Abstract
Transcriptional effectors of white adipocyte-selective gene expression have not been described. Here we show that TLE3 is a white-selective cofactor that acts reciprocally with the brown-selective cofactor Prdm16 to specify lipid storage and thermogenic gene programs. Occupancy of TLE3 and Prdm16 on certain promoters is mutually exclusive, due to the ability of TLE3 to disrupt the physical interaction between Prdm16 and PPARγ. When expressed at elevated levels in brown fat, TLE3 counters Prdm16, suppressing brown-selective genes and inducing white-selective genes, resulting in impaired fatty acid oxidation and thermogenesis. Conversely, mice lacking TLE3 in adipose tissue show enhanced thermogenesis in inguinal white adipose depots and are protected from age-dependent deterioration of brown adipose tissue function. Our results suggest that the establishment of distinct adipocyte phenotypes with different capacities for thermogenesis and lipid storage is accomplished in part through the cell type–selective recruitment of TLE3 or Prdm16 to key adipocyte target genes.
Introduction
White and brown adipocytes are specialized for distinct functions. White adipocytes are optimized to store energy as triglycerides in large unilocular lipid droplets. In times of metabolic need, white adipocytes mobilize energy through hydrolysis of triglycerides and release of free fatty acids into the circulation (Zechner et al., 2009). White adipocytes express a battery of genes involved in lipid handling, triglyceride biosynthesis, triglyceride mobilization, and endocrine signaling (Coleman and Bell, 1980; Cook et al., 1987; Halaas et al., 1995; Kawamura et al., 1981). The importance of white adipose tissue function for systemic energy metabolism is highlighted by human and mouse models of lipodystrophy, which develop severe insulin resistance and hepatic steatosis (Chao et al., 2000; Reitman et al., 2000; Reue and Peterfy, 2000). These metabolic derangements are largely due to the inability to store lipid and the failure to secrete adipokines important for lipid and carbohydrate homeostasis, such as leptin and adiponectin (Shimomura et al., 1999; Yamauchi et al., 2001).
Brown adipocytes derive their color from their high mitochondrial content. Unlike white adipocytes, brown adipocytes store energy primarily to provide an intracellular fuel source for thermogenesis (Smith and Roberts, 1964). Brown adipose tissue is found predominantly in the interscapular region in mice and is highly vascularized and innervated (Cannon and Nedergaard, 2004; Rauch and Hayward, 1969). Although human infants have long been recognized to have significant brown adipose tissue depots, it has been debated whether adults have brown adipose tissue and whether it plays a role in thermogenesis (Lean et al., 1986). Recent studies using 2-fluorodeoxyfluoroglucose coupled with PET scanning have shown that humans have substantial brown adipose tissue in supraclavicular and paraspinal depots (Cypess et al., 2009; Mirbolooki et al., 2012; van Marken Lichtenbelt et al., 2009; Virtanen et al., 2009). During cold exposure, brown adipose tissue executes a transcriptional program that promotes energy expenditure and thermogenesis. Induction of the mitochondrial uncoupling protein, UCP1, is critical for brown fat thermogenesis (Bouillaud et al., 1985; Jacobsson et al., 1985; Matthias et al., 2000).
Lineage-tracing studies suggest that subscapular brown adipose tissue and white adipose tissue are derived from distinct precursors (Atit et al., 2006; Seale et al., 2008). However, it has recently become clear that certain white adipose tissue depots exhibit plasticity with respect to their thermogenic capacity. Mice exposed to cold, β-adrenergic agonists, or thiazolidinediones adapt by increasing the expression of UCP1, Elovl3, Dio2, Cidea, and other brown adipocyte-selective factors in “brite” or “beige” cells, a discrete subpopulation of cells found in the white adipose tissue depots (Cousin et al., 1992; Ohno et al., 2012; Young et al., 1984). Understanding the processes involved in the adaptive reprogramming of white adipocytes to brown adipocytes has the potential to provide new therapeutic strategies to combat obesity and metabolic disease.
The transcriptional determinants of the white and brown adipocyte gene programs are incompletely understood. PPARγ is the master transcriptional regulator of white and brown fat differentiation, and mice deficient in PPARγ lack both types of adipose tissue (Barak et al., 1999; Rosen et al., 1999; Tontonoz et al., 1994a; Tontonoz et al., 1994b). Seminal studies by Spiegelman and colleagues identified the transcriptional cofactor Prdm16 as a key factor driving brown adipocyte lineage development (Kajimura et al., 2008; Seale et al., 2007). Prdm16 acts in concert with PPARγ and C/EBPs to promote the expression of brown-selective factors such as UCP-1, Cidea, Elovl3, and Dio2 (Kajimura et al., 2009; Seale et al., 2008). Another cofactor, PGC-1α is also preferentially expressed in brown adipose tissue and is highly induced in response to cold exposure (Puigserver et al., 1998). PGC-1α is particularly important for UCP-1 expression and mitochondrial oxidative metabolism in brown fat (Wu et al., 1999).
Relatively little is known about factors that specify white adipocyte-selective gene expression. In particular, it has been unclear whether white fat is a “default” transcriptional program that is executed by PPARγ in the absence of Prdm16 and PGC-1α, or whether there are white fat-selective counterparts to these brown-selective cofactors. Previously, we identified the groucho family member TLE3 as an adipogenic coregulator (Villanueva et al., 2011). Here we demonstrate that TLE3 is a white-selective PPARγ cofactor that enforces an opposing transcriptional program to Prdm16. TLE3 counters the brown fat program and promotes lipid storage by blocking the interaction of Prdm16 with PPARγ, thereby reducing the occupancy of Prdm16 on brown fat-selective genes. Mice expressing elevated levels of TLE3 in brown adipose tissue exhibit a phenotypic switch from brown to white, and mice deficient in TLE3 in adipose tissue show enhanced thermogenic capacity. Our results suggest that the brown and white fat phenotypes are accomplished in part through the cell type-selective recruitment of TLE3 or Prdm16 to key adipocyte target genes.
