Abstract
Purpose.
To investigate genetic determinants of macular pigment optical density in women from the Carotenoids in Age-Related Eye Disease Study (CAREDS), an ancillary study of the Women's Health Initiative Observational Study.
Methods.
1585 of 2005 CAREDS participants had macular pigment optical density (MPOD) measured noninvasively using customized heterochromatic flicker photometry and blood samples genotyped for 440 single nucleotide polymorphisms (SNPs) in 26 candidate genes related to absorption, transport, binding, and cleavage of carotenoids directly, or via lipid transport. SNPs were individually tested for associations with MPOD using least-squares linear regression.
Results.
Twenty-one SNPs from 11 genes were associated with MPOD (P ≤ 0.05) after adjusting for dietary intake of lutein and zeaxanthin. This includes variants in or near genes related to zeaxanthin binding in the macula (GSTP1), carotenoid cleavage (BCMO1), cholesterol transport or uptake (SCARB1, ABCA1, ABCG5, and LIPC), long-chain omega-3 fatty acid status (ELOVL2, FADS1, and FADS2), and various maculopathies (ALDH3A2 and RPE65). The strongest association was for rs11645428 near BCMO1 (βA = 0.029, P = 2.2 × 10−4). Conditional modeling within genes and further adjustment for other predictors of MPOD, including waist circumference, diabetes, and dietary intake of fiber, resulted in 13 SNPs from 10 genes maintaining independent association with MPOD. Variation in these single gene polymorphisms accounted for 5% of the variability in MPOD (P = 3.5 × 10−11).
Conclusions.
Our results support that MPOD is a multi-factorial phenotype associated with variation in genes related to carotenoid transport, uptake, and metabolism, independent of known dietary and health influences on MPOD.
In 1585 postmenopausal women of the Carotenoids in Age-Related Eye Disease Study sample, common genetic variants in or near genes involved in carotenoid transport, uptake, and metabolism were associated with density of lutein and zeaxanthin in the macula, independent of other known predictors, including dietary intake of carotenoids.
Introduction
The carotenoids lutein (L) and zeaxanthin (Z) from the diet, and meso-Z, synthesized in the retina from L,1 concentrate in the inner retinal layer of the macula2 where they comprise macular pigment. The density3 and concentration4,5 of these macular carotenoids vary more than 10-fold among individuals. There is interest in mechanisms to enhance macular pigment because of emerging evidence that they are important to ocular health. L and Z may lower the risk and/or delay onset of the common and costly condition of AMD.6–8 This may be due to the ability of macular pigment to absorb short wavelength (blue) light; depending on the density of the pigment, it is estimated that 40% to 90% of blue light is absorbed by these pigments.9 Blue light absorption may limit oxidative stress, photochemical damage, or damage due to the formation of photosensitizing pigments in the retina as byproducts of the visual cycle, which might otherwise act as inflammatory triggers. These carotenoid pigments are also thought to have other protective roles in the macula, including antioxidant and anti-inflammatory effects directly10,11 and indirectly.12 Independent of protection against development or worsening of AMD, macular pigment might also contribute to better visual performance.13–17
Understanding the predictors of macular pigment optical density (MPOD) in large samples is one way to understand conditions that favor the accretion of these carotenoids in the retina. Previous studies in the Carotenoids in Age-Related Eye Disease Study (CAREDS) indicated that women with higher levels of L and Z in diet or serum, have higher MPOD.3 These factors account for only 8% of the variability in MPOD, indicating the influence of other factors. Consistent with other factors playing an auxiliary role, the blood18–20 and retinal1,21–36 response to oral supplementation is known to be highly variable.
Results of a previous study in twins suggest that genetic factors may explain 67% of variability in MPOD37 and 27% of variability in response to supplementation over 6 months.34 Variation in candidate genes may influence the ability to accumulate macular pigments.38 Candidates include genes encoding proteins that (1) bind carotenoids in the retina,39–41 (2) cleave carotenoids or influence metabolism of vitamin A,38,42–44 (3) have joint roles in transport of carotenoids and lipids through plasma membranes in the retina45,46 and intestine,46,47 (4) influence the circulating levels of high density lipoprotein (HDL) molecules48,49 that carry approximately half of circulating macular carotenoids,50,51 (5) influence omega-3 fatty acid levels in the blood or retina,52,53 and/or (6) genes that have been previously related to both macular pigment density and maculopathies.54–56
The main objective of this paper is to examine and provide a broad overview of the hypothesized genetic determinants of MPOD in the CAREDS, an ancillary study of the Women's Health Initiative (WHI). CAREDS was designed to investigate the relationships of L and Z in the diet, serum, and retina to AMD57,58 and cataract presence and severity among postmenopausal women with low and high LZ intakes at three of the WHI clinical sites.59
Methods
All participants provided informed consent for participation in CAREDS, approval for research was granted by the institutional review boards at each participating university, and all procedures conformed to the Declaration of Helsinki. Detailed description of methods can be found in Supplementary Material S1 (see Supplementary Material and Supplementary S1, http://www.iovs.org/content/54/3/2333/suppl/DC1). In brief, 1585 women of the 2005 women in CAREDS with macular pigment measures and genotype data were included in the present study. Previous manuscripts provide detailed descriptions of CAREDS design and sampling,3 macular pigment measurement protocols,60 measures of serum carotenoids,3 and measures of dietary and lifestyle factors.3 A total of 473 SNPs tagging variation (minor allele frequency [MAF] ≥ 5%, r2 ≥ 0.80) from 26 candidate genes for carotenoid status, and 190 ancestry informative markers,61 were genotyped. After standard quality control procedures (see Supplementary Material and Supplementary S1, http://www.iovs.org/content/54/3/2333/suppl/DC1), 440 SNPs were tested for association with MPOD (see Supplementary Material and Supplementary Table S2, http://www.iovs.org/content/54/3/2333/suppl/DC1 for complete list of SNPs). The focus of this report is on MPOD at 0.5° from foveal center, the location at which the within to between variability of MPOD was the lowest60 and is most commonly measured across studies. Associations between SNPs and other eccentricities of MPOD (at 0.25°, 1°, and 1.75°) were examined, but were generally similar (data not shown) and will be the focus of future investigations. The ε2, ε3, and, ε4 alleles of apolipoprotein E (APOE) were measured as haplotypes of SNPs rs429358 and rs7412. Association analysis was performed using linear regression as implemented in PLINK version 1.07,62 assuming an additive genetic model and adjusting for global (genome-wide) ancestry, via the first two principal components, and LZ intake (from diet and supplements). Further adjustments for other determinants of MPOD were tested to ascertain direct SNP effects on MPOD. Because of correlation between SNPs within a gene, conditional modeling, using forward selection, was performed to ascertain SNPs with independent effects within the gene. SNPs associated with MPOD were tested for interactions with LZ intake and waist circumference, two strong, modifiable predictors of MPOD. Statistical interactions were tested using an interaction term in the linear regression model where the SNP is coded additively and the environmental factor continuous. When the one degree of freedom test for interaction was suggestive (P < 0.20), SNP–MPOD associations were stratified by tertile of environmental variable to qualitatively understand dependency of SNP effects by waist circumference or dietary intake.
Results
Sample Characteristics and Predictors of MPOD
In this sample of 1585 women, over 97% of which were self-reported Caucasian, the mean (±SD) MPOD at 0.5° from the foveal center in the right eye was 0.36 (±0.21). Women were, on average 69 years of age, had a body mass index of 28 kg/m2, and waist circumference of 88 cm. Six percent reported having diabetes, 45% were using hormone therapy at the time of MPOD measurement, 26% were using cholesterol lowering medication, and only 3% were self-reported current smokers. Correlates of MPOD in this subset of CAREDS participants, after adjustment for LZ intake, (Table 1) were consistent with previously reported predictors of MPOD in the CAREDS sample.3 Additional variables considered as potential physiological determinants of MPOD since our previous publication in 2006, informed either through new literature or the current analysis, included serum triglycerides (r = −0.07, P = 0.003), serum measures of β-carotene3 (r = 0.13, P < 0.001), C-reactive protein (r = −0.07, P = 0.03), and 25-hyrdoxyvitamin D63 (r = 0.05, P = 0.08), the Healthy Eating Index-2005 score (r = 0.08, P < 0.001) and Healthy Lifestyle Score64 (r = 0.008, P = 0.001).
Table 1. .
