Abstract
Challenge studies following passive immunization with neutralizing antibodies suggest that an HIV vaccine could be efficacious were it able to elicit broadly neutralizing antibodies (bNAbs4). To better understand the requirements for activation of B cells producing bNAbs, we generated cell lines expressing bNAbs or their germline-reverted versions (gl-bNAbs) as BCRs. We then tested the abilities of the bNAb-expressing cells to recognize HIV pseudovirions and vaccine candidate proteins by binding and activation assays. The results suggest that HIV Env antigen-expressing, infection-competent virions are poorly recognized by high affinity bNAb-expressing cells, as measured by the inability of antigens to induce rapid increases in intracellular calcium levels. Other antigen forms appear to be highly stimulatory: in particular, soluble gp140 trimers and a multimerized, scaffolded epitope protein. Virions failed to efficiently activate bNAb-expressing B cells owing to delayed or inefficient BCR recognition, most likely caused by the low density of Env spikes. Importantly, B cells carrying gl-bNAb BCRs were not stimulated by any of the tested vaccine candidates. These data provide insight into why many HIV immunogens, and natural HIV infections, fail to rapidly stimulate bNAb responses and suggest that bNAb-expressing cell lines might be useful tools in evaluation of vaccine antigens for infectious diseases. As soluble Env trimers or multimerized scaffolded epitopes are best at activating B cell expressing bNAbs, these antigenic forms should be considered as preferred vaccine components, though they should be modified to better target naïve gl-bNAb B cells.
Introduction
There is a growing consensus that an effective HIV vaccine should include a component that elicits bNAbs (reviewed in 1, 2–5). A growing number of bNAbs have been identified and characterized (6–18). Several bNAbs have been shown to afford protection in passive transfer studies in animals (19–28). However, eliciting significant levels of bNAbs through immunization has not yet been successful. B cells producing bNAbs may not be efficiently generated for several reasons. Precursor HIV Env-specific B cells may be rare because of immune tolerance (29) or because cells of the appropriate specificity are difficult to generate through the processes of gene diversification. For example, some bNAbs appear to require relatively unusual structures, such as very long H-chain CDR3s (6, 12) or domain exchange (30). Alternatively, bNAb precursor B cells may be abundant, but difficult to stimulate owing to topological reasons, e.g., because the epitope has poor accessibility, or because of the need for more powerful adjuvants or immunogens of a more stimulatory nature.
To elicit a bNAb response to HIV-1 Env, B cells with bNAb specificities must be activated. In this study, we have expressed in B cell lines a number of previously identified broadly neutralizing HIV antibodies (Table I) that recognize a variety of sites on Env, including the CD4 binding site (b12, VRC01, PGV04, PGV19, NIH45-46), the membrane-proximal external region (MPER) of gp41 (4E10), a V2/glycan dependent site on the trimer (PG9, PG16, PGT145), the high mannose rich face of gp120 (2G12), a V3/glycan site (PGT128), a V4/glycan site (PGT135) and another glycan dependent site still being defined (PGT121). We then tested the ability of different Env-containing antigens and virions to stimulate these cells. The results suggest that soluble Env trimer preparations are highly stimulatory for early calcium mobilization, whereas monomers and virion preparations, including infectious virions and pseudovirions, are generally non-stimulatory. However, internally labeled pseudovirions were shown to bind to mutated, but not germline-reverted bNAb-expressing B cells, and to stimulate the expression of the early activation marker CD69 upon prolonged exposure in vitro. These findings suggest that naturally expressed HIV-1 envelope glycoprotein is poorly stimulatory for bNAb-expressing B cells and that soluble trimers or multimeric scaffolded epitopes capable of binding gl-bNAbs may be more desirable components for an effective HIV-1 vaccine that elicits bNAbs.
Table I.
bNAb specificities in Tet-inducible lentivirus carrying 2A peptide-linked BCRs.
| bNAb name | References |
|---|---|
| b12 | (6, 60) |
| germline b12 | This study and (54) |
| 4E10 | (7, 8) |
| germline 4E10 | This study and (61) |
| PGT128 | (16) |
| PGT121 | (16) |
| PG9 | (10, 16) |
| PGT135 | (16) |
| PGV04 | (11) |
| PG16 | (12, 16) |
| PGT145 | (16) |
| VRC01 | (13, 14) |
| germline VRC01 | This study and (11) |
| PGV19 | VRC/IAVI, manuscript in preparation |
| NIH 45–46 | (15) |
Materials and Methods
Conventional B cell transfectants
For the heavy chain gene constructs, the mouse VHJ558.85.191 promoter and leader were fused to the b12 or 2G12 VDJ exon, yielding an Asc1-flanked promoter-L-VDJ segment, which was then extended to include the intronic enhancer using sequences from the natural interval starting from the end of mouse JH4 to the downstream EcoR1 site. An EcoR1 fragment carrying this construct, including splice donor sequences was cloned into the EcoRI site in the pSal genomic IgM expression vector. pSal is a modified version the plasmid 3–83μ (31) in which an irrelevant EcoR1 site was changed to Sal1 and the EcoR1 fragment carrying the natural VDJ was removed. For constitutively expressed L-chain constructs, VJ sequences were appended on the 5' end with a leader sequence from Vκ4-53 or the mouse IgG1 signal sequence and at the 3' end with hCκ cDNA (modified from a vector received as a kind gift from Patrick Wilson, U. Chicago). Coding sequences were inserted into the pBudCE4.1 expression plasmid (Invitrogen, Carlsbad, CA), which drives expression under the EF-1α promoter. The V(D)J sequences of b12 and 2G12 were previously described (32, 33). Electroporation of K46 mouse B cells was carried out as follows. 10 μg of both heavy and light chain expression vectors (linearized using Sal I and Nhe I, respectively) were added to 5×106 K46 cells in 500 μl DMEM and incubated in 4 mm cuvettes on ice for 5 min. Cells were subjected to electroporation with a Gene Pulser Xcell (Bio-Rad, Hercules, CA), set at 230 V, 960 μF, and ∞ Ω resulting in a time constant of ~ 30 ms. After incubation on ice for 10 min cells were cultured in 24 well plates complete medium (DMEM or RPMI, 10% FCS, 55 μM β-mercaptoethanol, 2 mM L-glutamine, 100 μg/ml streptomycin, 100 IU/ml penicillin, 100 μM non-essential amino acids). Selection was started after 0–2 days with 1 mg/ml G418 (Gibco, Carlsbad, CA) for heavy chain transfection and/or 1 mg/ml Zeocin (Invitrogen, Carlsbad, CA) for light chain transfections. Surviving cells were passaged as needed. After selection, single clones were generated by single cell sorting of heavy and/or light chain expressing K46 cells on a FACSAria (BD, San Jose, CA).
B cell lines transduced with lentivirus encoded antibody constructs
bNAb variable region containing cDNA plasmids were used as templates for PCR sewing using appropriate primers. bNAb LVDJ elements were linked with mouse IgM constant region cDNA sequence, then the resulting μH-chain gene was further linked with a Gly-Ser-Gly linker followed by the P2A peptide and the mature light chain sequences, which carry human Cκ or Cλ constant regions corresponding to the original L-chain used. PG9, PG16, PGV19, PGT121 and PGT128 carry λ L-chain, whereas the other bNAbs use κ. Fusion genes with the corresponding restriction sites were cloned into EcoRI/NotI digested modified gateway entry vector (Invitrogen), then transferred to pLenti CMVTRE3G Puro Dest plasmid (developed by Eric Campeau and obtained from Addgene) using LR Clonase II enzyme (Invitrogen). bNAb encoding lentiviruses were produced as previously described (34). Briefly, each bNAb TRE3G plasmid was co-transfected with psPax2 and MD2.G plasmids into 293T cells using FuGeneHD. Cell supernatants harvested on day 2 were used to transduce rtTA3G-expressing WEHI-231 or K46 B cell lines in the presence of 10 μg/ml polybrene (Millipore). Starting two days later, cells were selected with 2 μg/ml puromycin. bNAb expressing cells were induced by incubation with doxycycline (1μg/ml) overnight and stained with 200-fold diluted anti-hCκ-biotin (G20-193, BD Biosciences) or hCλ-biotin (JDC-12, BD Biosciences) in FACS buffer and further stained with streptavidin-phycoerythrin (SA-PE) (eBioscience). High-expressing cells were selected by sorting using FACS Aria II (BD Biosiences) or MoFlo (Beckman Coulter) machines. Single clones of some cell lines were obtained by limiting dilution. Anti-CD69-allophycocyanin (H1.2F3) was purchased from eBiosciences. MAbs against mouse IgM (M41;Dylight 650) were labeled in-house. All samples were read on an LSR-II instrument (BD) and analyzed using the FlowJo program (Tree Star, Inc.).