Results
Transient Expression of TLE3 During Brown Fat Differentiation
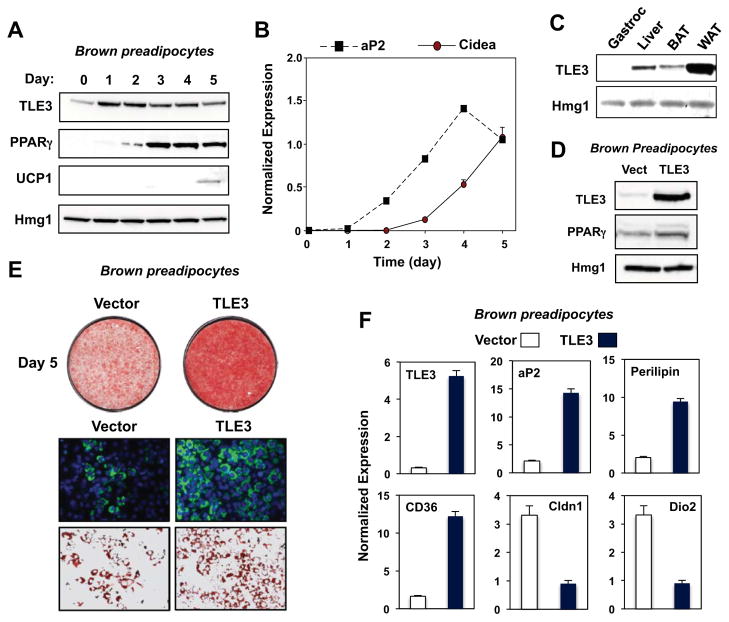
Previously, we identified TLE3 as cofactor for PPARγ that promotes the differentiation of white adipocytes (Villanueva et al., 2011). To investigate a potential role for TLE3 in brown adipose tissue gene expression, we analyzed protein expression during the differentiation of immortalized brown adipocytes. As expected, mRNA encoding PPARγ was induced during the first 2-days of differentiation, while that encoding UCP1 was induced later. TLE3 protein expression peaked on the first day of differentiation, but eventually declined as differentiation proceeded (Fig. 1A). The induction of TLE3 protein expression coincided with induction of the mRNA encoding aP2, an adipocyte marker highly induced in both white and brown fat differentiation (Fig. 1B). By contrast, mRNA encoding the brown adipocyte marker Cidea was induced later during the course of differentiation. As we had previously observed in 3T3-L1 white preadipocytes (Villanueva et al., 2011), Tle3 expression was also induced by the PPARγ agonist rosiglitazone in brown preadipocytes. Stimulators of brown fat-selective gene expression such as norepinephrine (NE), thyroid hormone (T3), bezafibrate (PPARα agonist), and (β-adrenergic agonists (CL-316,243, isoproterenol) did not alter TLE3 mRNA or protein expression (Fig. S1A, B). Interestingly, TLE3 protein expression in mice was markedly higher in white adipose tissue (WAT) than in brown adipose tissue (BAT) (Fig. 1C).
Figure 1. TLE3 modulates the differentiation program of brown preadipocytes.
A. Time course of TLE3 protein expression during the differentiation of immortalized primary brown preadipocytes determined by immunoblotting. B. Induction of aP2 precedes induction of the brown adipocyte marker Cidea in primary brown adipocytes as determined by realtime PCR. C. TLE3 protein expression in mouse tissues determined by immunoblotting. D. TLE3 protein expression in brown preadipocytes transduced with control or TLE3-expressing retroviral vectors. E. Oil red-O staining (top) and bodipy staining (bottom) of control and TLE3-expressing brown preadipocytes 5 days after induction of differentiation. F. Expression of WAT- and BAT-selective target genes in control and TLE3-expressing brown preadipocytes. Similar results were obtained as in E and F with multiple independent control and TLE3-expressing cell pools.
Forced TLE3 Expression in Brown Preadipocytes Promotes Differentiation and Lipid storage
To examine the role of TLE3 in brown fat cell differentiation, we generated stable cell pools by retroviral expression (Fig. 1D). Western blot analysis of brown preadipocytes confirmed overexpression of TLE3 protein (Fig. 1D). Forced expression of TLE3 in brown preadipocytes increased lipid storage as measured by Oil-Red-O (ORO) and BODIPY staining (Fig. 1E). Gene-expression analysis revealed that mRNAs encoding PPARγ and its downstream targets aP2, CD36, and perilipin were upregulated in TLE3-expressing cells, consistent with enhanced differentiation (Fig. 1F). Unexpectedly, however, the overall gene expression profile of TLE3-expressing brown adipocytes was more reminiscent of white adipocytes, with decreased expression of multiple brown fat-selective markers, including Dio2 and Cldn1 (Fig. 1F).
TLE3 Expression Promotes a Brown-to-White Phenotypic Switch
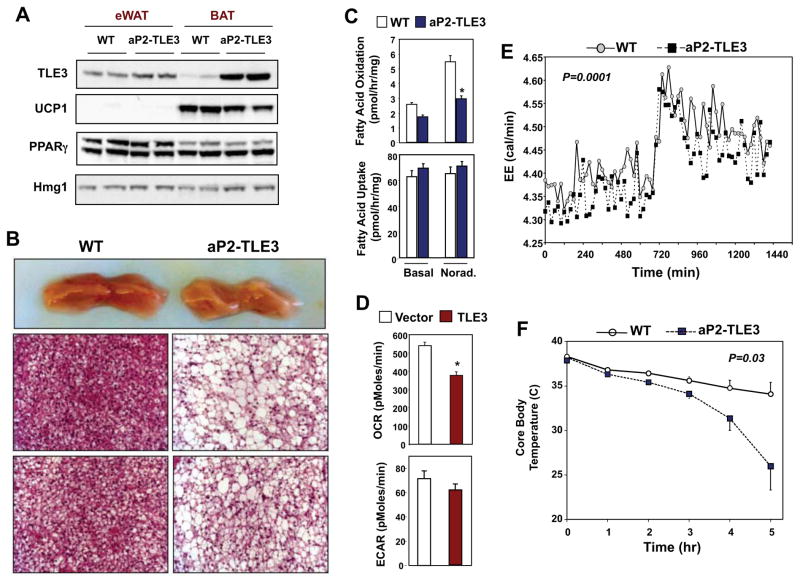
To test whether TLE3 expression might alter the brown adipose gene-expression program in vivo, we analyzed transgenic mice that selectively express TLE3 in adipose tissue (Villanueva et al., 2011). The level of TLE3 protein in the BAT of aP2-TLE3 Tg mice was substantially elevated, but comparable to the level normally observed in WAT (Fig. 2A). Interestingly, UCP-1 protein expression was correspondingly reduced in BAT from aP2-TLE3 Tg mice. Analysis of subscapular brown adipose tissue revealed a marked phenotypic change in the BAT from aP2-TLE3 Tg mice (Fig. 2B). Grossly, the tissue was paler than that from WT animals, suggesting higher lipid content (Fig. 2B). Histologically, BAT from aP2-TLE3 Tg mice showed increased numbers of cells with large unilocular lipid droplets, a feature more characteristic of WAT (Fig. 2B). We speculated that the increased lipid storage in BAT of aP2-TLE3 Tg mice might negatively impact thermogenic capacity. Indeed, BAT expressing WAT levels of TLE3 exhibited reduced rates of fatty acid oxidation, despite preserved rates of fatty acid uptake compared to controls (Fig. 2C). To determine whether cellular energetics were altered, we performed real time respirometry (using the Seahorse extracellular flux analyzer). Retroviral expression of TLE3 in brown preadipocytes led to a reduced rate of oxygen consumption and there was a trend towards a reduced rate of extracellular acidification (Fig. 2D). In accordance with these in vitro results, aP2-TLE3 Tg mice exhibited reduced energy expenditure when housed in metabolic cages (Fig. 2E). They also showed increased activity and reduced food intake (Fig. S2). To further test the functional consequences of the changes in BAT phenotype in aP2-TLE3 Tg mice, we challenged them with cold exposure at 4°C. aP2-TLE3 Tg mice had an impaired ability to respond to cold exposure compared to littermate control mice, as evidence by a marked drop in core body temperature (Fig. 2F). It is possible that the increased activity of the aP2-TLE3 mice may be a compensatory response to reduced thermogenesis.