Mean Macular Pigment Optical Density (MPOD) by Level of Various Potential Predictors
|
Variable |
N
of Subjects |
Adjusting for Dietary L and Z* |
Additional Adjustment* for Waist Circumference, Diabetes Status, Fiber Intake, and Serum L and Z and 13 SNPs Associated with MPOD |
||
|
Mean MPOD |
% Diff. from Ref. |
Mean MPOD |
% Diff. from Ref. |
||
| Demographics, anthropometrics, and medical history | |||||
| Age group, y | |||||
| ≤69 | 801 | 0.36 | Ref. | 0.36 | Ref. |
| 70 to 74 | 386 | 0.38 | 5 | 0.39 | 7 |
| ≥75 | 398 | 0.34 | −5 | 0.34 | −5 |
| P value† | 0.81 | 0.81 | |||
| Waist circumference, cm, quartile (median) | |||||
| 1 (73.7) | 440 | 0.40 | Ref. | 0.39 | Ref. |
| 2 (83.8) | 378 | 0.39 | −4 | 0.37 | −3 |
| 3 (91.4) | 361 | 0.34 | −14 | 0.35 | −10 |
| 4 (104.1) | 406 | 0.31 | −22 | 0.34 | −12 |
| P value† | <.0001 | <.0001 | |||
| Presence of diabetes | |||||
| No | 1482 | 0.37 | Ref. | 0.37 | Ref. |
| Yes | 103 | 0.28 | −24 | 0.30 | −17 |
| P value† | <.0001 | 0.003 | |||
| Study visit site | |||||
| Iowa | 535 | 0.38 | Ref. | 0.38 | Ref. |
| Oregon | 515 | 0.39 | 2 | 0.38 | 0 |
| Wisconsin | 535 | 0.32 | −15 | 0.32 | −16 |
| P value† | <.0001 | <.0001 | |||
| Education, WHI baseline | |||||
| High school | 355 | 0.34 | Ref | 0.35 | Ref. |
| College | 776 | 0.36 | 5 | 0.36 | 4 |
| Postcollege | 454 | 0.38 | 10 | 0.36 | 3 |
| P value† | 0.02 | 0.48 | |||
| Diet and lifestyle | |||||
| Dietary L+Z, μg, quintile (median) | |||||
| 1 (797.9) | 317 | 0.31 | Ref. | 0.33 | Ref. |
| 2 (1383.1) | 317 | 0.33 | 8 | 0.34 | 2 |
| 3 (2120.5) | 317 | 0.38 | 24 | 0.38 | 14 |
| 4 (3121) | 317 | 0.39 | 27 | 0.37 | 12 |
| 5 (5330.9) | 317 | 0.41 | 32 | 0.38 | 14 |
| P value† | <.0001 | 0.18 | |||
| Physical activity, MetHrs/wk‡ | |||||
| <3 | 369 | 0.34 | Ref. | 0.37 | Ref. |
| 3 to 10 | 348 | 0.36 | 5 | 0.37 | 0 |
| 10 to 21 | 440 | 0.36 | 4 | 0.35 | −5 |
| ≥21 | 417 | 0.39 | 12 | 0.36 | −3 |
| P value† | 0.14 | 0.09 | |||
| Dietary fiber, g/d, quintile (median) | |||||
| 1 (7.9) | 316 | 0.31 | Ref. | 0.33 | Ref. |
| 2 (12.1) | 317 | 0.36 | 14 | 0.36 | 10 |
| 3 (16.0) | 316 | 0.36 | 15 | 0.35 | 7 |
| 4 (20.1) | 317 | 0.39 | 25 | 0.38 | 16 |
| 5 (27.0) | 316 | 0.39 | 26 | 0.38 | 15 |
| P value† | <.0001 | 0.004 | |||
| Dietary saturated fat, % of energy, quintile (median) | |||||
| 1 (7.6) | 316 | 0.38 | Ref. | 0.35 | Ref. |
| 2 (9.8) | 317 | 0.38 | 0 | 0.37 | 5 |
| 3 (11.5) | 316 | 0.36 | −6 | 0.36 | 4 |
| 4 (13.6) | 317 | 0.36 | −6 | 0.36 | 3 |
| 5 (16.6) | 316 | 0.35 | −9 | 0.37 | 5 |
| P value† | 0.01 | 0.82 | |||
| Dietary polyunsaturated fat, % of energy, quintile (median) | |||||
| 1 (4.34) | 316 | 0.34 | Ref. | 0.33 | Ref. |
| 2 (5.66) | 317 | 0.37 | 11 | 0.37 | 13 |
| 3 (6.67) | 316 | 0.37 | 11 | 0.37 | 13 |
| 4 (7.82) | 317 | 0.37 | 11 | 0.38 | 15 |
| 5 (9.78) | 316 | 0.36 | 9 | 0.37 | 12 |
| P value† | 0.21 | 0.03 | |||
| Dietary total n-3 fatty acids, g/d, quintile (median) | |||||
| 1 (0.61) | 316 | 0.34 | Ref. | 0.35 | Ref. |
| 2 (0.91) | 317 | 0.36 | 5 | 0.36 | 5 |
| 3 (1.16) | 316 | 0.37 | 10 | 0.36 | 5 |
| 4 (1.49) | 317 | 0.37 | 8 | 0.36 | 4 |
| 5 (2.10) | 316 | 0.38 | 11 | 0.37 | 8 |
| P value† | 0.05 | 0.23 | |||
| Dietary retinol, μg/d, quintile (median) | |||||
| 1 (196.2) | 316 | 0.37 | Ref. | 0.37 | Ref. |
| 2 (338.8) | 317 | 0.37 | 1 | 0.37 | 0 |
| 3 (479.0) | 316 | 0.36 | −1 | 0.36 | −4 |
| 4 (641.4) | 317 | 0.36 | −2 | 0.36 | −4 |
| 5 (919.1) | 316 | 0.35 | −4 | 0.35 | −7 |
| P value† | 0.15 | 0.07 | |||
| Total HEI-2005 score, quintile (median) | |||||
| 1 (61.4) | 317 | 0.34 | Ref. | 0.37 | Ref. |
| 2 (67.0) | 317 | 0.36 | 4 | 0.36 | −2 |
| 3 (70.5) | 317 | 0.36 | 4 | 0.36 | −2 |
| 4 (73.5) | 317 | 0.37 | 7 | 0.36 | −2 |
| 5 (77.7) | 317 | 0.39 | 15 | 0.37 | 1 |
| P value† | 0.001 | 0.70 | |||
| Healthy lifestyle score | |||||
| 0 to 2 | 415 | 0.34 | Ref. | 0.37 | Ref. |
| 3 | 426 | 0.36 | 7 | 0.36 | −2 |
| 4 | 368 | 0.37 | 8 | 0.36 | −3 |
| 5 | 287 | 0.38 | 13 | 0.36 | −3 |
| 6 | 89 | 0.40 | 18 | 0.35 | −4 |
| P value† | 0.001 | 0.44 | |||
| Serum analytes | |||||
| L+Z, trans, μmol/L, quintile (median) | |||||
| 1 (0.15) | 313 | 0.25 | Ref. | 0.28 | Ref. |
| 2 (0.22) | 314 | 0.35 | 36 | 0.35 | 27 |
| 3 (0.29) | 313 | 0.37 | 46 | 0.37 | 32 |
| 4 (0.37) | 313 | 0.41 | 59 | 0.39 | 40 |
| 5 (0.50) | 314 | 0.44 | 71 | 0.42 | 51 |
| P value† | <.0001 | <.0001 | |||
| Retinol, μmol/L, quintile (median) | |||||
| 1 (1.84) | 313 | 0.36 | Ref. | 0.36 | Ref. |
| 2 (2.20) | 314 | 0.35 | −2 | 0.35 | −4 |
| 3 (2.50) | 313 | 0.38 | 8 | 0.39 | 7 |
| 4 (2.80) | 314 | 0.37 | 4 | 0.36 | −1 |
| 5 (3.31) | 313 | 0.36 | 0 | 0.36 | −1 |
| P value† | 0.96 | 0.70 | |||
| Retinyl palmitate, μmol/L, quintile (median) | |||||
| 1 (0.0004) | 195 | 0.37 | Ref. | 0.37 | Ref. |
| 2 (0.0010) | 193 | 0.36 | −3 | 0.36 | −2 |
| 3 (0.0019) | 191 | 0.38 | 3 | 0.38 | 2 |
| 4 (0.0032) | 194 | 0.40 | 8 | 0.40 | 7 |
| 5 (0.0070) | 193 | 0.34 | −7 | 0.33 | −10 |
| P value† | 0.13 | 0.03 | |||
| Beta-carotene, all trans, μmol/L, quintile (median) | |||||
| 1 (0.23) | 313 | 0.29 | Ref. | 0.34 | Ref. |
| 2 (0.40) | 314 | 0.36 | 21 | 0.37 | 9 |
| 3 (0.60) | 313 | 0.38 | 30 | 0.37 | 11 |
| 4 (0.90) | 314 | 0.39 | 31 | 0.36 | 8 |
| 5 (1.52) | 313 | 0.40 | 37 | 0.37 | 12 |
| P value† | <.0001 | 0.31 | |||
| Triglyceride, μmol/L, quintile (median) | |||||
| 1 (0.90) | 316 | 0.36 | Ref. | 0.34 | Ref. |
| 2 (1.25) | 315 | 0.39 | 8 | 0.38 | 12 |
| 3 (1.57) | 305 | 0.37 | 3 | 0.36 | 8 |
| 4 (2.07) | 319 | 0.38 | 7 | 0.39 | 17 |
| 5 (2.96) | 310 | 0.32 | −12 | 0.34 | 0 |
| P value† | 0.003 | 0.48 | |||
| Vitamin D, adjusted for blood drawn month, nmol/L, quintile (median) | |||||
| 1 (30.4) | 235 | 0.35 | Ref. | 0.37 | Ref. |
| 2 (44.5) | 236 | 0.36 | 2 | 0.37 | 0 |
| 3 (55.7) | 235 | 0.36 | 3 | 0.36 | −2 |
| 4 (67.2) | 236 | 0.38 | 9 | 0.37 | 1 |
| 5 (84.9) | 235 | 0.38 | 7 | 0.35 | −6 |
| P value† | 0.07 | 0.52 | |||
| C-reactive protein, mg/L, quintile (median) | |||||
| 1 (0.7) | 221 | 0.38 | Ref. | 0.35 | Ref. |
| 2 (1.5) | 219 | 0.38 | −1 | 0.35 | 0 |
| 3 (2.7) | 216 | 0.36 | −6 | 0.36 | 3 |
| 4 (4.8) | 225 | 0.36 | −5 | 0.37 | 5 |
| 5 (9.9) | 221 | 0.35 | −8 | 0.38 | 8 |
| P value† | 0.03 | 0.35 | |||
Diff., different.
Exception to adjustment variables are when the respective variable is that being tested.
P values are for trend of MPOD across continuous variable or categorical, where appropriate.
MetHrs/Week is h/wk spent doing an activity by the intensity (the fold increase the activity is estimated to increase metabolic activity [oxygen consumption] relative to the resting state).
Multivariable analyses of modifiable factors that might be physiologically related to lower MPOD resulted in the following model of independent predictors of MPOD (in order of model entry): LZ intake, serum LZ, waist circumference, dietary fiber, and diabetes status at CAREDS. This model explained 9.5% of the variability in MPOD and determined the full multivariable adjustment model used to test whether SNPs were associated with MPOD directly or indirectly via influence on these other determinants.