Generation of HIV virions
HIV-1 enveloped pseudoviruses were prepared by co-transfecting 293T cells with 15 μg of the backbone plasmid (pSG3ΔEnv or pNL4-3.Luc), 5 μg of a functional wild-type or b12-knockout mutant (D185A, P369Q) envelope clone, and Fugene reagent (Roche) as previously described (35, 36). Green fluorescent viruses were prepared by co-transfection of pEGFP-C3 (Clontech, Madison, WI) containing the entire Vpr (BRU) coding region fused to the COOH terminus of EGFP (GFP-Vpr) (37). Virus stocks were titrated on TZM-bl cells as described (35, 36) and the TCID50s were calculated according to Reed and Muench (38). The p24 concentration was determined by ELISA (Aalto Bio Reagents, Dublin, Ireland; DIY ELISA Protocol 2).
Envelope protein concentration
Concentrated JRFLΔCT-GFP virus was tested by ELISA for Env protein amount as follows. 1000× fold concentrated JRFLΔCT virus was first diluted in TBS, 1% Empigen BB (Sigma), 1% BSA, 0.1% Tween 20, then serially diluted in TBS 1% BSA, 0.1% Tween 20 and applied to ELISA plates (Nunc MaxiSorp) previously coated with sheep anti-gp120 (Aalto) at 3 μg/ml. JRFL gp140 trimer was used as standard. Bound envelope protein was detected with PGT121, followed by detection with biotinylated anti-human Lambda (BD) and streptavidin-HRP (BD). This approach revealed that 1000× fold concentrated JRFLΔCT virus stock contained 11.2 nM (4.7 μg/ml) Env trimer equivalents.
Flow cytometry
Binding of biotinylated antigens or antibodies to B cell lines was measured on a C6 Flow cytometer (Accuri, Ann Arbor, MI) or LSR II (BD, San Jose, CA) either using directly labeled antibodies (anti-mouse-IgM (M41); anti-human-kappa, Jackson) or using biotinylated antigens (biotin-gp120 JRFL/JRCSF/YU2) and a streptavidin-PE secondary probe. Antibodies used for detection of activation markers for K46 cell stimulations included IgM (M41) Alexa 488 or 647, CD69 (H1.2F3), CD83 (Michel-19), and CD19 (6D5). Antibodies, except IgM (M41) were purchased from Biolegend, San Diego, CA). Sheep anti-HIV-1 gp120 (Aalto Bio Reagents Ltd., Dublin, Ireland) was labeled with Alexa Fluor-647 (Life Technologies, Grand Island, NY) depleted for cross-reactivity against K46 b12 and 2G12 expressing K46 cells for 25 min at room temperature and filtered for detection of HIV proteins bound to the cells surface by flow cytometry. The His-tag MAb used in crosslinking and detection of the 2bodx-043 b12 scaffold was purchased from Genescript (Piscataway, NJ). FlowJo (Tree Star, Ashland, OR) and Prism 5.0 for Mac (GraphPad, La Jolla, CA) were used for analysis.
Ca++ flux assays
In all experiments, except for those shown in Fig 1, cells were suspended at 4 million cells/ml in Advanced DMEM, labeled with 1.5 μM Indo-1 (Invitrogen) for 30 min at 37°C and washed with 2mM CaCl2 HBSS, followed by another 30 min at 37°C. Aliquots of 2 × 106 cells in 0.5 mL were then stimulated at room temperature with BCR ligands. Ca++ signals were recorded for 180 seconds, measuring the 405/485-nm emission ratio of Indo-1 fluorescence upon UV excitation. Calcium flux analysis was performed on an LSRII cytometer (BD, San Jose, CA). BCR ligand concentrations used are indicated in the figures. Kinetic analysis was performed using FlowJo (Tree Star, Ashland, OR). For experiments shown in Fig 1, assays were performed using cells loaded with the Fluo-4 NW dye (Life Technologies, Grand Island, NY) per the manufacturer's recommendations.
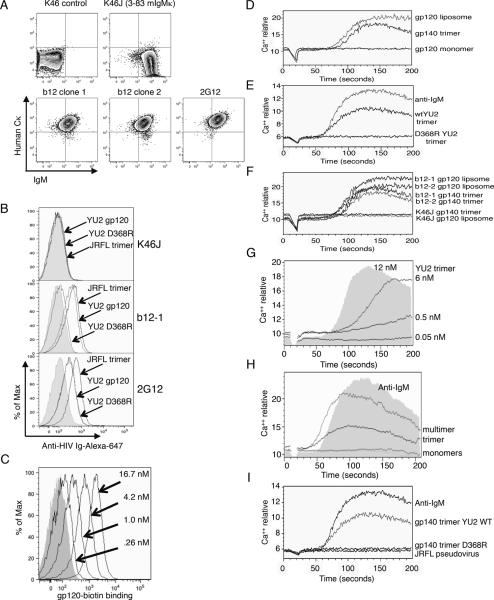
Figure 1.
BCR expression, Env protein binding, and Ca++ mobilization in B cell transfectants carrying bNAb specificities. (A) Heterologous antibody chain expression in K46 transfectants. Shown is two-color flow cytometry analysis of human Cκ and mouse IgM cell surface expression. (B,C) Env antigen binding of the indicated ligands. (B) Shows binding of cell lines carrying the indicated bNAbs to soluble YU2 gp120 monomer, mutated YU2 monomer with a substitution abrogating CD4 binding (YU2 D368R), or soluble JRFL trimer. Binding was detected by secondary staining with a polyclonal anti-HIV reagent. Grey fill shows background with secondary reagent alone. (C) Titration of YU2-biotin gp120 monomer binding to cells carrying the b12 BCR. (D–I) Recognition of BCR ligands was assessed by Ca++ mobilization assay. Time scale of D–G is indicated in the horizontal axis of (G). (D) Activation of b12 transfectant clone 1 (b12-1) with YU2 gp120-biotin monomer, gp140 trimer, or YU2-biotin monomer associated with avidin liposomes. Final concentrations of Env protein were 5 μg/ml. (E) Comparison of b12-1 activation using YU2 gp140 wild type trimers compared to D368R mutant trimers at a final concentration of 12 nM (5 μg/ml). Stimulation with F(ab')2 anti-IgM 2.5 μg/mL, was used as a positive control. (F) The indicated ligands were used to activate two independent b12-expressing clones or K46J cells, which carry an irrelevant BCR specificity. (G) Analysis of Ca++ responses in b12-1 cells as a function of YU2 trimer concentration. (H) Response of b12-1 cells to different forms of 2bodx-043 (44), which carries two discontinuous HIV peptides (365–372 and 472–476) on an unrelated protein scaffold. In this experiment we compared gp120 monomers (5 μg/ml, 41.7 nM) and anti-IgM (2.5 μg/ml) to 2bodx-043 trimers (10 nM) and a higher order multimer created by preincubation with anti-His antibody to further multimerize the 2bodx-043 trimer. Anti-His antibody alone failed to stimulate calcium in b12 or K46J cells. (I) Comparison of responses shown in E carried out contemporaneously with stimulation by a high titer, but unconcentrated, JRFL pseudovirion preparation (final concentration 25% by volume).