Figure 2. Transgenic TLE3 expression alters brown adipose tissue phenotype and function.
A. Immunoblot analysis of TLE3 and UCP-1 protein expression in epididymal WAT and subscapular BAT from control and aP2-TLE3 Tg mice. B. Gross and histological analysis (H&E staining) of subscapular BAT from control and aP2-TLE3 Tg mice. C. Fatty acid uptake and oxidation in primary brown adipose tissue from control and aP2-TLE3 Tg mice was determined as described in Experimental Procedures. D. Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) from vector or TLE3-expressing brown adipocytes were determined with the Seahorse extracellular flux analyzer. *P < 0.05 by Student’s t-test. E. Energy expenditure in control and aP2-TLE3 Tg mice (N=5/group) determined by indirect calorimetry with Oxymax metabolic cages. F. Time course of core body temperature of control (WT) and aP2-TLE3 Tg mice (N=5/group) housed at 4 °C. Data are presented as mean +/− SEM. Statistical significance was analyzed by repeated measures two-way ANOVA.
Transcriptional Reprogramming of BAT by TLE3
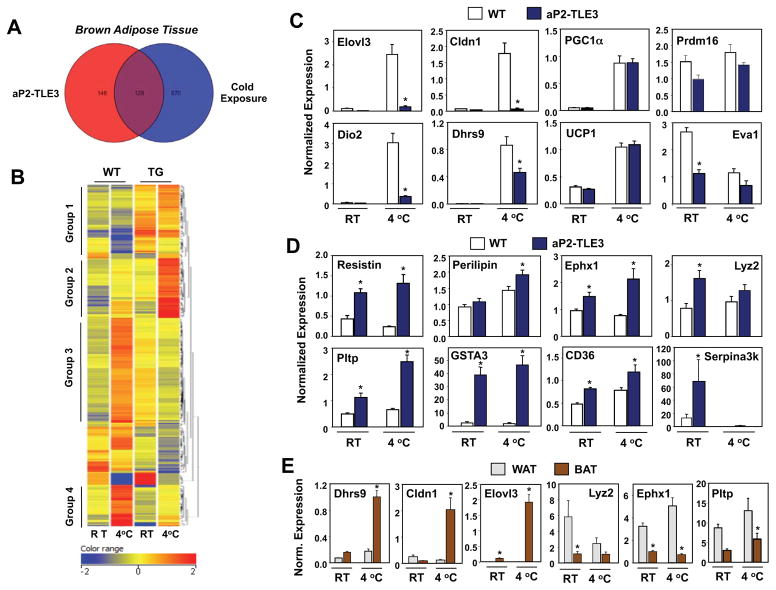
The blunted thermogenic response of aP2-TLE3 Tg mice led us to hypothesize that TLE3 might be altering the BAT transcriptional response to cold exposure. We performed transcriptional profiling using Affymetrix GeneChip Mouse Gene 1.0 ST Arrays with BAT RNA isolated from control and aP2-TLE3 Tg mice at ambient temperature or after 5 h at 4°C. Remarkably, as depicted by the Venn Diagram in Fig. 3A, 47% of the transcripts changed by more than twofold in response to TLE3 expression (aP2-TLE3 Tg versus WT) were also altered during cold exposure (WT RT versus WT 4 °C). Hierarchical clustering on entities revealed several classes of genes differentially expressed in BAT from aP2-TLE3 Tg and control mice (Fig. 3B; microarray data is available through the gene expression omnibus (GEO) on NCBI). Group 1 included genes minimally changed in WT mice in response to cold, but increased basally in aP2-TLE3 Tg mice. Group 2 included genes slightly upregulated in WT mice in response to cold in WT mice but strikingly upregulated in aP2-TLE3 Tg mice in response to cold. Groups 3 and 4 included genes upregulated by cold in WT mice that failed to be upregulated in aP2-TLE3 Tg mice. Overall, these data are consistent with a markedly altered gene-expression response to thermal challenge in aP2-TLE3 Tg mice.
Figure 3. Transcriptional reprogramming of the BAT thermogenic gene program by TLE3.