Overview of SNP Associations with MPOD
Adjusting for LZ intake and ancestry, 21 SNPs from 11 genes were associated with MPOD at P less than or equal to 0.05. Conditional modeling within each gene resulted in 14 SNPs from 11 genes maintaining independent associations with MPOD (Table 2). Further adjustment for other MPOD predictors had little or no influence. The few exceptions are noted below.
Table 2. .
SNPs Independently Associated with MPOD within Each Gene, in CAREDS (N = 1585)
|
Gene |
SNP |
Genotype |
N |
Adjusted for Dietary L and Z and Ancestry* |
Further Adjusted for Waist Circumference, Diabetes, and Fiber Intake |
Final Adjustment for Serum L and Z |
|||||
|
Mean MPOD |
% Change from Reference |
β† |
P
Value |
β† |
P
Value |
β† |
P
Value |
||||
| Xanthophyll binding in retina | |||||||||||
| GSTP1 | rs675679 | AA | 1307 | 0.36 | Ref. | 0.028 | 0.033 | 0.027 | 0.035 | 0.027 | 0.031 |
| AC | 263 | 0.39 | 8 | ||||||||
| CC | 14 | 0.40 | 13 | ||||||||
| Carotenoid cleavage | |||||||||||
| BCMO1 | rs11645428 | GG | 710 | 0.35 | Ref. | 0.029 | 2.2 × 10−4 | 0.031 | 6.3 × 10−5 | 0.017 | 0.027 |
| AG | 694 | 0.36 | 5 | ||||||||
| AA | 181 | 0.42 | 20 | ||||||||
| rs6564863 | GG | 686 | 0.35 | Ref. | 0.017 | 0.027 | 0.020 | 0.011 | 0.018 | 0.018 | |
| AG | 718 | 0.36 | 3 | ||||||||
| AA | 176 | 0.40 | 12 | ||||||||
| HDL transport or status | |||||||||||
| ABCA1 | rs1929841 | AA | 995 | 0.37 | Ref. | −0.020 | 0.029 | −0.024 | 0.007 | −0.022 | 0.011 |
| AC | 518 | 0.36 | −2 | ||||||||
| CC | 72 | 0.30 | −19 | ||||||||
| ABCG5 | rs10179921 | GG | 1426 | 0.37 | Ref. | −0.041 | 0.024 | −0.038 | 0.028 | −0.038 | 0.026 |
| AG or AA | 153 | 0.33 | −11 | ||||||||
| LIPC | rs6078 | GG | 1480 | 0.37 | Ref. | −0.044 | 0.041 | −0.049 | 0.020 | −0.043 | 0.038 |
| AG or AA | 105 | 0.32 | −12 | ||||||||
| Lipid and/or carotenoid absorption | |||||||||||
| SCARB1 | rs10744182 | AA | 525 | 0.38 | Ref. | −0.021 | 0.005 | −0.021 | 0.004 | −0.020 | 0.005 |
| AG | 760 | 0.36 | −5 | ||||||||
| GG | 298 | 0.34 | −11 | ||||||||
| rs838879 | AA | 768 | 0.38 | Ref. | −0.018 | 0.020 | −0.020 | 0.010 | −0.018 | 0.016 | |
| AG | 647 | 0.35 | −7 | ||||||||
| GG | 169 | 0.35 | −8 | ||||||||
| rs4379922 | AA | 651 | 0.37 | Ref. | −0.016 | 0.036 | −0.019 | 0.010 | −0.016 | 0.025 | |
| AG | 711 | 0.36 | −3 | ||||||||
| GG | 220 | 0.34 | −10 | ||||||||
| Long-chain fatty acid status | |||||||||||
| ELOVL2 | rs1150561 | GG | 1407 | 0.36 | Ref. | 0.040 | 0.016 | 0.045 | 0.006 | 0.040 | 0.012 |
| AG or AA | 178 | 0.40 | 11 | ||||||||
| FADS1 | rs174534 | AA | 711 | 0.35 | Ref. | 0.017 | 0.031 | 0.015 | 0.056 | 0.015 | 0.041 |
| AG | 695 | 0.38 | 8 | ||||||||
| GG | 177 | 0.37 | 7 | ||||||||
| FADS2 | rs2727271 | AA | 1244 | 0.36 | Ref. | 0.024 | 0.050 | 0.019 | 0.113 | 0.017 | 0.136 |
| AT | 323 | 0.38 | 7 | ||||||||
| TT | 15 | 0.40 | 12 | ||||||||
| Genes previously related to maculopathies | |||||||||||
| RPE65 | rs4926339 | GG | 556 | 0.34 | Ref. | 0.018 | 0.020 | 0.017 | 0.021 | 0.015 | 0.038 |
| AG | 784 | 0.37 | 10 | ||||||||
| AA | 244 | 0.37 | 8 | ||||||||
| ALDH3A2 | rs8069576 | GG | 495 | 0.38 | Ref. | −0.017 | 0.027 | −0.016 | 0.026 | −0.015 | 0.037 |
| AG | 740 | 0.36 | −6 | ||||||||
| AA | 290 | 0.35 | −9 | ||||||||
First two principal components from principal component analysis using 176 ancestry informative markers.
Beta is increase in MPOD per additional minor allele (additive genetic model).
A multivariable SNP model containing 13 SNPs significantly associated with MPOD independent of other known predictors was able to explain 5.1% of the variability in MPOD (Table 3). SNPs contributing to the most variability included two in or near BCMO1 and three in SCARB1, the gene for scavenger receptor class B, member 1. Jointly, these BCMO1 and SCARB1 SNPs explained approximately 2.3% of the variability in MPOD. Detailed results for SNPs from each class of candidate gene are described below. SNP associations were consistent across tertile of the largest two modifiable determinants of MPOD, LZ intake and waist circumference, with no evidence for statistical interactions between SNPs and either LZ intake or waist circumference (Pinteraction > 0.20), except where noted below.
Table 3. .
Final Multivariable SNP Model for SNPs Associated with MPOD in the CAREDS Population
|
Gene, SNP |
β |
P
Value |
Partial
R2 |
| BCMO1, rs11645428 | 0.025 | 0.001 | 0.7% |
| BCMO1, rs6564863 | 0.018 | 0.021 | 0.3% |
| SCARB1, rs10744182 | −0.024 | 0.002 | 0.6% |
| SCARB1, rs4379922 | −0.014 | 0.061 | 0.3% |
| SCARB1, rs838879 | −0.020 | 0.014 | 0.4% |
| GSTP1, rs675679 | 0.032 | 0.015 | 0.4% |
| ABCA1, rs1929841 | −0.018 | 0.048 | 0.3% |
| ABCG5, rs10179921 | −0.041 | 0.023 | 0.3% |
| LIPC, rs6078 | −0.039 | 0.061 | 0.2% |
| ELOVL2, rs1150561 | 0.046 | 0.006 | 0.5% |
| FADS1, rs174534 | 0.020 | 0.011 | 0.4% |
| RPE65, rs4926339 | 0.019 | 0.014 | 0.4% |
| ALDH3A2, rs8069576 | −0.018 | 0.018 | 0.4% |
| Full model R2 | 5.1% | ||
| Full model P value | 3.5 × 10−11 |
Xanthophyll-Specific Carotenoid Binding Proteins.
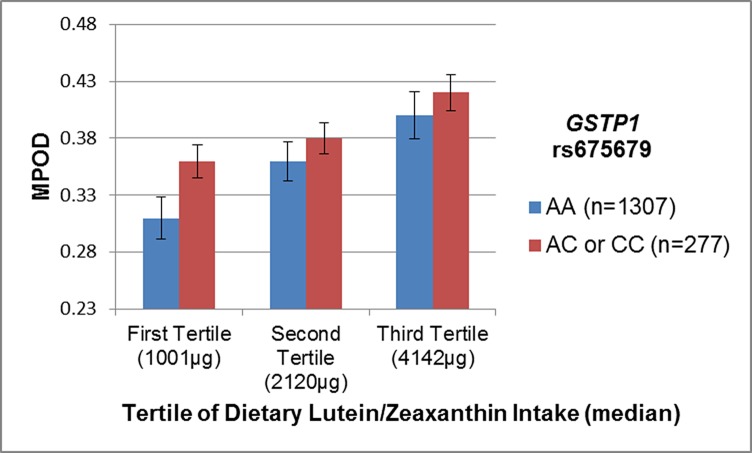
One SNP, rs675679, near GSTP1, the gene for a human ocular Z-specific binding protein40 was associated with MPOD independent of LZ intake (P = 0.03). The test for statistical interaction between rs675679 and LZ intake was suggestive (Pinteraction = 0.04). Stratifying women by tertile of LZ intake demonstrated that while the direction of SNP effects were consistent, the SNP association was most significant at the lowest level of intake (Fig. 1). A combination of the two risk effects (low LZ intake and homozygosity for the common allele, AA) resulted in significant deviation from additivity; these women had 26% lower MPOD compared with women with at least one C allele and highest intake (Fig. 1). No significant relationships with MPOD were identified for SNPs in STARD3, the gene for StAR-related lipid transfer domain containing 3, a human ocular lutein specific binding protein,39 even though the major allele (A) of rs9892427 was related to higher levels of LZ in serum (P = 0.006).
Figure 1.
Joint effects of dietary lutein and zeaxanthin intake and GSTP1 rs675679 on MPOD in CAREDS. The minor allele, C, of rs675679 was associated with higher mean MPOD in CAREDS. The direction of this relationship is consistent across all levels of dietary lutein and zeaxanthin intake, but significantly stronger at the lowest level of intake (Pinteraction = 0.04).
Carotenoid Cleavage and Retinoid Related Genes.