HIV antigens
YU2 gp120-His6 AviTag was cloned into pIRES-hrGFP Zeocin plasmid and cotransfected with BirA-ER plasmid to 293F cells. Stable drug-resistant cells were obtained and cultured in 5% FCS containing Advanced DMEM (Invitrogen), penicillin, streptomycin, Glu-Max and 0.2 mM Biotin. Expressed protein was purified over HisPur Cobalt Resin (Thermo Fisher), eluted with imidazole and dialyzed against PBS. YU2 gp120 biotinylation was confirmed by western blot using HRP anti-biotin antibody (Jackson). YU2 gp140-GCN4-bio and JRFL trimers were prepared as described (39, 40). 92BR020 gp120 monomer sequences were cloned into the phCMV vector and the plasmid was used to transfect 293F cells. gp120 protein was purified from supernatants by Galanthus nivalis lectin chromatography followed by size exclusion chromatography. 293F cells were grown in 293F expression media (Invitrogen).
Results
Conventional B cell transfectants carrying b12 or 2G12
In initial experiments, we expressed H- and L-chains of b12 and 2G12 in the B cell line K46 using mouse genomic constructs encoding H-chains carrying IgM constant (C) regions and pEF1α-driven cDNA plasmids for the light chains, which carried human Cκ. Coexpression of the desired immunoglobulin chains was verified by detection of antibodies to human Cκ and to IgM (Fig 1A, lower panels). As controls, we also included untransfected K46 cells and a K46 transfectant (K46J) expressing the IgM antibody 3–83 (41, 42) (Fig 1A, upper panels). B cell lines coexpressing IgM H-chain and human Cκ were obtained (Fig 1A), indicating that they carried high levels of the desired antibodies. To assess specificity, we measured the binding of gp120 monomers to the cells and detected binding specifically to b12- and 2G12-expressing cells. We were also able to detect binding by the cells to YU2 gp140 and JRFL gp140 trimer preparations (Fig 1B and data not shown) (40). The fine specificity of b12 cells for gp120 was further confirmed by their inability to react with the CD4 binding site mutant (43) YU2 D368R (Fig 1B, middle panel). By contrast, 2G12, which binds to a distinct site on gp120, bound both mutant and WT versions similarly (lower panel). Binding of JRFL gp120 monomer by b12 cells was detected using as little as 0.26 nM (31 ng/mL) antigen (Fig 1C). These data show one can generate B cell lines expressing bNAbs as a BCR, providing a new tool to assess vaccine candidate antigens for their ability to stimulate anti-HIV B cells.
Ca++ flux analysis
To measure antigen driven cellular activation, we stimulated B cell lines expressing or lacking anti-HIV BCRs with various Env proteins, protein scaffolds carrying selected epitopes, or virion preparations, and monitored increases in intracellular Ca++ levels (Fig 1D–I). We found that Ca++ responses in b12 or 2G12 cells could be stimulated only with multimeric gp120 preparations. Env monomer preparations were non-stimulatory unless they were multimerized on the surface of avidin-carrying liposomes, whereas soluble gp140 trimers, generated using heterologous C-terminal foldon trimerization motifs, gave robust stimulation for b12-expressing cells (Fig 1D). Trimers also triggered 2G12 cells, but required higher concentrations (not shown). Activation was strictly specific for antigen, as YU2 trimers carrying the D368R CD4-binding site mutation, which abrogates b12 binding and neutralization, were unable to activate b12 cells (Fig 1E). Anti-IgM was used in these experiments as a positive control. Env preparations that were stimulatory for two independently generated clones of b12 cells (b12-1 and b12-2) failed to activate control K46J cells, which carry an irrelevant surface IgM (Fig 1F). Soluble trimers generated using gp140 expression constructs were able to stimulate b12 cells at concentrations as low as 0.5 nM (Fig 1G). We also found that the b12 epitope scaffold 2bodx-043 (44) was stimulatory provided that it was presented in a multimeric form (Fig 1H). Thus, soluble multimeric Env proteins or their mimics are stimulatory for these bNAb expressing cell lines.
Having established the ability of the B cell lines coexpressing HIV antibodies to be stimulated by various soluble HIV Env preparations, we next assessed their ability to recognize pseudovirions (JRCSF, JRFL, BaL, SF162) or virions (NL4-3) carrying distinct, well-characterized Env. These viruses were chosen because of their known sensitivity to neutralization by the antibodies in question. These infection-competent preparations failed to promote a Ca++ flux. Fig 1I shows that freshly prepared JRFL pseudovirus was inactive, in contrast to gp140 trimer, in triggering a Ca++ flux. Similar negative results were obtained when we tested the ΔCT mutant version of JRFL, which is predicted to carry ~5–10-fold higher-than-normal levels of surface envelope proteins (45) (Fig S1A). The highest doses of pseudovirions corresponded to a multiplicity of virions to cells of ~104. We verified that the pseudovirions tested carried native, functional Env because they could infect target cells. Collectively, these data indicated that live, functional viruses or pseudoviruses could not appreciably stimulate K46 B cells carrying 2G12 or b12. These results suggest that, in many cases, virions carrying Env proteins capable of being neutralized by free b12 or 2G12 antibodies fail to stimulate B cell lines carrying these same antibodies as B cell receptors.
These results led us to wonder if virus preparations might inhibit B cell activation. We first tested if free monomers could suppress the activation induced by soluble trimers. Indeed, high levels of monomer led to an appreciable loss of Ca++ flux in response to soluble trimer (Fig S1B), presumably by simple competition. In contrast, pseudovirions were unable to suppress the response to soluble trimers (Fig S1C). Hence the inability of pseudovirions to stimulate B cell lines carrying bNAb BCRs could not be attributed to contamination with non-stimulatory antigen forms or to an independent inhibitory activity of virions.
Generation of B cell lines carrying P2A linked and inducible anti-HIV BCRs
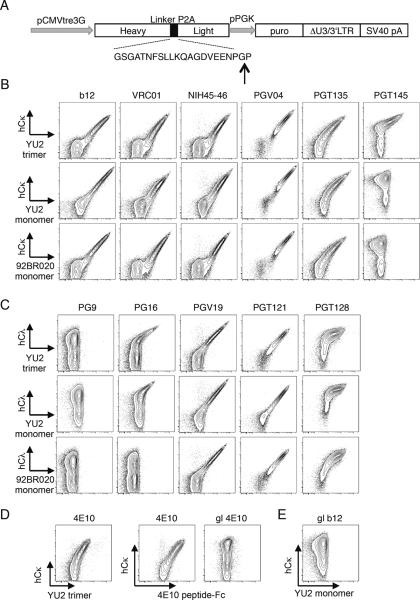
In the course of the above-mentioned studies we attempted to generate stable B cell lines carrying a number of other broadly neutralizing HIV antibodies. However, we had some difficulties in doing so, in part because of the inefficiency of conventional electroporation methods, and possibly also because their expression was toxic to the cells, perhaps because of BCR autoreactivity. To facilitate expression, we engineered antibody cDNAs into lentivirus vectors that drove expression of both H and L genes from a single cistron separated by the picornavirus 2A peptide. The 22-residue 2A peptide prevents the formation of a normal peptide bond between glycine and the last proline, effectively “cleaving” the peptide in two during translation (Fig 2A). This allows one to express Ig H- and L-chains at equal molar ratio using just one compact gene. In this case, we used constant region sequences from the membrane form of mouse μ H-chain and human κ or λ L-chain. Although initial experiments were done using lentivirus vectors driving bNAb constitutive expression, we show here studies using doxycycline-inducible bNAb expression in WEHI-231 and K46 B cells expressing the Tet transactivator rtTA3G (Figs 2 and S2, respectively). WEHI-231 cell lines expressing several bNAb BCRs were assessed by flow cytometry for the ability to bind to cognate antigen and for levels of human κ or λ L-chains (Fig 2 B–E). Based on the expression of one antibody chain or another, all 16 tested bNAb specificities were expressed. Since the constructs encoded both chains off one cistron, it is assumed that lentivirus-encoded H and L-chain protein expression levels were similar. Importantly, transductants of most bNAbs exhibited demonstrable binding to soluble Env proteins (Fig 2B–E). In contrast, however, cell lines expressing germline-reverted (given prefix “gl”) versions of 4E10, b12, and VRC01 (sequences given in Table II), exhibited high-level surface expression of both transduced chains, but failed to bind soluble Env proteins (Fig 2D,E and data not shown). This indicated that the receptors were expressed, but had too low an affinity to detectably bind to soluble antigen. We conclude that the induced expression of transduced bNAb BCRs was generally achieved, but that gl-bNAb receptors bound antigen poorly despite high level expression.