A. BAT cDNA from control and aP2-TLE3 Tg mice house at RT or 4 °C was analyzed by hybridization to Affymetrix GeneChip Mouse Gene 1.0 ST Arrays. Data was processed using GeneSpring software as described in Experimental Procedures. Venn diagram depicting overlap between genes altered greater than 2-fold by TLE3 or cold exposure. B. Heat map representation and cluster analysis of genes altered greater than 2-fold between control and aP2-TLE3 Tg BAT as determined by Affymetrix arrays. A complete list of genes in each grouping is provided in Supplemental Table 1. C. Realtime PCR analysis of the expression of genes selective for BAT in control and aP2-TLE3 Tg BAT (N=4–7/group). D. Realtime PCR analysis of the expression of genes selective for WAT in control and aP2-TLE3 Tg BAT (N=4–7/group). E. Realtime PCR analysis of gene expression in WAT and BAT from C57BL/6 mice (N=6–8/group). Data are presented as mean +/− SEM. *P < 0.05. Significance was analyzed by two-way ANOVA followed by Bonferroni post-hoc test.
Subsequent validation of these results focused primarily on brown fat-selective genes implicated in thermogenesis, many of which were represented in Groups 3 and 4. Real-time PCR showed that mRNAs encoding Elovl3 and Dio2, enzymes involved in fatty acid elongation and thyroid hormone (T3) synthesis, respectively, failed to be induced in aP2-TLE3 Tg mice in response to cold challenge (Fig. 3C). We also identified a number of genes with no previous link to thermogenesis such as Cldn1 and Dhrs9 that were highly induced during cold exposure in WT mice but showed diminished response in the setting of TLE3 expression. Notably, however, not all thermogenic genes were altered between the genotypes; WT and aP2-TLE3 Tg mice exhibited similar levels of mRNA encoding PGC-1α at room temperature and during cold exposure (Fig. 3C).
We also validated a number of genes from Group 1 whose expression was unresponsive to cold, but generally increased in aP2-TLE3 Tg mice (Fig. 3D). Interestingly, many of these genes are known to be preferentially expressed in WAT compared to BAT, including resistin, Gsta3, Lyz2, Ephx1, and Pltp (Siersbaek et al., 2012). In fact, many of the genes most differentially regulated between WT and aP2-TLE3 Tg mice are also differentially expressed in WAT and BAT (Fig. 3E and data not shown). In general, WAT-selective transcripts were upregulated in BAT of aP2-TLE3 mice, whereas BAT-selective transcripts (e.g., Elovl3, Cldn1, and Dhrs9) were reduced.
Finally, analysis of genes in Group 2 (genes strongly upregulated by TLE3 in the setting of cold exposure) revealed multiple targets of PPARα and PPARδ, such as Cyp4b1, Egr1, Cyp4a32, Cyp4a14, and Cyp4a10 (Table S1). These findings are consistent with our previous observation that TLE3 may co-activate PPARαand PPARδ as well as PPARγ (Villanueva et al., 2011). Overall, these data suggest that the expression of TLE3 in BAT affects transcriptional reprogramming towards a gene-expression profile that is more consistent with the lipid storage function of WAT rather than the thermogenic function of BAT.
Conditional Deletion of TLE3 in Adipose Tissue Improves Thermogenic Response
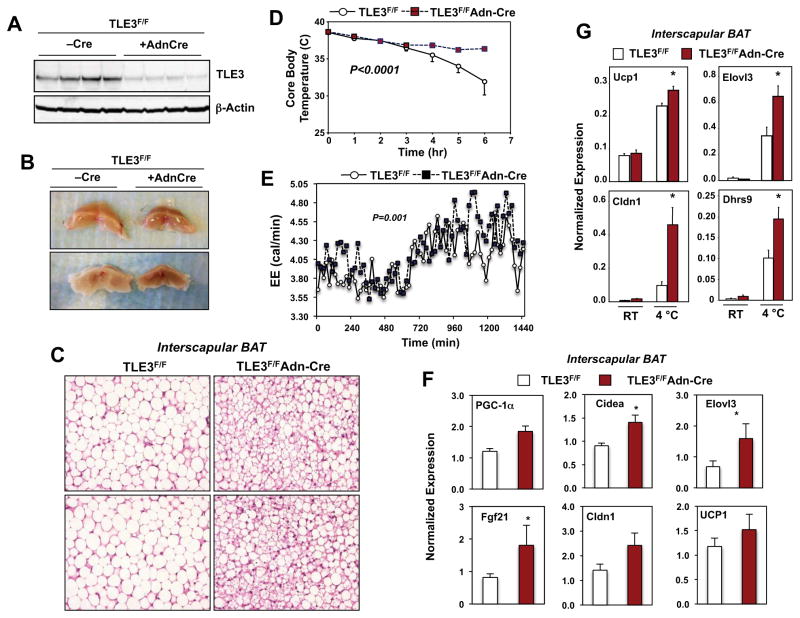
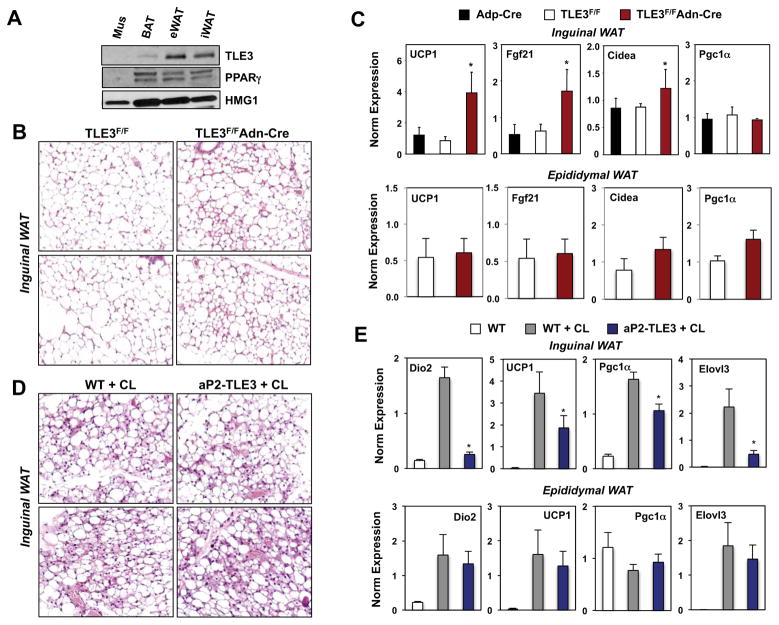
To further examine the role of TLE3 in brown adipose tissue biology, we generated adipose tissue-selective TLE3 KO (Ad-TLE3 KO) mice by crossing Tle3fl/fl mice (Fig. S3; see Experimental Procedures) with adiponectin-Cre transgenic mice (Eguchi et al., 2011). In BAT, Tle3fl/fl mice expressing adiponectin-Cre had markedly diminished levels of TLE3 protein compared to Cre-negative littermate controls (Fig. 4A). Adiponectin is expressed only in mature adipocytes, and the residual expression observed in unfractionated adipose tissue is due to TLE3 expression in stromal cells (data not shown). As mice age, the thermogenic capacity of their BAT deteriorates and the tissue acquires WAT-like characteristics, including unilocular lipid droplets and reduced mitochondrial content. To test whether deletion of TLE3 could prevent the age-dependent whitening of brown adipose tissue, we examined brown adipose tissue at 1.5 yrs of age. BAT from Ad-TLE3 KO mice was obviously browner in color compared to BAT from control animals (Fig. 4B). Furthermore, histological analysis revealed that, whereas BAT from control mice was composed primarily of cells with large unilocular lipid droplets, BAT from Ad-TLE3 KO mice contained more cells with multilocular lipid droplets characteristic of young brown adipose tissue (Fig. 4C).