Five SNPs in or near BCMO1 were associated with MPOD (rs11645428, rs6564851, rs16955008, rs6564863, and rs9924371). The strongest associations were for two SNPs in weak linkage disequilibrium (r2 = 0.51) upstream of BCMO1, rs11645428 (P = 2.2 × 10−4), and rs6564851 (P = 0.003). After adjustment for serum LZ concentration, the effects of these two SNPs on MPOD were modestly attenuated because both rs11645428 and rs6564851 were strongly associated with serum LZ in CAREDS (P = 8.5 × 10−25 and P = 7.2 × 10−23, respectively). Three other SNPs in BCMO1 (rs9924371, rs6564863, and rs16955008) were associated with MPOD independent of serum LZ. However, conditional modeling of all five BCMO1 SNPs suggests correlation of effects, as only rs11645428 and rs6564863 maintain independent associations (Table 2) and jointly explained 1% of the variability in MPOD (Table 3).
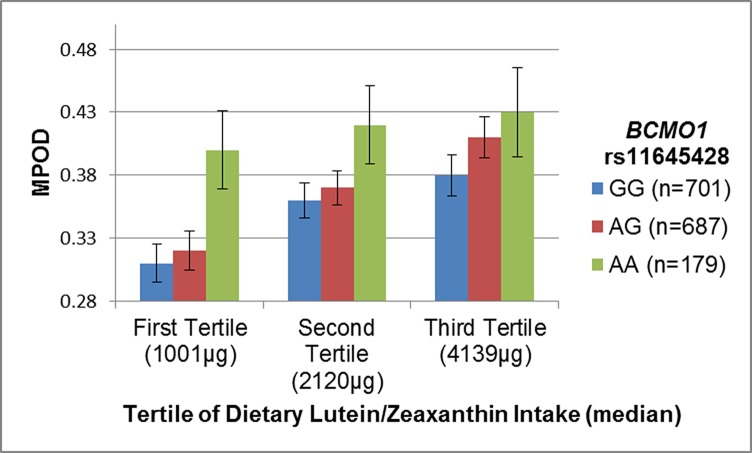
There was suggestive evidence for an interaction between rs1164528 and LZ intake (Pinteraction = 0.14). The trend for increasing MPOD with increasing number of minor alleles was consistent across all levels of LZ intake; however, the effect of the risk allele was greatest at the lowest level of intake (Fig. 2). Women both homozygous for the minor allele and in the highest tertile of LZ intake had a 32% higher MPOD than those with no minor alleles and intake in the lowest tertile.
Figure 2.
Joint effects of dietary lutein and zeaxanthin intake and BCMO1 rs11645428. The minor allele, A, of rs11645428 was associated with higher mean MPOD in CAREDS. This relationship is consistent across all levels of dietary lutein and zeaxanthin intake, but less marked with increasing lutein intake (Pinteraction = 0.15).
Three SNPs (rs4926339, rs6688807, and rs11581095) in RPE65, the gene for retinal pigment epithelium (RPE)–specific protein 65 kDa, were associated with MPOD in CAREDS. The effects of these SNPs were correlated, only rs4926339 maintained independent associations with MPOD (Table 2) and explained 0.4% of the variability in MPOD (Table 3). No associations were found between MPOD and SNPs in either RORA, the RAR-related orphan receptor A gene, or BCO2, the beta carotene dioxygenase gene.
Lipid and Carotenoid Transport and Metabolism Genes.
Four SNPs within SCARB1 were associated with MPOD: rs10744182, rs4379922, rs838879, and rs4238001. The most significant of these was rs10744182 (P = 0.005). When the conditional independence of these four SNPs was tested three (rs10744182, rs4379922, and rs838879) maintained independent effects within a multivariable SNP model (Table 2), and jointly explained 1.3% of the variability in MPOD in CAREDS (Table 3). There were no associations identified for SNPs in SCARB2, scavenger receptor class B, member 2, or CD36, cluster determinant 36.
Genes Related to HDL or Cholesterol Status.
Variants from the following genes related to HDL or cholesterol status were genotyped: ATP-binding cassette, subfamily A, member 1 (ABCA1), ATP-binding cassette, subfamily G, member 5 (ABCG5), and ATP-binding cassette, subfamily G, member 8 (ABCG8), hepatic lipase (LIPC), cholesteryl ester transfer protein, plasma (CETP), Niemann-Pick C1-like protein 1 (NPC1L1), and APOE. From these genes, one SNP each from ABCA1 (rs1929841), ABCG5 (rs10179921), and LIPC (rs6078) was associated with MPOD (Table 2). These three variants were able to explain 0.8% of the variability in MPOD (Table 3).
Synthesis or Metabolism of Long-Chain Fatty Acids in Blood or Retina.
Tag SNPs from five genes related to status of long-chain fatty acids in the blood or retina including elongation of very long chain fatty acid proteins 2, 4, and 5 (ELOVL2, ELOVL4, and ELOVL5) and fatty acid desaturase 1 and 2 (FADS1 and FADS2), were genotyped. After adjustment for LZ intake, two SNPs within ELOVL2 (rs1150561 and rs3846851), two SNPs in FADS1 (rs174534 and rs2727270), and one SNP in FADS2 (rs2727271) were associated with MPOD. Further adjustment for other predictors of MPOD slightly attenuated the FADS2 relationship (Table 2). After testing independence of the SNPs in each gene, only rs1150561 (ELOVL2) and rs174534 (FADS1) maintained independent effects and explained 0.5% and 0.4% of the variability in MPOD, respectively (Table 3).
Genes with Known Variants Related to Macular Pigment and Maculopathies.
We genotyped six tag SNPs within aldehyde dehydrogenase 3 family, member A2 (ALDH3A2), a gene with rare mutations known to cause the autosomal recessive condition Sjorgren-Larsson Syndrome, which among other clinical presentations, results in a complete lack of macular pigment.54 Of these, rs8069576 was associated with MPOD in CAREDS, independent of other predictors such as waist circumference, diabetes, LZ intake, and fiber (P = 0.047; Table 2). We were not able to identify any single associations between complement factor H (CFH), age-related maculopathy susceptibility 2 (ARMS2), HtrA serine peptidase 1 (HTRA1), or APOE and MPOD (data not shown).
Discussion
This study, conducted in 1585 postmenopausal women, is the most extensive investigation of genetic predictors of MPOD, to date. Results from the present cross-sectional study support the previously proposed hypothesis,38,65 and results of twin studies,34,37,66 suggesting a role of genetic variation in macular pigment density.
The genetic variation underlying MPOD appears to be polygenic involving common variation in numerous genes related to (1) carotenoid transport, uptake, and metabolism, (2) omega-3 fatty acid metabolism, and (3) some inherited maculopathies (discussed in detail below). Single genes that explain the greatest variability in MPOD were SCARB1, a scavenger receptor protein responsible for lipid and carotenoid uptake into gut and retina, and BCMO1, a carotenoid cleavage enzyme with preferred affinity for provitamin A carotenoids for synthesis of retinoids relative to xanthophyll carotenoids such as L and Z. BCMO1 and SCARB1 SNPs independently explained 2.3% of the variability in MPOD.
Evidence from this study suggests that the low percentage of variability in MPOD explained by dietary LZ in the present sample3 may be, in part, explained by genetic factors. For example, women in the lowest tertile of LZ intake who had two alleles of certain variants in GSTP1 and BCMO1, had levels of macular pigment, which were higher than or nearly as high as women with intakes in the highest tertile (Figs. 1, 2). This suggests the possibility that having certain genotypes may compensate, to some degree, for low intake of macular carotenoids. It also indicates that LZ intake may explain a higher percentage of variability in MPOD in persons with certain genotypes. It is important to note that the low estimate of variability explained by LZ intake reflects between person variability of individual LZ intake; thus, does not constitute evidence that the level of LZ intake is unimportant on an individual level. The persistence of a strong relationship of serum LZ to MPOD, even after adjusting for dietary, genetic, and phenotype characteristics might reflect a residual influence of variability in levels of dietary intake of these carotenoids. LZ concentration in serum is not subject to variability in composition of LZ in the particular foods as dietary estimates are; however, serum LZ might also reflect additional unknown and unaccounted for genetic, phenotypic, or dietary factors that influence the update of LZ into the body and transport into the blood.
The variable response to supplementation observed across many studies might be, in part, due to genetic factors. Consistent with this, results of a recent study of 161 mono- and dizygotic twin pairs estimated that 27% of the response to supplementation with 18 mg L and 2.4 mg Z for 6 months was due to heritable factors. Dietary supplementation with macular carotenoids has been observed to increase MPOD in 50%28 to 95%67 of subjects with many studies suggesting estimates between these two extremes.1,21–34 However, the magnitude of individual response in these studies is quite variable. We also note that the definition of macular “response” to intake of macular carotenoids is not standard; the magnitude of response in different regions of the macula needed to result in measurable differences in vision function or clinical outcomes remains to be further evaluated.
Variants in Genes Related to Carotenoid Uptake and Metabolism
We hypothesized variation in genes related to L and Z–specific binding proteins in the retina, StARD3,39 and GSTP1,40 respectively, might regulate retinal uptake or intracellular trafficking, thereby influencing MPOD. Variants in these genes have not yet been studied in relation to MPOD. We observed that one SNP, rs675679, near GSTP1, was associated with MPOD independent of known predictors of MPOD. No relationships with MPOD were identified for SNPs in STARD3, although variants in this gene were related to levels of LZ in serum.