Figure 2.
Design and expression of bNAb antigen receptors by lentivirus transduction. (A) Schematic arrangement of vector showing gene elements encoding antibody genes and adjacent elements. The H-chain genes carry a leader exon followed by VDJ codons and constant region for membrane IgM. This is followed in frame by the picornavirus P2A peptide, shown below, and the light chain VJ and C codons. Also shown is the puromycin resistance gene (puro). The vertical arrow shows the P2A “cleavage” site, which lacks a peptide bond upon translation. (B–E) Expression and soluble Env binding by the indicated bNAb BCRs in transduced WEHI231 cells. Final concentration of antigens used in staining was 25 μg/ml, except for 98BR020 monomer, which was at 20 μg/ml.
Table II.
Sequences of germline-reverted (gl) variable regions used in this study.
| gl-b12 H |
| QVQLVQSGAEVKKPGASVKVSCKASGYTFTSYAMHWVRQAPGQRLEWMGWINAGNGNTKYSQKFQGRVTITRDTSASTAYMELSSLRSEDTAVYYCARVGPYCGGDSPQDNYYMDVWGKGTTVTVSS |
| gl-b12 L |
| EIVLTQSPGTLSLSPGERATLSCRASQSVSSSYLAWYQQKPGQAPRLLIYGASSRATGIPDRFSGSGSGTDFTLTISRLEPEDFAVYYCQQYGSSSYTFGQGTKVEIKR |
| gl-4E10 H |
| QVQLVQSGAEVKKPGSSVKVSCKASGGTFSSYAISWVRQAPGQGLEWMGGIIPIFGTANYAQKFQGRVTITADKSTSTAYMELSSLRSEDTAVYYCAREGTTGWGWLGKPIGAFDYWGQGTLVTVSS |
| gl-4E10L (gl-b12 L was used) |
| gl-VRC01 H |
| QVQLVQSGAEVKKPGASVKVSCKASGYTFTGYYMHWVRQAPGQGLEWMGWINPNSGGTNYAQKFQGRVTMTRDTSISTAYMELSRLRSDDTAVYYCARGKNCDYNWDFQHWGQGTLVTVSS |
| gl-VRC01 L |
| EIVLTQSPATLSLSPGERATLSCRASQSVSSYLAWYQQKPGQAPRLLIYDASNRATGIPARFSGSGSGTDFTLTISSLEPEDFAVYYCQQYEFFGQGTKLEIKR |
Binding and activation by ligands in cells with inducible bNAb BCRs
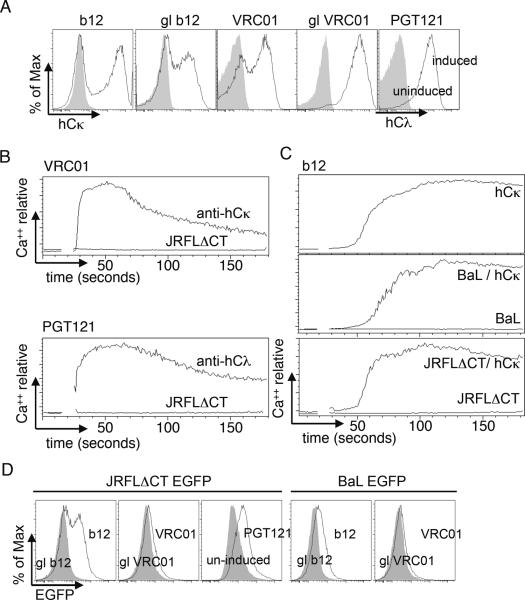
In these cell lines, bNAb expression was strictly doxycycline-dependent and readily induced upon overnight incubation with 1 μg/ml of drug (Fig 3A), allowing us to use uninduced cells as negative controls for binding and activation studies. Accordingly, cells carrying or lacking VRC01, PGT121, or b12 BCRs were compared for their abilities to bind and to be activated by BCR ligands, soluble antigens and pseudovirions. As with the b12 and 2G12 cell lines described above, we were unable to detect a Ca++ mobilization response to pseudovirions, while the response to anti-human L-chain was robust (Fig 3B,C). After challenge with pseudovirions, b12 cells were fully able to respond to anti-hCκ, again arguing that pseudovirions were not suppressive in this assay (Fig 3C, BaL/hCκ, JRFLΔCT/hCκ).
Figure 3.
Analysis of inducible expression and the ability of GFP-labeled pseudovirions to activate bNAb-expressing WEHI231 cells. (A) Comparison of human L-chain expression in induced (black line) versus non-induced (grey fill) cells carrying the indicated bNAb genes. (B,C) Ca++ mobilization analysis of the indicated bNAb-expressing cell lines upon stimulation with anti-hCκ (10 μg/ml) or the indicated pseudovirions (100× concentrated stock final concentration, approximately 3×109/ml for JRFLΔCT and 2×1012/ml for BaL; the JRFLΔCT concentration used corresponded to 1 nM Env trimer equivalents as established by ELISA). (C) b12 cells were stimulated with anti-hCκ alone (top panel). In the lower two panels activation by the indicated pseudovirions (lower traces), was followed by challenge with anti-hCκ (upper traces). (D) Analysis of pseudovirion binding in the same experiment, using the same virus particles and concentrations as in B and C, but incubating on ice for 1 hour. Filled traces shown binding to uninduced cells or cells carrying germline reverted (gl) bNAbs.
We reasoned that the inability of virions or pseudovirions to stimulate B cells could be the result either of an inability of the viruses to bind to the bNAb BCRs or to a failure to trigger upon binding. To address these issues, we assessed the binding of virions by either generating virus particles that incorporate green fluorescent protein (GFP) or by detection of bound particles with secondary antibodies. In studies using stably transfected K46 cells, we often had difficulty detecting specific binding, as defined by binding that was dependent upon the presence of an anti-HIV BCR and could be inhibited by soluble Env protein (data not shown). By contrast, as shown above, binding to soluble monomeric or trimeric Env proteins was readily demonstrable. To extend these experiments, we incubated B cell lines on ice with infectious virus preparations, followed by low speed centrifugation in order to test their ability to absorb infectious activity. These studies failed to demonstrate specific binding to cells carrying anti-HIV BCRs. By contrast, however, use of highly concentrated, internally GFP-labeled pseudovirions derived from JRFLΔCT virus, allowed clear documentation of BCR-specific binding (Fig 3D). (Similar results were obtained with the b12 and 2G12 K46 clones described above (not shown)). Under these conditions, however, no Ca++ mobilization was seen (Fig 3B,C). Similar results were obtained with BaL pseudovirions (Fig 3C,D). We conclude that pseudovirions, and by inference, native viruses, fail to efficiently trigger cognate B cell lines, even those potentially capable of neutralization, and even under conditions where virions are bound. By contrast, soluble trimers and other multimeric presentations of Env can be quite stimulatory.
