Peter ME. Targeting of mRNAs by multiple miRNAs: the next step. Oncogene. 2010; 29: 2161–2164 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.