In addition to the uptake in the retina, we hypothesized that variants in genes coding for enzymes involved in carotenoid cleavage and vitamin A metabolism might also influence the levels of macular carotenoids. BCMO1 encodes an enzyme active in the intestine and other tissues of the body that oxidatively cleave provitamin carotenoids to make vitamin A (retinal and retinol) and many biologically active retinoids, which are active in the visual cycle.68 Variants in BCMO1, including the SNPs reported here, rs11645428 and rs6564851, have been associated with serum carotenoid levels in a large meta-analysis, genome-wide association study,69 a result we were able to replicate in CAREDS. One previous report in a small sample of middle-aged men suggested rs7501331 in BCMO1 was associated with MPOD.70 In the CAREDS, we were unable to replicate this association (P = 0.56). However, we did identify five other SNPs in BCMO1 to be associated with MPOD. Although there is no evidence that L and Z are substrates for BCMO1,71 it has been previously suggested that low functioning of this enzyme might lead to higher circulating levels of levels of beta-carotene, which then might compete with L and Z for entry into tissues.72 Variation in BCMO1 function may also have broader physiological effects, including influence on the accumulation of liver lipids and subsequent metabolic effects.42,73,74
RPE65 belongs to the family of carotenoid cleavage enzymes75; it isomerizes all-trans retinyl ester to 11-cis-retinal in the visual cycle.76,77 Mutations in RPE65 are associated with Leber's congenital amaurosis and a recessive form of retinitis pigmentosa.78 Variants in RPE65 have not been previously studied in relation to MPOD; here, we identified rs4926339 as associated with MPOD. Increased RPE65 activity results in the accumulation of the fluorescent bis-retinoid A2E, which is a byproduct of this cycle. A2E, a major component of retinal lipofuscin, is nondegradable, accumulates with age and in Stargardt's disease76,79 and has a variety of toxic effects on RPE cells, including an increase in blue light photo damage and oxidative damage, which is thought to trigger the complement system and chronic inflammation in the macula.80 Thus, common variation in RPE genes that increase its activity may lower MPOD by increasing A2E accumulation and oxidative stress.
Variants in Genes Related to Carotenoid and Cholesterol Transport
A larger proportion of L, Z, and other xanthophylls, are carried on HDLs (>50%), unlike other less polar carotenoids, such as lycopene and alpha- and beta-carotene, which are mainly carried on LDLs,50,51 suggesting that blood lipoprotein distribution may be an important indicator of xanthophyll transport and, ultimately, availability to tissue.
A large number of proteins with roles in HDL formation or metabolism, or that are related to HDL levels in the blood, are suspected of being able to influence the accumulation of L and Z in tissues. This includes ABCA1, sterol transporters, ABCG5 and ABCG8, LIPC, CETP, APOE, and SR-B1. From the genes for these proteins, one SNP each from ABCA1, ABCG5, and LIPC was related to MPOD (Table 2). While variants in some of these genes have been previously characterized for importance in cholesterol status and/or AMD status,49,81–84 this is the first report describing the relationship between these genes and MPOD. Previous reports have supported the role of these genes in carotenoid status, as carriers of a dysfunctional allele in ABCA1 have low levels and unstable HDL particles in humans with Tangiers disease,85 as well as deficiencies in tissue carotenoids in chickens.65,86 Polymorphisms in ABCG5, which encodes a protein that directs cholesterol back into the intestinal lumen rather than into chylomicrons, which would carry cholesterol to the liver, have also been suggested to influence plasma response to both dietary cholesterol and carotenoids.87
We observed three SNPs in SCARB1, the gene encoding for scavenger receptor class B type I (SR-B1) lipid transporter, independently related to MPOD. SR-B1 knock-out mice consuming an atherogenic diet display retinal abnormalities similar to dry AMD88 and in humans, heterozygosity for a specific variant within SCARB1 was related to AMD in one sample of people without high risk variants in the well-known AMD susceptibility genes CFH and ARMS2.89 SR-B1, a plasma membrane receptor for HDL, mediates cholesterol efflux and carotenoid uptake in the RPE45,46 and intestine.46,47 The SR-B1 receptor, together with the Z-specific receptor GSTP1, may explain the selective accumulation of xanthophylls in the retina, and saturation of these receptors might explain the plateau in MPOD as LZ intake increases in this sample.3
The gene for SR-B1 (SCARB1) has high homology to both the ninaD gene that, when mutated in drosophila, results in blindness, and to cluster determinant 36 (CD36),90 expressed in both human gut and retina. Each of these is related to the joint transport of lipids and carotenoids in these tissues.45,91 In the retina, the CD36 protein is involved in phagocytosis of rod outer segments and lipid transport.46 Also, the silkworm analog of human CD36, Cameo2, is essential for LZ uptake into the insect's silk gland.92 In the present study, variants in the CD36 gene were related to levels of LZ in the serum (data not shown), but not to MPOD, as had been previously reported in one small study.70 However, expression of CD36 is also influenced by diet,93 which might further modify the association between LZ status and MPOD.
Contrary to one previous report in 302 persons reporting higher MPOD with increasing ε4 count,56 we observed no statistically significant differences in MPOD relative to APOE genotype. In fact, MPOD was lower, rather than higher, among women with ε4/ε4 genotype in our sample. However, the lower MPOD with higher ε4 count relative to ε3/ε3 was not significant (P = 0.25). Overall, there were no statistically significant differences of MPOD in any APOE genotype relative to ε3/ε3 (see Supplementary Material and Supplementary Table S3, http://www.iovs.org/content/54/3/2333/suppl/DC1). As a positive control for APOE-phenotype correlations in CAREDS, we observed the expected association between at least one copy of the ε4 allele and higher total cholesterol (P = 0.01).
Variants in Genes Related to Synthesis or Metabolism of Long-Chain Omega-3 Fatty Acids in Blood or Retina
Previous evidence suggested that long chain (LC) omega-3 (n-3) fatty acids in the diet,3 serum,52 and in supplements53 were related to MPOD. Docosahexaenoic acid (DHA) is the most abundant LC n-3 fatty acid in rod outer segment membranes94,95 at a concentration that exceeds levels found elsewhere in the body96 and is critical to retinal health. Furthermore, monkeys fed low n-3 fatty acid diets since birth have differences in foveal architecture relative to those fed adequate levels.97 For this reason, and because low intake of LC n-3 fatty acids may be related to maculopathies, which subsequently result in lowering of MPOD, we hypothesized that common variants in genes related to better LC n-3 status were related to MPOD. Relationships between genetic determinants of LC n-3 status and MPOD had not been previously studied.
Variants in FADS1 and ELOVL2, two genes that encode proteins related to the metabolism of LC n-3 fatty acids, have been related to levels of n-3 fatty acids in red blood cell membrane phospholipids in previous large genome-wide association studies.98,99 SNPs tagging variation for FADS1 and ELOVL2 were independently related to MPOD in the present study. FADS1 encodes a desaturase involved in converting the plant-derived essential fatty acid α-linolenic acid (ALA, 18:3n3) to the LC n-3 fatty acids eicosapentaenoic acid (EPA, 20:5n3) and the LC n-3 metabolite of EPA (docosapentaenoic acid- DPA) to docosahexaenoic acid (DHA, 22:6n3), the primary LC n-3 in the retina.98,99 The SNP in FADS1 related to lower MPOD in the present study (rs174534), is related to lower levels of ALA and higher levels of EPA in circulating phospholipids in a large genome-wide association study in European populations.99
FADS2 is an essential enzyme for DHA synthesis, as FADS2 knock-out mice do not have DHA.100 In the present study, FADS2 was related to MPOD, but not significantly after further adjusting for other determinants of MPOD, suggesting the association with MPOD may, in part, reflect an influence of other determinants.
ELOVL2 encodes an enzyme responsible for the elongation of the very LCn-3 fatty acid EPA to DPA and, subsequently, DHA. In CAREDS, rs1150561, which tags variation downstream of ELOVL2, was associated with MPOD.
Variants in Known Genes for Maculopathies
Finally, we considered variants in genes related to either AMD or other maculopathies, which were also related to the status of carotenoids or retinoids in previous studies. Associations with RPE were discussed above. We also hypothesized that common variants within ALDH3A2 may impact MPOD in the general population. Rare mutations within ALDH3A2, an aldehyde dehydrogenase, are known to cause Sjorgren-Larsson Syndrome, which results in numerous phenotypic effects including macular dystrophy, photophobia, and a complete lack of macular pigment.54 In the first report of common variants and MPOD, we found the minor allele of rs8069576 to be associated with lower mean MPOD.
Similar to a recent report,55 we were not able to identify any single associations between CFH or ARMS2 variants and MPOD. However, this same report by Loane and colleagues suggested individuals homozygous for the risk allele at both Y402H and A69S may be at higher risk for lower MPOD.55 Six women in CAREDS were homozygous at both these SNPs and their mean MPOD was 0.25. While lower mean MPOD relative to the rest of our sample (0.36) and consistent with the report by Loane et al,55 it was not statistically significant (P = 0.14), likely due to low number of women homozygous at both genes.
Limitations
Overall, SNPs identified through our current candidate gene association study explained 5.1% of the variability in MPOD (Table 3), which falls short of the 67% heritability predicted in a previous twin study.37 “Missing heritability” is a common phenomenon in human genetics research,101,102 and in CAREDS is likely reflective of (1) the small number of genetic variants tested (relative to genome-wide association studies), (2) the focus on variants with a minor allele frequency greater than 5%, when the majority of variation within the genome occurs at frequencies less than 5%,103 (3) the exclusion of other types of genetic variants such as copy number variants, and/or (4) the exclusion of the complex interplay of physiological factors likely to influence MPOD such as gene–gene and gene–environment interactions. Evidence of an interaction between LZ intake and genetic factors (Figs. 1, 2) was detected, suggesting that while the genetic factors measured here only explained 5% of the variability in MPOD in the CAREDS population, the relevance of certain genes may shift under various environmental conditions, importantly in this case dietary intake of carotenoids. More complex statistical modeling of interactions and joint effects may reveal additional insights.
Some of the SNPs related to MPOD in the present study may be chance findings, given the large number of associations studied. Under the Bonferroni multiple testing correction, a P value less than 1 × 10−4 would be required to claim statistical significance. Only rs11645428 in BCMO1 could be considered in this scenario. However, this correction is overly conservative as it assumes independence of tests, which is violated given the correlation of SNPs within genes, and results of conditional SNP modeling within genes. Therefore, the consistency of associations across levels of other major determinants of MPOD (dietary intake and waist circumference), and support of biological plausibility of relationships with previous evidence, lessens the possibility of false positives. Replication in additional, independent cohorts, which are as, or better, powered to detect genetic effects will aid in understanding the nature of these results. The sample in CAREDS is comprised of largely white women from the United States (US), older than 55 years of age, who have high socioeconomic status, relative to the US population. It is unknown the extent to which these genotypes would be related to MPOD among men or among persons with ancestry that evolved under conditions of higher light exposure. Studies of genetic determinants of MPOD in additional study samples of diverse composition will aid in better understanding the genetic architecture underlying MPOD.