Analysis of induction of early activation markers
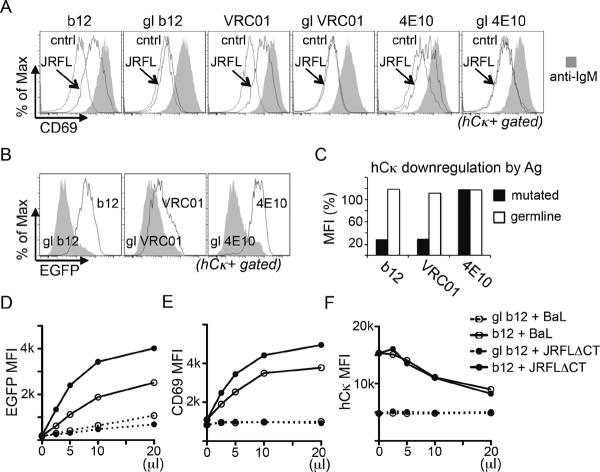
We considered the possibility that virus:B cell interactions or virion-induced activation were too weak or slow to be detected by Ca++ flux assays, but might occur upon extended incubation. To assess this, we took advantage of the fact that B cells upregulate activation markers after 12–24 hours of BCR signaling. CD83 and CD69 were increased in expression upon anti-BCR stimulation of K46 transfectants (data not shown), whereas in WEHI231 cells, CD69 expression alone was well induced. We then assessed the ability of 18 hr incubation with virions to induce expression. Although experiments using K46 transfectants were all negative, we were able to observe, in several WEHI231 bNAb transductants, a virion-mediated and BCR-dependent induction of CD69 (Fig 4A). Consistent with the soluble trimer binding data of Fig 2, cells carrying germline reverted versions of these bNAbs failed to upregulate CD69 (Fig 4A). Interestingly, under these conditions bNAb-expressing cells clearly bound or took up GFP+ virions, as they scored GFP+ when assessed by flow cytometry (Fig 4B) and binding to b12 and VRC01, but not 4E10, was correlated with BCR downmodulation (Fig 4C). Stimulation of CD69 expression, hCκ downregulation, and binding of BaL or JRFLΔCT was dose-dependent in b12 WEHI231 cells, but was weak or negative in cells carrying germline reverted b12 BCR (Fig 4D–F). These data indicate that induction of CD69 expression was likely caused by binding of virus particles, rather than shedding of Env trimers, which would lack associated GFP. We conclude that pseudovirions were poorly stimulatory for, but not invisible to, bNAb-expressing B cell lines, possibly owing to degradation in culture or to a slower pace of cell activation.
Figure 4.
Flow cytometry analysis of bNAb-expressing WEHI231 B cells cultured with JRFLΔCT-EGFP pseudovirions. bNAb BCR genes were induced one day before stimulation and then cells were treated for 18 hr as indicated. (A) Analysis of CD69 upregulation in cells stimulated with doxycycline medium alone (control), anti-IgM (MAb M41, 2μg/ml) or 100-fold concentrated JRFLΔCT-EGFP pseudovirus (final concentration 3×109/ml). (B) Cell associated virion binding/internalization as measured by EGFP level. (C) Downregulation of hCκ expression in virion-treated compared to untreated cells. (D–F) Dose/response analysis in b12 and gl-b12 WEHI-231 cell lines after pseudovirion stimulus (highest final concentrations of pseudovirions were approximately 3×109/ml for JRFLΔCT and 2×1012/ml for BaL, corresponding to 1 and 67 nM Env, respectively): (D) Virus binding measured by GFP fluorescence, (E) CD69 induction, (F) Human Igκ downregulation.
Discussion
In order to understand how to generate desired antibody responses eliciting broadly neutralizing HIV antibodies, it is important to have models of bNAb-expressing B cells. Here, we have addressed this goal by introducing bNAbs into B cell lines and assessing their binding and activation by pseudovirions and other predicted bNAb ligands. We find that soluble gp140 trimers are especially efficient at binding and cell activation, whereas gp120 monomers and pseudovirions fail to efficiently activate these cells. Simple multimerization of biotin-tagged gp120 monomers on liposomes carrying avidin restored activation of b12-expressing cells. Activation in this study was measured by assessing rapid Ca++ mobilization upon antigen exposure. We also assessed upregulation of surface markers such as CD69 and CD83, when antigen was included in B cell line cultures for 18 hours. Infection-competent pseudovirions, which should display Env trimers in a multimeric, albeit paucivalent, array failed to activate obvious Ca++ mobilization, but at high concentrations they did bind to bNAb-expressing cells and promoted CD69 upregulation upon prolonged exposure. Importantly, virions at superphysiological concentrations failed to rapidly stimulate even high affinity B cell lines with bNAb specificity. Based on these in vitro features, our data suggest that soluble trimers (or perhaps higher order soluble multimers) should be most effective as vaccine components in driving B cell activation responses, while unmultimerized monomer gp120 and lentivirus-like particles are likely to be less effective. Immunization studies in animals support the notion that trimers may be more efficient for antibody induction than monomers (40, 46, 47).
A surprising finding in our studies was the inability of infection-competent HIV Env-expressing pseudovirions to activate Ca++ responses in anti-HIV B cell lines, even though these cells carried cognate antibodies capable of neutralizing the same virus and responding to its Env when presented in soluble trimeric form. This general result was obtained with cell lines carrying bNAb BCRs specific for at least three distinct epitopes on Env: the CD4 binding site (b12, VRC01), the carbohydrate-dependent epitope defined by PGT121, and the high mannose epitope defined by 2G12. These findings, along with the detection of some virus binding to cells, suggest that the BCR had access to Env epitopes on the virus surface, and that virions were available at sufficient concentration, but that these interactions failed to generate the necessary conformational changes or crosslinking required for strong immediate biochemical signal transduction. Alternatively or in addition, soluble Env trimers might be less conformationally constrained than virion-bound spikes, which would favor bNAb BCR crosslinking by trimers. Multimeric particulate antigen is typically more stimulatory for B cells than paucivalent antigens (48, 49), but T cell dependent antibody responses are not believed to be strictly dependent upon antigen multimerization. Cryoelectron microscopy tomography studies have indicated that HIV-1 particles carry on average only 14 spikes, which are widely dispersed (50). Spike paucivalency might reduce the BCR occupancy possible even at saturating virion concentrations. It should be noted that the concentration of virions used for B cell line stimulations here exceeded by several orders of magnitude the virus titers typically seen at the peak of acute HIV infection and in many experiments we used JRFLΔCT pseudovirions, which have an elevated number of spikes. (In addition, similarly constructed bNAb B cell lines reactive to influenza virus responded robustly to those virions even at low virion concentrations (data not shown).) Although bNAb B cell lines may be less sensitive to virion antigens than primary B cells, given that even B cell lines carrying high affinity receptors failed to be well activated by virions, it seems likely that the unhypermutated precursors of bNAb B cells should not be efficiently activated during natural infection. These difficulties perhaps explain why bNAbs typically appear in patients only after years of infection. Initiating the gl-bNAb response might require one or more fortuitous events, such as the appearance during infection of virion variants carrying high epitope densities of Env, along with fortuitous mutations in gl-bNAb B cells (whilst responding to non-HIV antigens) that allow them to subsequently see HIV.