Figure 4. Adipose-specific deletion of TLE3 potentiates BAT thermogenic gene expression and function.
A. Immunoblot analysis of TLE3 protein expression in whole brown adipose tissue from floxed TLE3 mice (TLE3F/F) mice in the presence or absence of an adiponectin (Adn) Cre transgene. B. Gross appearance of BAT from Cre-negative and Cre-positive TLE3F/F mice. C. H&E staining of BAT sections from Cre-negative and Cre-positive TLE3F/F mice. D. Time course of core body temperature from Cre-negative and Cre-positive TLE3F/F mice (N=4–7/group) housed at 4 °C. Statistical significance was analyzed by repeated measures two-way ANOVA. E. Energy expenditure in Cre-negative and Cre-positive TLE3F/F mice (N=6–10/group) determined by indirect calorimetry with Oxymax metabolic cages. F. Realtime PCR analysis of BAT-selective gene expression from Cre-negative and Cre-positive TLE3F/F mice (N=6–8/group). G. Realtime PCR analysis of BAT-selective gene expression in BAT from Cre-negative and Cre-positive TLE3F/F mice housed at RT or 4 °C for 6 h. N=4–7/group. *P < 0.05. Data are presented as mean +/− SEM.
These morphological changes translated to altered thermogenic capacity when mice were challenged with cold exposure at 4°C. Ad-TLE3 KO mice showed an improved thermogenic response when compared to controls, as evidenced by a better ability to maintain core body temperature during cold exposure (Fig. 4D). Consistent with these results, Ad-TLE3 KO mice showed increased energy expenditure when housed in metabolic cages (Fig. 4E). Body weight and adipose tissue mass determined by MRI were similar between groups (Fig. S4A). Although the difference did not reach statistical significance, there was a trend towards decreased activity in Ad-TLE3 KO mice (Fig. S4B). Food intake was not different between control and Ad-TLE3 KO mice in these metabolic cage studies (Fig. S4C).
To determine the transcriptional basis for this improved BAT function we analyzed gene expression. Ad-TLE3 KO mice exhibited increased basal expression of multiple genes characteristic of BAT, including Cidea, Elovl3, Cldn1 and FGF-21 (Fig. 4F). Furthermore, the expression of cold-responsive genes, including UCP-1, Elovl3, Dhrs9, and Cldn1 was enhanced.
Recent studies have reported that mice exposed to cold, β-adrenergic agonists, or thiozolidinediones, induce the expression of UCP-1, Elovl3, Dio2, Cidea, and other BAT-selective proteins in a subpopulation of cells present in inguinal WAT depots termed “brite” or “beige” cells (Cousin et al., 1992; Ohno et al., 2012; Wu et al., 2012; Young et al., 1984). Given the suppressive effect of TLE3 on BAT-selective gene expression, we hypothesized that TLE3 deficiency might affect BAT-selective gene expression in WAT depots. Interestingly, the expression of TLE3 protein was higher in epididymal fat compared to inguinal fat (Fig. 5A). Furthermore, inguinal adipose depots of Ad-TLE3 KO mice showed evidence of browning by histology, with increased numbers of cells with multilocular lipid droplets characteristic of thermogenic beige adipocytes (Fig. 5B). Furthermore, the expression of UCP1, FGF-21, and Cidea was elevated in inguinal WAT compared to littermate controls (Fig. 5C). However, we observed no induction of these genes in epididymal WAT from Ad-TLE3 KO mice, indicating that the consequences of TLE3 loss are depot-specific.
Figure 5. TLE3 modulates the induction of the thermogenic gene program in inguinal WAT.
A. Immunoblot analysis of TLE3 protein levels in various adipose depots from WT C57Bl/6 mice. B. H&E staining of representative inguinal WAT sections from Cre-negative and Cre-positive TLE3F/F mice. C. Realtime PCR analysis of gene expression in inguinal and epididymal WAT from control and Adn-Cre/TLE3F/F mice fed high-fat diet for 13 wks. N=7–11. D. H&E staining of representative inguinal WAT sections from aP2-TLE Tg and littermate controls (N=3–4) treated for 3 days with 1 mg/kg/day CL-316,243 (CL). E. Real-time PCR analysis of gene expression from inguinal WAT from aP2-TLE Tg and littermate controls treated for 3 days with 10 μM CL-316,243. *P < 0.05. Data are presented as mean +/− SEM (N=3).
We also analyzed the effect of transgenic TLE3 expression on the histology and gene expression of inguinal WAT. Since WT C57Bl/6 mice contain few beige cells at baseline, we treated aP2-TLE3 Tg and littermate control mice with the β3-adrenergic agonist CL-316,243 (CL) to induce browning(Seale et al., 2011). As expected, CL treatment had a striking effect on inguinal WAT histology (Fig. 5D). We did not appreciate a consistent morphological difference between control and aP2-TLE3 Tg mice (Fig. 5D); however, the gene expression profile of the adipocytes in this depot was clearly different, despite the comparable morphology. The expression of genes linked to thermogenesis, including Dio2, UCP-1, Pgc1a and Elovl3, was decreased in aP2-TLE3 Tg mice compared to controls (Fig. 5E). As expected the degree of induction of brown adipocyte genes by CL was lower in epididymal fat compared to inguinal fat (Fig. 5E). We did not observe a significant effect of transgenic TLE3 expression on the induction of these genes in epididymal fat. Collectively, the observations that similar target genes are dysregulated in both aP2-TLE3 Tg mice and Ad-TLE3 KO mice, and that these changes correlate with altered thermogenesis, implicate TLE3 as a physiological modulator of thermogenic gene expression.