Another possible limitation is the focus on MPOD at 0.5° from the foveal center. The SNPs reported here may not reflect differences in the spatial distribution of macular pigment between individuals. An enhanced macular pigment at approximately 0.7°, which gives the appearance of a ring structure, has been related to lower occurrence of macular degeneration in some past studies.104,105 Persons with this ring have lower MPOD at 0.25° and 0.5° and higher at 1° and 2°.105 Results of a recent twin study indicated macular pigment peak density to be more strongly related to genetic factors than macular pigment spatial distribution, which was more strongly related to environmental factors.66 Although results in CAREDS were generally similar at all four eccentricities measured (not shown), further exploration of genetic determinants of the spatial distribution of MPOD in this sample are warranted.
Conclusions
In conclusion, this study in 1585 postmenopausal women is the most extensive investigation of genetic predictors of MPOD to date and suggests variation in genes related to the metabolism of carotenoids, LC n-3 fatty acids and a maculopathy related to absence of macular pigment is related to variation in the density of macular pigments. These findings, together with nondietary determinants of MPOD identified in this and other studies, likely contribute to the high variability in MPOD levels across different people and variable response to the intake of macular carotenoids. Further investigation of complex gene–gene and gene–environment joint effects and interactions might explain additional variability in MPOD. If macular pigment is found to protect against age-related maculopathies and optimize vision, this information about determinants of MPOD could be used to design intervention trials aimed at directly studying strategies to enhance macular pigment.
Supplementary Material
Acknowledgments
The authors thank the women who generously contributed their time to participate in the CAREDS and all CAREDS and WHI Investigators who have contributed over the years. They also thank Bill Wooten of Brown University for his input into the design of CAREDS and assessment of MPOD. A short list of investigators who have contributed to WHI science can be found in Supplementary Material S4 (see Supplementary Material and Supplementary S4, http://www.iovs.org/content/54/3/2333/suppl/DC1).
Footnotes
Supported by grants from the National Institutes of Health, National Eye Institute (Grants EY013018, EY016886), the Research to Prevent Blindness, the Retina Research Foundation, and the Carl and Mildred Reeves Foundation. Additional support from the National Eye Institute (EY011600, PSB), the National Heart, Lung, and Blood Institute (The Women's Health Initiative), National Institutes of Health, United States Department of Health and Human Services through contracts HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HHSN268201100003C, HHSN268201100004C, and HHSN271201100004C.
Disclosure: K.J. Meyers, None; E.J. Johnson, Alcon (F); P.S. Bernstein, None; S.K. Iyengar, None; C.D. Engelman, None; C.K. Karki, None; Z. Liu, None; R.P. Igo Jr, None; B. Truitt, None; M.L. Klein, None; D.M. Snodderly, None; B.A. Blodi, None; K.M. Gehrs, None; G.E. Sarto, None; R.B. Wallace, None; J. Robinson, None; E.S. LeBlanc, None; G. Hageman, Alcon (F), Optherion (F), Sequenom (C); L. Tinker, None; J.A. Mares, None
References
- 1. Bone RA, Landrum JT, Cao Y, Howard AN, Alvarez-Calderon F. Macular pigment response to a supplement containing meso-zeaxanthin, lutein and zeaxanthin. Nutr Metab (Lond). 2007; 4: 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Snodderly DM, Brown PK, Delori FC, Auran JD. The macular pigment. I. Absorbance spectra, localization, and discrimination from other yellow pigments in primate retinas. Invest Ophthalmol Vis Sci. 1984; 25: 660–673 [PubMed] [Google Scholar]
- 3. Mares JA, LaRowe TL, Snodderly DM, et al. Predictors of optical density of lutein and zeaxanthin in retinas of older women in the Carotenoids in Age-Related Eye Disease Study, an ancillary study of the Women's Health Initiative. Am J Clin Nutr. 2006; 84: 1107–1122 [DOI] [PubMed] [Google Scholar]
- 4. Bhosale P. Zhao da Y, Bernstein PS. HPLC measurement of ocular carotenoid levels in human donor eyes in the lutein supplementation era. Invest Ophthalmol Vis Sci. 2007; 48: 543–549 [DOI] [PubMed] [Google Scholar]
- 5. Bone RA, Landrum JT, Fernandez L, Tarsis SL. Analysis of the macular pigment by HPLC: retinal distribution and age study. Invest Ophthalmol Vis Sci. 1988; 29: 843–849 [PubMed] [Google Scholar]
- 6. Sangiovanni JP, Neuringer M. The putative role of lutein and zeaxanthin as protective agents against age-related macular degeneration: promise of molecular genetics for guiding mechanistic and translational research in the field. Am J Clin Nutr. 2012; 96: 1223S–1233S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mares JA, Millen AE, Meyers K. Diet and Supplements and the Prevention and Treatment of Eye Diseases. In: Coulston A, Boushey C, Ferruzzi M. eds Nutrition in the Prevention and Treatment of Disease. 3rd ed. San Diego, CA: Elsevier, Inc.; 2013: 341–371 [Google Scholar]
- 8. Sabour-Pickett S, Nolan JM, Loughman J, Beatty S. A review of the evidence germane to the putative protective role of the macular carotenoids for age-related macular degeneration. Mol Nutr Food Res. 2012; 56: 270–286 [DOI] [PubMed] [Google Scholar]
- 9. Landrum JT, Bone RA. Mechanistic Evidence for Eye Diseases and Carotenoids. In: Krinsky NI, Mayne ST, Sies H. eds Carotenoids in Health and Disease. New York: Marcel Dekker, Inc.; 2004; 445–472 [Google Scholar]
- 10. Li SY, Fung FK, Fu ZJ, Wong D, Chan HH, Lo AC. Anti-inflammatory effects of lutein in retinal ischemic/hypoxic injury: in vivo and in vitro studies. Invest Ophthalmol Vis Sci. 2012; 53: 5976–5984 [DOI] [PubMed] [Google Scholar]
- 11. Sasaki M, Ozawa Y, Kurihara T, et al. Neuroprotective effect of an antioxidant, lutein, during retinal inflammation. Invest Ophthalmol Vis Sci. 2009; 50: 1433–1439 [DOI] [PubMed] [Google Scholar]
- 12. Bian Q, Gao S, Zhou J, et al. Lutein and zeaxanthin supplementation reduces photooxidative damage and modulates the expression of inflammation-related genes in retinal pigment epithelial cells. Free Radic Biol Med. 2012; 53: 1298–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stringham JM, Garcia PV, Smith PA, McLin LN, Foutch BK. Macular pigment and visual performance in glare: benefits for photostress recovery, disability glare, and visual discomfort. Invest Ophthalmol Vis Sci. 2011; 52: 7406–7415 [DOI] [PubMed] [Google Scholar]
- 14. Stringham JM, Hammond BR. Macular pigment and visual performance under glare conditions. Optom Vis Sci. 2008; 85: 82–88 [DOI] [PubMed] [Google Scholar]
- 15. Renzi LM, Hammond BR Jr. The relation between the macular carotenoids, lutein and zeaxanthin, and temporal vision. Ophthalmic Physiol Opt. 2010; 30: 351–357 [DOI] [PubMed] [Google Scholar]
- 16. Ma L, Dou HL, Huang YM, et al. Improvement of retinal function in early age-related macular degeneration after lutein and zeaxanthin supplementation: a randomized, double-masked, placebo-controlled trial. Am J Ophthalmol. 2012; 154: 625–634 e621 [DOI] [PubMed] [Google Scholar]
- 17. Loughman J, Nolan JM, Howard AN, Connolly E, Meagher K, Beatty S. The impact of macular pigment augmentation on visual performance using different carotenoid formulations. Invest Ophthalmol Vis Sci. 2012; 53: 7871–7880 [DOI] [PubMed] [Google Scholar]
- 18. Bowen PE, Garg V, Stacewicz-Sapuntzakis M, Yelton L, Schreiner RS. Variability of serum carotenoids in response to controlled diets containing six servings of fruits and vegetables per day. Ann N Y Acad Sci. 1993; 691: 241–243 [DOI] [PubMed] [Google Scholar]
- 19. Bowen PE, Herbst-Espinosa SM, Hussain EA, Stacewicz-Sapuntzakis M. Esterification does not impair lutein bioavailability in humans. J Nutr. 2002; 132: 3668–3673 [DOI] [PubMed] [Google Scholar]
- 20. Rosenthal JM, Kim J, de Monasterio F, et al. Dose-ranging study of lutein supplementation in persons aged 60 years or older. Invest Ophthalmol Vis Sci. 2006; 47: 5227–5233 [DOI] [PubMed] [Google Scholar]
- 21. Hammond BR Jr, Johnson EJ, Russell RM, et al. Dietary modification of human macular pigment density. Invest Ophthalmol Vis Sci. 1997; 38: 1795–1801 [PubMed] [Google Scholar]
- 22. Bone RA, Landrum JT, Guerra LH, Ruiz CA. Lutein and zeaxanthin dietary supplements raise macular pigment density and serum concentrations of these carotenoids in humans. J Nutr. 2003; 133: 992–998 [DOI] [PubMed] [Google Scholar]
- 23. Johnson EJ, Hammond BR, Yeum KJ, et al. Relation among serum and tissue concentrations of lutein and zeaxanthin and macular pigment density. Am J Clin Nutr. 2000; 71: 1555–1562 [DOI] [PubMed] [Google Scholar]
- 24. Berendschot TT, Goldbohm RA, Klopping WA, van de Kraats J, van Norel J, van Norren D. Influence of lutein supplementation on macular pigment, assessed with two objective techniques. Invest Ophthalmol Vis Sci. 2000; 41: 3322–3326 [PubMed] [Google Scholar]