We considered the possibility that the virions engage inhibitory receptors on B cells to actively suppress triggering. However, this seems unlikely for three reasons. First, we tested, and failed to detect, any inhibitory effect of pseudovirions on Ca++ mobilization responses to Env trimers given simultaneously. Second, even the most robust inhibitory signals typically fail to completely prevent a Ca++ response, but rather blunt and redirect initial signaling (51, 52). Finally, incubation of B cells with high concentrations of viruses was able to reveal some activation at 18 hours, as indicated by early activation marker expression. We conclude that the low density of Env or an adapted inability to cluster BCRs accounts for the weak stimulatory activity of virions. However, we cannot exclude that virions change conformation or shed soluble multimeric receptors upon cell culture.
Anti-HIV B cell lines could have many practical uses. One is to rank and categorize different HIV antigens and isolates for relevant structural features that impact on interactions with responding B cells. This application is non-trivial because the arrangement of the antigen receptor on the B cell membrane involves tight association with the CD79a/b complex and other elements that likely sterically restrict some approaches of the BCR to virions or vaccine candidates. bNAb BCR expression in B cell lines may also facilitate analysis of receptor reactivity to ligands that mimic HIV. For example, we have been able to validate the ability of HIV epitopes engineered into heterologous scaffold proteins to activate cognate B cells. This approach should be most beneficial when used in conjunction with gl-bNAb specificities, which have essentially undetectable affinity for the HIV antigens (this study and 13, 53, 54–56). bNAb B cells might also be helpful in defining conditions to detect B cells with similar specificities in HIV infected patients. In addition, bNAb-expressing B cell lines could facilitate analysis of receptor autoreactivity. We have consistently observed that 4E10-transduced B cells poorly express their BCRs, and such cells selectively lose that expression with time in culture, whereas cells carrying distinct BCRs, including germline-reverted 4E10, are quite stable. 4E10 and the anti-MPER MAb 2F5 have been reported to be, at least weakly, self-reactive owing in part to the hydrophobicity of their CDRH3 elements (29, 57). Consistent with this, mice carrying the 4E10 BCR show clear signs of B cell central tolerance (CDC, DN, manuscript in preparation, Laurent Verkoczy, Duke U., pers. comm.). However, our germline reverted 4E10 BCR that retains CDRH3 sequences appears to be well expressed. PG9, PG16 and VRC01 BCRs also appeared to be selected against in B cell lines, though the significance of this is unclear. In the case of PG9, BCR expression was too low to evaluate responses to Env ligands.
While self-reactivity may preclude some B cell specificities from contributing to the normal repertoire through tolerance mechanisms, polyreactivity has been argued to contribute to the anti-HIV response by promoting bivalent heteroligation between one high-affinity anti-HIV-gp140 combining site and a second low-affinity site on another molecular structure on HIV (15). That study evaluated the effects of polyreactivity on the effector phase of the anti-HIV response, but did not address the induction phase. In the present study, B cell lines carrying BCRs with specificity for distinct Env epitopes (b12, VRC01, 2G12, PGT121), failed to mobilize Ca++ upon short term incubation with infection-competent pseudovirions, despite the ability of these same BCRs to neutralize the same virus when provided as free immunoglobulin, and despite the ability of these BCRs to be triggered by soluble trimers. Binding of bNAb cell lines to pseudovirions, though poor, was detectable by direct flow cytometry analysis and by CD69 upregulation upon overnight cell culture. Thus, antibody polyreactivity might conceivably contribute to late, but not to early, B cell activation. However, it is unclear if activation induced with extended in vitro incubation is of relevance in vivo in HIV infection because the plasma half life of virions is remarkably short, on the order of minutes (58). In any case, the analysis of early activation markers on the bNAb B cell lines provides a readout that may be useful in ranking the potency of candidate vaccine antigens for HIV.
The binding patterns of bNAb B cell lines to soluble Env antigens were not entirely as predicted (9, 12, 14, 16, 59). As expected, all five CD4 binding site bNAbs along with three glycan-dependent bNAbs PGT121, PGT128 and PGT135 reacted to both gp120 monomers and trimers. However, PGT145 and PG16 were not expected to bind to soluble YU2 trimers, whereas PG9 has weak affinity (9). PG9, PG16 and PGT145 are believed to be relatively specific for membrane trimers of YU2 (9), but PG9 and PG16 can see monomers of certain other isolates (12). PG9 cell lines likely fail to bind trimer owing to poor surface expression or receptor downregulation (Fig. S2). Surprisingly, the PG16 B cell line bound not only soluble YU2 gp120 trimers, but also weakly to YU2 monomer. As comparable results for PG9, PG16 and PGT145 were obtained in K46 and WEHI231 cell lines transduced with these sequence verified BCRs (not shown), it is unlikely that the endogenous Ig-chains expressed by the cell lines paired with transduced Ig-chains to generate novel anti-Env specificities, or that these specificities arose from V region mutations occurring after transduction. One possibility for these discrepancies is that the B cell lines alter bNAb affinity by post-translational modification. PG9 and PG16 are partially sulfated on CDRH3 tyrosine contact residues, which improves neutralization 10–100-fold (10, 12). These modifications might be distinct in quality or quantity in B cell lines compared to the 293 cells used to generate recombinant antibodies for binding studies. Most importantly, however, for vaccine research studies, all gl-bNAb B cell lines tested in the present study failed to bind to, or be activated by, any antigen preparation tested, supporting studies indicating that gl-bNAbs lack detectable affinity for Env antigens (13, 53–56). Our gl-bNAb B cell lines should be useful in modeling vaccine candidates intended to target gl-bNAb B cells.
We have studied B cells carrying the IgM form of BCR. IgM is thought to extend farther from the plasma membrane than most other Ig classes owing to its additional CH domain, but it lacks a flexible hinge region. It may be that other Ig classes are better adapted to dimerization by Env spikes, or heteroligation to other epitopes. For the purposes of vaccine design, however, the naïve B cells that would presumably be targeted initially by any HIV vaccine are likely to express IgM and to lack class-switched isotypes. In this regard, B cell lines carrying the germline versions of several bNAbs will potentially be useful in probing vaccine candidates designed to initiate the bNAb response.
Supplementary Material
Acknowledgments
The authors thank Krystalyn Hudson for critical reading of the manuscript, Patrick Skog and Michael Kubitz for technical assistance, and Leo Stamatatos for discussions.
Footnotes
Disclosures The authors have no financial conflicts of interest.
Abbreviations: bNAb, broadly neutralizing antibody; MPER, membrane-proximal external region;
The online version of this article contains supplemental material.