Reciprocal Roles for Prdm16 and TLE3 in WAT- and BAT-Selective Gene Expression
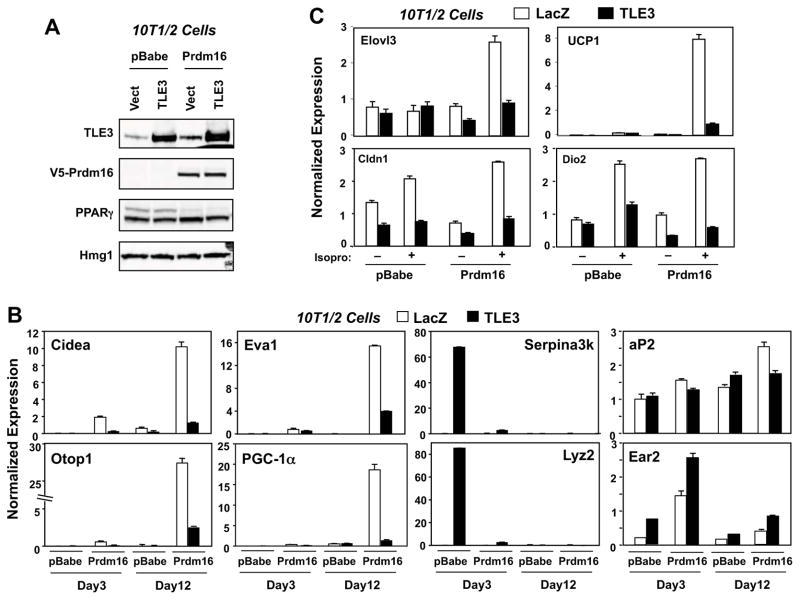
Prdm16 is a transcriptional coregulator that drives the brown adipocyte program (Seale et al., 2007). Transgenic overexpression of Prdm16 leads to the browning of white adipocytes (Seale et al., 2011). Interestingly, many of the genes whose expression was most strongly affected by gain or loss of TLE3 expression in BAT in our studies (e.g., UCP1, Elovl3, Eva1, and Dio2) were previously reported to be responsive to Prdm16 (Seale et al., 2007). We therefore hypothesized that Prdm16 and TLE3 might be mechanistically linked through differential interactions with PPARγ on the promoters of common adipose target genes. To test this hypothesis we employed viral vectors to express TLE3 and/or Prdm16 in 10T1/2 preadipocytes, differentiated the cells with a brown fat-permissive cocktail, and analyzed the impact on differentiation-dependent gene expression (Fig. 6A). Expression of TLE3 alone induced the level of representative WAT-selective transcripts including Serpina3K and Lyz2, whereas expression of Prdm16 alone had no effect on these transcripts (Fig. 6B). Expression of these genes was prominent on Day 3 of differentiation, but declined by the time they were differentiated on Day12. Coexpression of Prdm16 blunted the induction of WAT-selective genes by TLE3. By contrast, expression of Prdm16, but not TLE3, potently induced the expression of BAT genes, including mRNAs encoding Cidea, Otop-1, Eva1 and PGC-1α, consistent with prior reports (Seale et al., 2011). Furthermore, co-expression of TLE3 antagonized the Prdm16-mediated induction of the brown fat program at both Day 3 and Day 12 of differentiation (Fig. 6B). In addition, TLE3 expression inhibited the ability of the β-adrenergic agonist isoproterenol to induce Elovl3, Dio2, Cldn1, and Ucp1 expression in fully differentiated Prdm16-expressing cells (Fig. 6C). Other PPAR-responsive transcripts, such as the mRNAs encoding aP2 and Ear2, did not show reciprocal regulation by TLE3 and Prdm16, emphasizing the gene-specific nature of the crosstalk between these factors (Fig. 6C). Together with the results of Figs 2 and 3, our data indicate that these two PPARγ cofactors, TLE3 and Prdm16, promote distinct adipocyte gene programs in vitro and in vivo.
Figure 6. TLE3 and Prdm16 reciprocally regulate the expression of BAT and WAT-selective gene expression.
A. Immunoblot analysis of TLE3 and Prdm16 expression in virally transduced 10T1/2-CAR cells. Stable pools of vector or Prdm16-expressing cells were differentiated with BAT-induction cocktail for 9 days and then transduced with control or TLE3 adenoviral vectors for 3 days. Lysates were collected on Day 12. B. Realtime PCR analysis of BAT- and WAT-selective gene expression in virally-transduced 10T1/2-CAR cells on Day 3 and Day 12 after induction with BAT differentiation cocktail. RNA isolated 3-days after transduction with control or TLE3 adenovirus. C. Realtime PCR analysis of cold-inducible genes in virally-transduced 10T1/2-CAR cells on Day 10 after induction with BAT differentiation cocktail. Cells were also treated with vehicle or 5 μM isoproterenol for 4 h. RNA was isolated 3-days after adenoviral infection.