- 25. Koh HH, Murray IJ, Nolan D, Carden D, Feather J, Beatty S. Plasma and macular responses to lutein supplement in subjects with and without age-related maculopathy: a pilot study. Exp Eye Res. 2004; 79: 21–27 [DOI] [PubMed] [Google Scholar]
- 26. Kvansakul J, Rodriguez-Carmona M, Edgar D, et al. Supplementation with the carotenoids lutein or zeaxanthin improves human visual performance. Ophthalmic Physiol Opt. 2006; 26: 362–371 [DOI] [PubMed] [Google Scholar]
- 27. Duncan DD, Munoz B, West SK. Assessment of ocular exposure to visible light for population studies. Dev Ophthalmol. 2002; 35: 76–92 [DOI] [PubMed] [Google Scholar]
- 28. Aleman TS, Duncan JL, Bieber ML, et al. Macular pigment and lutein supplementation in retinitis pigmentosa and Usher syndrome. Invest Ophthalmol Vis Sci. 2001; 42: 1873–1881 [PubMed] [Google Scholar]
- 29. Aleman TS, Cideciyan AV, Windsor EA, et al. Macular pigment and lutein supplementation in ABCA4-associated retinal degenerations. Invest Ophthalmol Vis Sci. 2007; 48: 1319–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Trieschmann M, Beatty S, Nolan JM, et al. Changes in macular pigment optical density and serum concentrations of its constituent carotenoids following supplemental lutein and zeaxanthin: the LUNA study. Exp Eye Res. 2007; 84: 718–728 [DOI] [PubMed] [Google Scholar]
- 31. Schalch W, Cohn W, Barker FM, et al. Xanthophyll accumulation in the human retina during supplementation with lutein or zeaxanthin - the LUXEA (LUtein Xanthophyll Eye Accumulation) study. Arch Biochem Biophys. 2007; 458: 128–135 [DOI] [PubMed] [Google Scholar]
- 32. Ma L, Yan SF, Huang YM, et al. Effect of lutein and zeaxanthin on macular pigment and visual function in patients with early age-related macular degeneration. Ophthalmology. 2012; 119: 2290–2297 [DOI] [PubMed] [Google Scholar]
- 33. Richer SP, Stiles W, Graham-Hoffman K, et al. Randomized, double-blind, placebo-controlled study of zeaxanthin and visual function in patients with atrophic age-related macular degeneration: the Zeaxanthin and Visual Function Study (ZVF) FDA IND #78, 973. Optometry. 2011; 82: 667–680 e666 [DOI] [PubMed] [Google Scholar]
- 34. Hammond CJ, Liew SM, Van Kuijk FJ, et al. The heritability of macular response to supplemental lutein and zeaxanthin: a classical twin study. Invest Ophthalmol Vis Sci. 2012; 53: 4963–4968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nolan JM, Akkali MC, Loughman J, Howard AN, Beatty S. Macular carotenoid supplementation in subjects with atypical spatial profiles of macular pigment. Exp Eye Res. 2012; 101: 9–15 [DOI] [PubMed] [Google Scholar]
- 36. Connolly EE, Beatty S, Thurnham DI, et al. Augmentation of macular pigment following supplementation with all three macular carotenoids: an exploratory study. Curr Eye Res. 2010; 35: 335–351 [DOI] [PubMed] [Google Scholar]
- 37. Liew SHM, Gilbert CE, Spector TD, et al. Heritability of macular pigment: a twin study. Invest Ophthalmol Vis Sci. 2005; 46: 4430–4436 [DOI] [PubMed] [Google Scholar]
- 38. Borel P. Genetic variations involved in interindividual variability in carotenoid status. Mol Nutr Food Res. 2011; 56: 228–240 [DOI] [PubMed] [Google Scholar]
- 39. Li B, Vachali P, Frederick JM, Bernstein PS. Identification of StARD3 as a lutein-binding protein in the macula of the primate retina. Biochemistry. 2011; 50: 2541–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bhosale P, Larson AJ, Frederick JM, Southwick K, Thulin CD, Bernstein PS. Identification and characterization of a Pi isoform of glutathione S-transferase (GSTP1) as a zeaxanthin-binding protein in the macula of the human eye. J Biol Chem. 2004; 279: 49447–49454 [DOI] [PubMed] [Google Scholar]
- 41. Vachali P, Li B, Nelson K, Bernstein PS. Surface plasmon resonance (SPR) studies on the interactions of carotenoids and their binding proteins. Arch Biochem Biophys. 2012; 519: 32–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. von Lintig J. Colors with functions: elucidating the biochemical and molecular basis of carotenoid metabolism. Annu Rev Nutr. 2010; 30: 35–56 [DOI] [PubMed] [Google Scholar]
- 43. Thompson DA, Gyurus P, Fleischer LL, et al. Genetics and phenotypes of RPE65 mutations in inherited retinal degeneration. Invest Ophthalmol Vis Sci. 2000; 41: 4293–4299 [PubMed] [Google Scholar]
- 44. Bowne SJ, Humphries MM, Sullivan LS, et al. A dominant mutation in RPE65 identified by whole-exome sequencing causes retinitis pigmentosa with choroidal involvement. Eur J Hum Genet. 2011; 19: 1074–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. During A, Doraiswamy S, Harrison EH. Xanthophylls are preferentially taken up compared with beta-carotene by retinal cells via a SRBI-dependent mechanism. J Lipid Res. 2008; 49: 1715–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tserentsoodol N, Sztein J, Campos M, et al. Uptake of cholesterol by the retina occurs primarily via a low density lipoprotein receptor-mediated process. Mol Vis. 2006; 12: 1306–1318 [PubMed] [Google Scholar]
- 47. Reboul E, Abou L, Mikail C, et al. Lutein transport by Caco-2 TC-7 cells occurs partly by a facilitated process involving the scavenger receptor class B type I (SR-BI). Biochem J. 2005; 387: 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen W, Stambolian D, Edwards AO, et al. Genetic variants near TIMP3 and high-density lipoprotein-associated loci influence susceptibility to age-related macular degeneration. Proc Natl Acad Sci U S A. 2010; 107: 7401–7406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Neale BM, Fagerness J, Reynolds R, et al. Genome-wide association study of advanced age-related macular degeneration identifies a role of the hepatic lipase gene (LIPC). Proc Natl Acad Sci U S A. 2010; 107: 7395–7400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Parker RS. Absorption, metabolism, and transport of carotenoids. FASEB J. 1996; 10: 542–551 [PubMed] [Google Scholar]
- 51. Wang W, Connor SL, Johnson EJ, Klein ML, Hughes S, Connor WE. Effect of dietary lutein and zeaxanthin on plasma carotenoids and their transport in lipoproteins in age-related macular degeneration. Am J Clin Nutr. 2007; 85: 762–769 [DOI] [PubMed] [Google Scholar]
- 52. Delyfer MN, Buaud B, Korobelnik JF, et al. Association of macular pigment density with plasma omega-3 fatty acids: the PIMAVOSA Study. Invest Ophthalmol Vis Sci. 2012; 53: 1204–1210 [DOI] [PubMed] [Google Scholar]
- 53. Johnson EJ, Chung HY, Caldarella SM, Snodderly DM. The influence of supplemental lutein and docosahexaenoic acid on serum, lipoproteins, and macular pigmentation. Am J Clin Nutr. 2008; 87: 1521–1529 [DOI] [PubMed] [Google Scholar]
- 54. van der Veen RL, Fuijkschot J, Willemsen MA, Cruysberg JR, Berendschot TT, Theelen T. Patients with Sjogren-Larsson syndrome lack macular pigment. Ophthalmology. 2010; 117: 966–971 [DOI] [PubMed] [Google Scholar]
- 55. Loane E, Nolan JM, McKay GJ, Beatty S. The association between macular pigment optical density and CFH, ARMS2, C2/BF, and C3 genotype. Exp Eye Res. 2011; 93: 592–598 [DOI] [PubMed] [Google Scholar]
- 56. Loane E, McKay GJ, Nolan JM, Beatty S, Apolipoprotein E. Genotype is associated with macular pigment optical density. Invest Ophthalmol Vis Sci. 2010; 51: 2636–2643 [DOI] [PubMed] [Google Scholar]
- 57. Moeller SM, Mehta NR, Tinker LF, et al. Associations between intermediate age-related macular degeneration and lutein and zeaxanthin in the Carotenoids in Age-Related Eye Disease Study (CAREDS), an ancillary study of the Women's Health Initiative. Arch Ophthalmol. 2006; 124: 1–24 [DOI] [PubMed] [Google Scholar]
- 58. LaRowe TL, Mares JA, Snodderly DM, Klein ML, Wooten BR, Chappell R. Macular pigment density and age-related maculopathy in the Carotenoids in Age-Related Eye Disease Study. An ancillary study of the women's health initiative. Ophthalmology. 2008; 115: 876–883 e871 [DOI] [PubMed] [Google Scholar]
- 59. Moeller SM, Voland R, Tinker L, et al. Associations between age-related nuclear cataract and lutein and zeaxanthin in the diet and serum in the Carotenoids in the Age-Related Eye Disease Study, an Ancillary Study of the Women's Health Initiative. Arch Ophthalmol. 2008; 126: 354–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Snodderly DM, Mares JA, Wooten BR, et al. Macular pigment measurement by heterochromatic flicker photometry in older subjects: the carotenoids and age-related eye disease study. Invest Ophthalmol Vis Sci. 2004; 45: 531–538 [DOI] [PubMed] [Google Scholar]
- 61. Price AL, Butler J, Patterson N, et al. Discerning the ancestry of European Americans in genetic association studies. PLoS Genet. 2008; 4: e236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007; 81: 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Millen AE, Voland R, Sondel SA, et al. Vitamin D status and early age-related macular degeneration in postmenopausal women. Arch Ophthalmol. 2011; 129: 481–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mares JA, Voland RP, Sondel SA, et al. Healthy lifestyles related to subsequent prevalence of age-related macular degeneration. Arch Ophthalmol. 2011; 129: 470–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Connor WE, Duell PB, Kean R, Wang Y. The prime role of HDL to transport lutein into the retina: evidence from HDL-deficient WHAM chicks having a mutant ABCA1 transporter. Invest Ophthalmol Vis Sci. 2007; 48: 4226–4231 [DOI] [PubMed] [Google Scholar]