References
- 1.Walker LM, Burton DR. Rational antibody-based HIV-1 vaccine design: current approaches and future directions. Curr Opin Immunol. 2010;22:358–366. doi: 10.1016/j.coi.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hessell AJ, Haigwood NL. Neutralizing antibodies and control of HIV: moves and countermoves. Current HIV/AIDS reports. 2012;9:64–72. doi: 10.1007/s11904-011-0105-5. [DOI] [PubMed] [Google Scholar]
- 3.Mascola JR, Montefiori DC. The role of antibodies in HIV vaccines. Annu Rev Immunol. 2010;28:413–444. doi: 10.1146/annurev-immunol-030409-101256. [DOI] [PubMed] [Google Scholar]
- 4.Verkoczy L, Kelsoe G, Moody MA, Haynes BF. Role of immune mechanisms in induction of HIV-1 broadly neutralizing antibodies. Curr Opin Immunol. 2011;23:383–390. doi: 10.1016/j.coi.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moir S, Malaspina A, Fauci AS. Prospects for an HIV vaccine: leading B cells down the right path. Nat Struct Mol Biol. 2011;18:1317–1321. doi: 10.1038/nsmb.2194. [DOI] [PubMed] [Google Scholar]
- 6.Saphire EO, Parren PW, Pantophlet R, Zwick MB, Morris GM, Rudd PM, Dwek RA, Stanfield RL, Burton DR, Wilson IA. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science. 2001;293:1155–1159. doi: 10.1126/science.1061692. [DOI] [PubMed] [Google Scholar]
- 7.Cardoso RM, Zwick MB, Stanfield RL, Kunert R, Binley JM, Katinger H, Burton DR, Wilson IA. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity. 2005;22:163–173. doi: 10.1016/j.immuni.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 8.Zwick MB, Labrijn AF, Wang M, Spenlehauer C, Saphire EO, Binley JM, Moore JP, Stiegler G, Katinger H, Burton DR, Parren PW. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 2001;75:10892–10905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, O'Dell S, Patel N, Shahzad-ul-Hussan S, Yang Y, Zhang B, Zhou T, Zhu J, Boyington JC, Chuang GY, Diwanji D, Georgiev I, Kwon YD, Lee D, Louder MK, Moquin S, Schmidt SD, Yang ZY, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Burton DR, Koff WC, Walker LM, Phogat S, Wyatt R, Orwenyo J, Wang LX, Arthos J, Bewley CA, Mascola JR, Nabel GJ, Schief WR, Ward AB, Wilson IA, Kwong PD. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, O'Dell S, Perfetto S, Schmidt SD, Shi W, Wu L, Yang Y, Yang ZY, Yang Z, Zhang Z, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Simek M, Burton DR, Koff WC, Doria-Rose NA, Connors M, Mullikin JC, Nabel GJ, Roederer M, Shapiro L, Kwong PD, Mascola JR. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pejchal R, Walker LM, Stanfield RL, Phogat SK, Koff WC, Poignard P, Burton DR, Wilson IA. Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proc Natl Acad Sci U S A. 2010;107:11483–11488. doi: 10.1073/pnas.1004600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mouquet H, Scheid JF, Zoller MJ, Krogsgaard M, Ott RG, Shukair S, Artyomov MN, Pietzsch J, Connors M, Pereyra F, Walker BD, Ho DD, Wilson PC, Seaman MS, Eisen HN, Chakraborty AK, Hope TJ, Ravetch JV, Wardemann H, Nussenzweig MC. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature. 2010;467:591–595. doi: 10.1038/nature09385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Koff WC, Wilson IA, Burton DR, Poignard P. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwong PD, Mascola JR, Nabel GJ. Rational Design of Vaccines to Elicit Broadly Neutralizing Antibodies to HIV-1. Cold Spring Harbor perspectives in medicine. 2011;1:a007278. doi: 10.1101/cshperspect.a007278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, Sawyer LS, Hendry RM, Dunlop N, Nara PL, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 19.Baba TW, Liska V, Hofmann-Lehmann R, Vlasak J, Xu W, Ayehunie S, Cavacini LA, Posner MR, Katinger H, Stiegler G, Bernacky BJ, Rizvi TA, Schmidt R, Hill LR, Keeling ME, Lu Y, Wright JE, Chou TC, Ruprecht RM. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat Med. 2000;6:200–206. doi: 10.1038/72309. [DOI] [PubMed] [Google Scholar]
- 20.Conley AJ, Kessler JA, 2nd, Boots LJ, McKenna PM, Schleif WA, Emini EA, Mark GE, 3rd, Katinger H, Cobb EK, Lunceford SM, Rouse SR, Murthy KK. The consequence of passive administration of an anti-human immunodeficiency virus type 1 neutralizing monoclonal antibody before challenge of chimpanzees with a primary virus isolate. J Virol. 1996;70:6751–6758. doi: 10.1128/jvi.70.10.6751-6758.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gauduin MC, Parren PW, Weir R, Barbas CF, Burton DR, Koup RA. Passive immunization with a human monoclonal antibody protects hu-PBL-SCID mice against challenge by primary isolates of HIV-1. Nat Med. 1997;3:1389–1393. doi: 10.1038/nm1297-1389. [DOI] [PubMed] [Google Scholar]
- 22.Mascola JR, Stiegler G, VanCott TC, Katinger H, Carpenter CB, Hanson CE, Beary H, Hayes D, Frankel SS, Birx DL, Lewis MG. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat Med. 2000;6:207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 23.Mascola JR, Lewis MG, Stiegler G, Harris D, VanCott TC, Hayes D, Louder MK, Brown CR, Sapan CV, Frankel SS, Lu Y, Robb ML, Katinger H, Birx DL. Protection of Macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J Virol. 1999;73:4009–4018. doi: 10.1128/jvi.73.5.4009-4018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parren PW, Ditzel HJ, Gulizia RJ, Binley JM, Barbas CF, 3rd, Burton DR, Mosier DE. Protection against HIV-1 infection in hu-PBL-SCID mice by passive immunization with a neutralizing human monoclonal antibody against the gp120 CD4-binding site. AIDS. 1995;9:F1–6. doi: 10.1097/00002030-199506000-00001. [DOI] [PubMed] [Google Scholar]
- 25.Parren PW, Marx PA, Hessell AJ, Luckay A, Harouse J, Cheng-Mayer C, Moore JP, Burton DR. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J Virol. 2001;75:8340–8347. doi: 10.1128/JVI.75.17.8340-8347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hessell AJ, Poignard P, Hunter M, Hangartner L, Tehrani DM, Bleeker WK, Parren PW, Marx PA, Burton DR. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat Med. 2009;15:951–954. doi: 10.1038/nm.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hessell AJ, Hangartner L, Hunter M, Havenith CE, Beurskens FJ, Bakker JM, Lanigan CM, Landucci G, Forthal DN, Parren PW, Marx PA, Burton DR. Fc receptor but not complement binding is important in antibody protection against HIV. Nature. 2007;449:101–104. doi: 10.1038/nature06106. [DOI] [PubMed] [Google Scholar]
- 28.Hessell AJ, Rakasz EG, Poignard P, Hangartner L, Landucci G, Forthal DN, Koff WC, Watkins DI, Burton DR. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog. 2009;5:e1000433. doi: 10.1371/journal.ppat.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haynes BF, Fleming J, St Clair EW, Katinger H, Stiegler G, Kunert R, Robinson J, Scearce RM, Plonk K, Staats HF, Ortel TL, Liao HX, Alam SM. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 2005;308:1906–1908. doi: 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- 30.Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, Mimura Y, Kunert R, Zhu P, Wormald MR, Stanfield RL, Roux KH, Kelly JW, Rudd PM, Dwek RA, Katinger H, Burton DR, Wilson IA. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 2003;300:2065–2071. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- 31.Nemazee DA, Burki K. Clonal deletion of B lymphocytes in a transgenic mouse bearing anti-MHC class I antibody genes. Nature. 1989;337:562–566. doi: 10.1038/337562a0. [DOI] [PubMed] [Google Scholar]
- 32.Barbas CF, 3rd, Collet TA, Amberg W, Roben P, Binley JM, Hoekstra D, Cababa D, Jones TM, Williamson RA, Pilkington GR, et al. Molecular profile of an antibody response to HIV-1 as probed by combinatorial libraries. J Mol Biol. 1993;230:812–823. doi: 10.1006/jmbi.1993.1203. [DOI] [PubMed] [Google Scholar]
- 33.Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore JP, Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salmon P, Trono D. Production and titration of lentiviral vectors. Current protocols in human genetics / editorial board, Jonathan L. Haines … [et al.] 2007;Chapter 12(Unit 12):10. doi: 10.1002/0471142905.hg1210s54. [DOI] [PubMed] [Google Scholar]