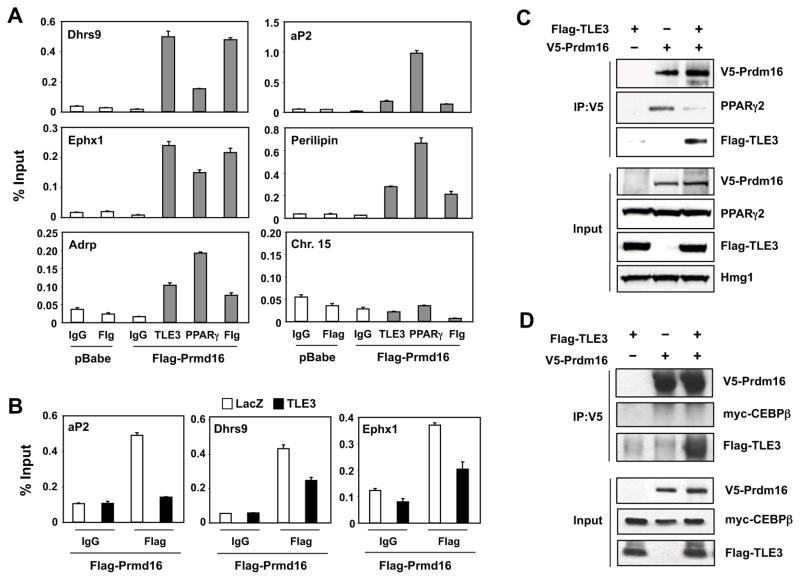
To determine whether the reciprocal effects of TLE3 and Prdm16 on adipocyte genes are due to their interactions with adipocyte gene promoters, we analyzed the occupancy of TLE3, PPARγ, and Flag-tagged Prdm16 on the regulatory regions of BAT- and WAT-selective genes. ChIP qPCR revealed that TLE3, Prdm16, and PPARγ occupied the promoters of Dhrs9, Cldn1, Ephx1, aP2, perilipin, Adrp, Agpat2, and Acsl1 in differentiated 10T1/2 cells (Fig. 7A and data not shown). We hypothesized that the differential effects of TLE3 and Prdm16 on the expression of certain adipocyte genes might be due to their mutually-exclusive occupancy at PPARγ binding sites. In support of this idea, we found that expression of TLE3 reduced the occupancy of Prdm16 on the aP2, Dhrs9, and Ephx1 promoters (Fig. 7B). Since Prdm16 interacts directly with PPARγ and because TLE3 is also present in PPARγ-containing complexes (Seale et al., 2008; Villanueva et al., 2011), we hypothesized that the interaction of Prdm16 and TLE3 with PPARγ might be mutually exclusive. To test this idea, we immunoprecipitated V5-Prdm16 from cells in the presence or absence of TLE3 and then immunoblotted for PPARγ. Remarkably, TLE3 expression disrupted the interaction of PPARγ with Prdm16 (Fig. 7C). This was most likely due to competitive physical interaction of Prdm16 with TLE3, as Prdm16 and TLE3 coimmunoprecipated (Fig. 7C). Prdm16 has also been reported to interact with C/EBPβ to regulate BAT gene expression(Kajimura et al., 2009). In our system we observed weak interaction between Prdm16 with C/EBPβ and this was not affected by the presence of TLE3 (Fig. 7D). Together, our results demonstrate that direct mechanistic interactions between TLE3 and Prdm16 on adipocyte promoters dictate differential adipocyte gene expression programs.
Figure 7. TLE3 binds to Prdm16 and competes for its interaction with PPARγ.<.
br>A. TLE3 and Prdm16 localize to common target genes. ChIP assays were performed on 10T1/2-CAR cells stably transduced with Flag-tagged Prdm16 as described in Experimental Procedures. Cells were crosslinked on Day 4 after induction with BAT differentiation cocktail, and proteins precipitated with control IgG or antibodies to Flag, endogenous PPARγ, and endogenous TLE3 as indicated. A non-specific region of chromosome 15 was used as a negative control for binding. Promoter occupancy was determined by realtime PCR. B. Expression of TLE3 displaces Prdm16 from BAT- and WAT-selective gene promoters. Stable 10T1/2-CAR cells expressing vector or Flag-Prdm16 were differentiated for 7 days with BAT induction cocktail and then transduced with control or TLE3-expressing adenoviral vectors for 3-days. ChIP assays were performed using control IgG or an antibody to Flag as in A. C. 293 cells were transfected with V5-Prdm16, Flag-PPARγ2 and/or Flag-TLE3. Lysates were immunoprecipitated with anti-V5 antibody, and the precipitates were analyzed by immunoblotting. Similar results were obtained in multiple independent experiments. D. 293 cells were transfected with V5-Prdm16, myc-C/EBPβ and/or Flag-TLE3. Lysates were immunoprecipitated with anti-V5 antibody, and the precipitates were analyzed by immunoblotting. Similar results were obtained in multiple independent experiments.
Discussion
White and brown adipocytes have shared but distinct transcriptional programs that facilitate long-term lipid storage and thermogenesis, respectively. The transcriptional determinants that specify white versus brown fat-selective gene expression are incompletely understood. We have shown here that adipose subtype-selective gene expression is accomplished in part through differential recruitment of the PPARγ cofactors TLE3 and Prdm16. TLE3, which is preferentially expressed in white fat, inhibits the expression of brown-selective Prdm16 target genes while promoting the expression of white-selective genes. Expression of TLE3 in vivo favors the “whitening” of brown adipose tissue and the suppression of adaptive thermogenesis, whereas the loss of TLE3 leads to the “browning” of inguinal WAT. The interaction of Prdm16 with TLE3 and PPARγ is mutually exclusive and TLE3 antagonizes thermogenic gene expression by inhibiting the co-occupancy of Prdm16 and PPARγ on adipocyte promoters. Although PPARγ is well known to interact with a variety of cofactors, to our knowledge this is the first study to directly correlate differential promoter occupancy of PPARγ coactivators with cell type-selective gene expression.
Although the vast majority of adipocyte genes are expressed at similar levels in brown and white adipocytes, a subset is preferentially expressed in one adipocyte type or the other. PPARγ drives a transcriptional cascade common to both white and brown adipocytes, regulating the expression of shared markers of the differentiated phenotype, for example aP2, CD36, Plin, and adiponectin (Tontonoz and Spiegelman, 2008). But PPARγ also promotes the expression of brown-selective markers such as UCP1 and Elovl3, and white-selective markers such as Ephx1. The mechanistic basis for the tissue-selective actions of PPARγ on brown and white adipocyte promoters is still unclear.
Several auxiliary factors have been identified that work in concert with PPARγ to facilitate brown fat-selective gene expression. The first to be characterized was PGC-1α, a cold-inducible coactivator of PPARγ that drives the expression of UCP1 in brown adipose tissue (Puigserver et al., 1998). PGC-1α promotes the expression of cold-inducible genes and those linked to mitochondrial oxidative metabolism, but has little effect on the expression of a number of other BAT-selective genes (Uldry et al., 2006). In addition to promoting adaptive thermogenesis, PGC-1α also plays important roles in oxidative metabolism and mitochondrial biogenesis in heart and skeletal muscle (Lehman et al., 2000; Wu et al., 1999). More recently, Prdm16 was identified as a critical factor that drives the entire brown fat-selective transcriptional program through direct interaction with PPARγ (Seale et al., 2008). Prdm16 is highly enriched in BAT and its forced expression in white preadipocytes or myoblasts promotes a lineage switch to brown fat (Seale et al., 2008; Seale et al., 2007). Mice overexpressing Prdm16 in adipose tissue show increased expression of BAT-selective transcripts in certain white adipose tissue depots, such as inguinal WAT (Seale et al., 2011), and Prdm16 also suppresses the expression of certain WAT-selective transcripts (Kajimura et al., 2008).