- 66. Hogg RE, Ong EL, Chamberlain M, et al. Heritability of the spatial distribution and peak density of macular pigment: a classical twin study. Eye (Lond). 2012; 26: 1217–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Nolan JM, Loughman J, Akkali MC, et al. The impact of macular pigment augmentation on visual performance in normal subjects: COMPASS. Vision Res. 2011; 51: 459–469 [DOI] [PubMed] [Google Scholar]
- 68. Kim CH. Retinoic acid, immunity, and inflammation. Vitam Horm. 2011; 86: 83–101 [DOI] [PubMed] [Google Scholar]
- 69. Ferrucci L, Perry JR, Matteini A, et al. Common variation in the beta-carotene 15,15′-monooxygenase 1 gene affects circulating levels of carotenoids: a genome-wide association study. Am J Hum Genet. 2009; 84: 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Borel P, de Edelenyi FS, Vincent-Baudry S, et al. Genetic variants in BCMO1 and CD36 are associated with plasma lutein concentrations and macular pigment optical density in humans. Ann Med. 2011; 43: 47–59 [DOI] [PubMed] [Google Scholar]
- 71. Kim YS, Oh DK. Substrate specificity of a recombinant chicken beta-carotene 15,15′-monooxygenase that converts beta-carotene into retinal. Biotechnol Lett. 2009; 31: 403–408 [DOI] [PubMed] [Google Scholar]
- 72. Lietz G, Oxley A, Leung W, Hesketh J. Single nucleotide polymorphisms upstream from the beta-carotene 15,15′-monoxygenase gene influence provitamin A conversion efficiency in female volunteers. J Nutr. 2012; 142: 161S–165S [DOI] [PubMed] [Google Scholar]
- 73. Hessel S, Eichinger A, Isken A, et al. CMO1 deficiency abolishes vitamin A production from beta-carotene and alters lipid metabolism in mice. J Biol Chem. 2007; 282: 33553–33561 [DOI] [PubMed] [Google Scholar]
- 74. Lietz G, Oxley A, Boesch-Saadatmandi C, Kobayashi D. Importance of beta,beta-carotene 15,15′-monooxygenase 1 (BCMO1) and beta,beta-carotene 9′,10′-dioxygenase 2 (BCDO2) in nutrition and health. Mol Nutr Food Res. 2012; 56: 241–250 [DOI] [PubMed] [Google Scholar]
- 75. Cai X, Conley SM, Naash MI. RPE65: role in the visual cycle, human retinal disease, and gene therapy. Ophthalmic Genet. 2009; 30: 57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Maiti P, Kong J, Kim SR, Sparrow JR, Allikmets R, Rando RR. Small molecule RPE65 antagonists limit the visual cycle and prevent lipofuscin formation. Biochemistry. 2006; 45: 852–860 [DOI] [PubMed] [Google Scholar]
- 77. Moiseyev G, Nikolaeva O, Chen Y, Farjo K, Takahashi Y, Ma JX. Inhibition of the visual cycle by A2E through direct interaction with RPE65 and implications in Stargardt disease. Proc Natl Acad Sci U S A. 2010; 107: 17551–17556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Thompson DA, Gal A. Genetic defects in vitamin A metabolism of the retinal pigment epithelium. Dev Ophthalmol. 2003; 37: 141–154 [DOI] [PubMed] [Google Scholar]
- 79. Delori FC, Staurenghi G, Arend O, Dorey CK, Goger DG, Weiter JJ. In vivo measurement of lipofuscin in Stargardt's disease–Fundus flavimaculatus. Invest Ophthalmol Vis Sci. 1995; 36: 2327–2331 [PubMed] [Google Scholar]
- 80. Zhou J, Jang YP, Kim SR, Sparrow JR. Complement activation by photooxidation products of A2E, a lipofuscin constituent of the retinal pigment epithelium. Proc Natl Acad Sci U S A. 2006; 103: 16182–16187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Chen W, Stambolian D, Edwards AO, et al. Genetic variants near TIMP3 and HDL-associated loci influence susceptibility to age-related macular degeneration. Proc Natl Acad Sci U S A. 2010; 107: 7401–7406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zareparsi S, Branham KE, Li M, et al. Strong association of the Y402H variant in complement factor H at 1q32 with susceptibility to age-related macular degeneration. Am J Hum Genet. 2005; 77: 149–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tikellis G, Sun C, Gorin MB, et al. Apolipoprotein e gene and age-related maculopathy in older individuals: the cardiovascular health study. Arch Ophthalmol. 2007; 125: 68–73 [DOI] [PubMed] [Google Scholar]
- 84. San Giovanni JP, Mehta S Variation in lipid-associated genes as they relate to risk of advanced age-related macular degeneration. World Rev Nutr Diet. 2009; 99: 105–158 [DOI] [PubMed] [Google Scholar]
- 85. Schaefer EJ, Santos RD, Asztalos BF. Marked HDL deficiency and premature coronary heart disease. Curr Opin Lipidol. 2010; 21: 289–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Attie AD, Hamon Y, Brooks-Wilson AR, et al. Identification and functional analysis of a naturally occurring E89K mutation in the ABCA1 gene of the WHAM chicken. J Lipid Res. 2002; 43: 1610–1617 [DOI] [PubMed] [Google Scholar]
- 87. Herron KL, McGrane MM, Waters D, et al. The ABCG5 polymorphism contributes to individual responses to dietary cholesterol and carotenoids in eggs. J Nutr. 2006; 136: 1161–1165 [DOI] [PubMed] [Google Scholar]
- 88. Provost AC, Vede L, Bigot K, et al. Morphologic and electroretinographic phenotype of SR-BI knockout mice after a long-term atherogenic diet. Invest Ophthalmol Vis Sci. 2009; 50: 3931–3942 [DOI] [PubMed] [Google Scholar]
- 89. Zerbib J, Seddon JM, Richard F, et al. rs5888 variant of SCARB1 gene is a possible susceptibility factor for age-related macular degeneration. PLoS One. 2009; 4: e7341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kiefer C, Sumser E, Wernet MF, Von Lintig J. A class B scavenger receptor mediates the cellular uptake of carotenoids in Drosophila. Proc Natl Acad Sci U S A. 2002; 99: 10581–10586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. During A, Dawson HD, Harrison EH. Carotenoid transport is decreased and expression of the lipid transporters SR-BI, NPC1L1, and ABCA1 is downregulated in Caco-2 cells treated with ezetimibe. J Nutr. 2005; 135: 2305–2312 [DOI] [PubMed] [Google Scholar]
- 92. Sakudoh T, Iizuka T, Narukawa J, et al. A CD36-related transmembrane protein is coordinated with an intracellular lipid-binding protein in selective carotenoid transport for cocoon coloration. J Biol Chem. 2010; 285: 7739–7751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Llorente-Cortes V, Estruch R, Mena MP, et al. Effect of Mediterranean diet on the expression of pro-atherogenic genes in a population at high cardiovascular risk. Atherosclerosis. 2010; 208: 442–450 [DOI] [PubMed] [Google Scholar]
- 94. Bazan NG, Scott BL. Dietary omega-3 fatty acids and accumulation of docosahexaenoic acid in rod photoreceptor cells of the retina and at synapses. Ups J Med Sci Suppl. 1990; 48: 97–107 [PubMed] [Google Scholar]
- 95. Fliesler SJ, Anderson RE. Chemistry and metabolism of lipids in the vertebrate retina. Prog Lipid Res. 1983; 22: 79–131 [DOI] [PubMed] [Google Scholar]
- 96. Arterburn LM, Hall EB, Oken H. Distribution, interconversion, and dose response of n-3 fatty acids in humans. Am J Clin Nutr. 2006; 83: S1467–1476 [DOI] [PubMed] [Google Scholar]
- 97. Leung IYF, Sandstrom MM, Zucker CL, Neuringer M, Max Snodderly D. Nutritional manipulation of primate retinas. IV. Effects of n-3 fatty acids, lutein, and zeaxanthin on S-cones and rods in the foveal region. Exp Eye Res. 2005; 81: 513–529 [DOI] [PubMed] [Google Scholar]
- 98. Tanaka T, Shen J, Abecasis GR, et al. Genome-wide association study of plasma polyunsaturated fatty acids in the InCHIANTI Study. PLoS Genet. 2009; 5: e1000338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Lemaitre RN, Tanaka T, Tang W, et al. Genetic loci associated with plasma phospholipid n-3 fatty acids: a meta-analysis of genome-wide association studies from the CHARGE Consortium. PLoS Genet. 2011; 7: e1002193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Stoffel W, Holz B, Jenke B, et al. Delta6-desaturase (FADS2) deficiency unveils the role of omega3- and omega6-polyunsaturated fatty acids. EMBO J. 2008; 27: 2281–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009; 461: 747–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Eichler EE, Flint J, Gibson G, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010; 11: 446–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Gorlov IP, Gorlova OY, Sunyaev SR, Spitz MR, Amos CI. Shifting paradigm of association studies: value of rare single-nucleotide polymorphisms. Am J Hum Genet. 2008; 82: 100–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Wustemeyer H, Jahn C, Nestler A, Barth T, Wolf S. A new instrument for the quantification of macular pigment density: first results in patients with AMD and healthy subjects. Graefes Arch Clin Exp Ophthalmol. 2002; 240: 666–671 [DOI] [PubMed] [Google Scholar]
- 105. Dietzel M, Zeimer M, Heimes B, Pauleikhoff D, Hense HW. The ringlike structure of macular pigment in age-related maculopathy: results from the Muenster Aging and Retina Study (MARS). Invest Ophthalmol Vis Sci. 2011; 52: 8016–8024 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.