- 35.Li M, Gao F, Mascola JR, Stamatatos L, Polonis V, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene K, Bilska M, Kothe D, Salazar-Gonzalez J, Wei X, Decker JM, Hahn BH, Montefiori DC. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manrique A, Rusert P, Joos B, Fischer M, Kuster H, Leemann C, Niederost B, Weber R, Stiegler G, Katinger H, Günthard HF, Trkola A. In vivo and in vitro escape from neutralizing antibodies 2G12, 2F5, and 4E10. J Virol. 2007;81:8793–8808. doi: 10.1128/JVI.00598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDonald D, Vodicka MA, Lucero G, Svitkina TM, Borisy GG, Emerman M, Hope TJ. Visualization of the intracellular behavior of HIV in living cells. J Cell Biol. 2002;159:441–452. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reed L, Muench H. A simple method of estimating fifty per cent endpoints. Am J Hygiene. 1938;27:493–497. [Google Scholar]
- 39.Li Y, Svehla K, Mathy NL, Voss G, Mascola JR, Wyatt R. Characterization of antibody responses elicited by human immunodeficiency virus type 1 primary isolate trimeric and monomeric envelope glycoproteins in selected adjuvants. J Virol. 2006;80:1414–1426. doi: 10.1128/JVI.80.3.1414-1426.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng Y, McKee K, Tran K, O'Dell S, Schmidt SD, Phogat A, Forsell MN, Karlsson Hedestam GB, Mascola JR, Wyatt RT. Biochemically defined HIV-1 envelope glycoprotein variant immunogens display differential binding and neutralizing specificities to the CD4-binding site. J Biol Chem. 2012;287:5673–5686. doi: 10.1074/jbc.M111.317776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nemazee D. In: The Tolerance Workshop. Editiones “Roche” Matzinger P, Flajnik M, Nemazee D, Rammensee HG, Rolink T, Stockinger G, Nicklin L, editors. Basle, Switzerland: 1986. pp. 58–72. [Google Scholar]
- 42.Kouskoff V, Famiglietti S, Lacaud G, Lang P, Rider JE, Kay BK, Cambier JC, Nemazee D. Antigens varying in affinity for the B cell receptor induce differential B lymphocyte responses. J. Exp. Med. 1998;188:1453–1464. doi: 10.1084/jem.188.8.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thali M, Olshevsky U, Furman C, Gabuzda D, Li J, Sodroski J. Effects of changes in gp120-CD4 binding affinity on human immunodeficiency virus type 1 envelope glycoprotein function and soluble CD4 sensitivity. J Virol. 1991;65:5007–5012. doi: 10.1128/jvi.65.9.5007-5012.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Azoitei ML, Correia BE, Ban YE, Carrico C, Kalyuzhniy O, Chen L, Schroeter A, Huang PS, McLellan JS, Kwong PD, Baker D, Strong RK, Schief WR. Computation-guided backbone grafting of a discontinuous motif onto a protein scaffold. Science. 2011;334:373–376. doi: 10.1126/science.1209368. [DOI] [PubMed] [Google Scholar]
- 45.Abrahamyan LG, Mkrtchyan SR, Binley J, Lu M, Melikyan GB, Cohen FS. The cytoplasmic tail slows the folding of human immunodeficiency virus type 1 Env from a late prebundle configuration into the six-helix bundle. J Virol. 2005;79:106–115. doi: 10.1128/JVI.79.1.106-115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, Svehla K, Mathy NL, Voss G, Mascola JR, Wyatt R. Characterization of antibody responses elicited by human immunodeficiency virus type 1 primary isolate trimeric and monomeric envelope glycoproteins in selected adjuvants. J. Virol. 2006;80:1414–1426. doi: 10.1128/JVI.80.3.1414-1426.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang X, Wyatt R, Sodroski J. Improved elicitation of neutralizing antibodies against primary human immunodeficiency viruses by soluble stabilized envelope glycoprotein trimers. J Virol. 2001;75:1165–1171. doi: 10.1128/JVI.75.3.1165-1171.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Snapper CM, Kehry MR, Castle BE, Mond JJ. Multivalent, but not divalent, antigen receptor cross-linkers synergize with CD40 ligand for induction of Ig synthesis and class switching in normal murine B cells. A redefinition of the TI-2 vs T cell-dependent antigen dichotomy. J. Immunol. 1995;154:1177–1187. [PubMed] [Google Scholar]
- 49.Dintzis HM, Dintzis RZ, Vogelstein B. Molecular determinants of immunogenicity: The immunon model of immune response. Proc.Natl.Acad.Sci.USA. 1976;73:3671–3675. doi: 10.1073/pnas.73.10.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu P, Liu J, Bess J, Jr., Chertova E, Lifson JD, Grise H, Ofek GA, Taylor KA, Roux KH. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 51.Ravetch JV, Lanier LL. Immune inhibitory receptors. Science. 2000;290:84–89. doi: 10.1126/science.290.5489.84. [DOI] [PubMed] [Google Scholar]
- 52.Duong BH, Tian H, Ota T, Completo G, Han S, Vela JL, Ota M, Kubitz M, Bovin N, Paulson J, Nemazee D. Decoration of T-independent antigen with ligands for CD22 and Siglec-G can suppress immunity and induce B cell tolerance in vivo. J. Exp. Med. 2010;207:173–174. doi: 10.1084/jem.20091873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alam SM, Liao HX, Dennison SM, Jaeger F, Parks R, Anasti K, Foulger A, Donathan M, Lucas J, Verkoczy L, Nicely N, Tomaras GD, Kelsoe G, Chen B, Kepler TB, Haynes BF. Differential reactivity of germ line allelic variants of a broadly neutralizing HIV-1 antibody to a gp41 fusion intermediate conformation. J Virol. 2011;85:11725–11731. doi: 10.1128/JVI.05680-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiao X, Chen W, Feng Y, Zhu Z, Prabakaran P, Wang Y, Zhang MY, Longo NS, Dimitrov DS. Germline-like predecessors of broadly neutralizing antibodies lack measurable binding to HIV-1 envelope glycoproteins: implications for evasion of immune responses and design of vaccine immunogens. Biochem Biophys Res Commun. 2009;390:404–409. doi: 10.1016/j.bbrc.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pancera M, McLellan JS, Wu X, Zhu J, Changela A, Schmidt SD, Yang Y, Zhou T, Phogat S, Mascola JR, Kwong PD. Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1. J Virol. 2010;84:8098–8110. doi: 10.1128/JVI.00966-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huber M, Le KM, Doores KJ, Fulton Z, Stanfield RL, Wilson IA, Burton DR. Very few substitutions in a germ line antibody are required to initiate significant domain exchange. J Virol. 2010;84:10700–10707. doi: 10.1128/JVI.01111-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Verkoczy L, Chen Y, Bouton-Verville H, Zhang J, Diaz M, Hutchinson J, Ouyang YB, Alam SM, Holl TM, Hwang KK, Kelsoe G, Haynes BF. Rescue of HIV-1 broad neutralizing antibody-expressing B cells in 2F5 VH × VL knockin mice reveals multiple tolerance controls. J Immunol. 2011;187:3785–3797. doi: 10.4049/jimmunol.1101633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murray JM, Kelleher AD, Cooper DA. Timing of the components of the HIV life cycle in productively infected CD4+ T cells in a population of HIV-infected individuals. J Virol. 2011;85:10798–10805. doi: 10.1128/JVI.05095-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, Hurley A, Myung S, Boulad F, Poignard P, Burton DR, Pereyra F, Ho DD, Walker BD, Seaman MS, Bjorkman PJ, Chait BT, Nussenzweig MC. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kessler JA, McKenna PM, Emini EA, Chan CP, Patel MD, Gupta SK, Mark GE, III, Barbas CF, III, Burton DR, Conley AJ. Recombinant human monoclonal antibody IgG1b12 neutralizes diverse human immunodeficiency virus type 1 primary isolates. AIDS Res.Hum.Retroviruses. 1997;13:575–582. doi: 10.1089/aid.1997.13.575. [DOI] [PubMed] [Google Scholar]
- 61.Kunert R, Wolbank S, Stiegler G, Weik R, Katinger H. Characterization of molecular features, antigen-binding, and in vitro properties of IgG and IgM variants of 4E10, an anti-HIV type 1 neutralizing monoclonal antibody. AIDS Res.Hum.Retroviruses. 2004;20:755–762. doi: 10.1089/0889222041524571. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.