By comparison, relatively little is known about factors that specify white adipocyte-selective gene expression. It has been unclear whether white fat is simply a “default” transcriptional program that is executed by PPARγ in the absence of Prdm16 and PGC-1α, or whether there are white fat-selective counterparts to these brown-selective cofactors. Our data suggest that TLE3 is one such factor, enforcing an opposing transcriptional program to Prdm16.
We initially identified TLE3 as an adipogenic coregulator in a high-throughput cDNA screen (Villanueva et al., 2011). TLE3 is induced early in the course of adipocyte differentiation and its expression is directly regulated by PPARγ. TLE3 and PPARγ act synergistically to drive adipocyte gene expression and differentiation, and colocalize on PPAR response elements within adipogenic promoters. PPARγ likely employs some common cofactors to promote gene expression common to both WAT and BAT, perhaps including p300, SRCs and mediator complex, and we initially suspected that TLE3 would promote the expression of all PPARγ target genes equally. Unexpectedly, however, we found that TLE3 exerts differential effects on BAT- and WAT-selective gene expression. Brown adipose tissue from aP2-TLE3 Tg mice showed histological features of white fat and impaired adaptive thermogenesis, and these changes correlated with an impaired capacity to induce thermogenic gene expression during cold challenge. Conversely, mice lacking TLE3 showed enhanced expression of thermogenic genes in both WAT and BAT and improved cold-tolerance. These findings suggest that TLE3 plays a specific role in helping to define the differential brown, white and perhaps beige adipocyte gene programs.
During brown preadipocyte differentiation, the induction of genes involved in lipid biosynthesis and lipid droplet formation occurs prior to the induction of most BAT-selective transcripts such as UCP-1. TLE3 is also induced early during the course of brown adipocyte differentiation, suggesting that TLE3 may be help to establish the lipid-storage and lipid-synthesis capacity common to all adipocytes, and may prevent the induction in brown fat transcripts during early differentiation. During the later stages of brown preadipocyte differentiation, TLE3 expression declines, allowing the brown fat program to proceed. This observation is consistent with the relatively low level of expression of TLE3 in mature BAT compared to mature WAT.
Together with earlier work (Kajimura et al., 2008), our data suggest that the mechanistic basis for the differential actions of PPARγ in the WAT and BAT gene expression programs involves the formation of cell-type selective transcriptional complexes on the promoters of key adipocyte genes. Prdm16 binds directly to PPARγ, and studies have established that the ability of Prdm16 to interact with PPARγ and C/EBPβ is critical for its ability to drive BAT development (Kajimura et al., 2009; Seale et al., 2008). TLE3 is also present in PPARγ-containing transcriptional complexes and can be localized to PPREs by ChIP, although the two proteins do not appear to interact directly (Villanueva et al., 2011). We have shown that the choice of PPARγ to form complexes with Prdm16 and TLE3 is mutually exclusive. Prdm16 binds to TLE3, and TLE3 competes for the interaction between Prdm16 and PPARγ. Elevated TLE3 expression antagonizes thermogenic gene expression by competing for the occupancy of Prdm16 on adipocyte promoters. Conversely, the loss of TLE3 expression in BAT provides a more permissive context for the actions of Prdm16. It is likely that in cell types where both TLE3 and Prdm16 are present, such as beige adipocytes in WAT depots, PPARγ-TLE3 and PPARγ-Prdm16 complexes exist in equilibrium, and the precise balance between these complexes may help to determine the level of activation for particular promoters. The Prdm16-C/EBP interaction, which does not appear to be affected by TLE3, is also likely to be an important determinant in the activation of certain adipocyte genes.
Overall, these studies highlight an intricate balance between overlapping transcriptional programs that specifies adipose cell-type specific gene expression. Ultimately, a better understanding of the mechanisms by which PPARγ controls the lipid storage and thermogenic gene programs may aid in the development of pharmacologic modulators of adipocyte phenotype. Optimization of PPARγ agonists for the their ability to affect the balance between TLE3 and Prdm16 recruitment may facilitate the therapeutic manipulation of thermogenic gene expression and energy expenditure in humans.
EXPERIMENTAL PROCEDURES
Cell Culture
10T1/2 cells and brown preadipocytes were plated in DMEM containing 10% FBS, 20 nM insulin, and 1 nM T3. After confluence cells were stimulated to differentiate with DMEM, 10% FBS, 20 nM insulin, 1 nM T3, 0.5 mM isobutylmethylxanthine, 0.5 μM dexamethasone, and 0.125 mM indomethacin for 2 days, followed by DMEM containing 10% FBS, 20 nM insulin, and 1 nM T3. Brown preadipocytes were isolated as previously described (Rodriguez-Cuenca et al., 2007); cells were immortalized by retroviral expression of the SV40 Large T-antigen using a hygromycin selection marker. Gain of function studies for 10T1/2 cells or brown preadipocytes were completed using the pBabe expression system for TLE3 (puromycin), V5-Prdm16 (puromycin) or Flag-Prdm16 (puromycin). Acute overexpression studies were completed using adenoviral expression system using pAd gateway destination vectors (Invitrogen) and adenovirus was amplified using 293 cells. Adenovirus was purified by CsCl gradients and titered for potency (Viraquest Inc., North Liberty, IA). Cells were infected using an MOI of 50.
Gene Expression and Microarray Analysis
Total RNA was isolated using Trizol reagents (Invitrogen) and reverse transcribed using iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). Gene expression was quantified by real-time PCR using SYBR Green (Diagenode, Denville, NJ) and an ABI 7900 instrument. Primer pairs were designed using the Universal Probe Library (Roche). A standard curve was generated for each primer pair to quantify gene expression and a list of primer pairs is included in Table S1. For microarray experiments of brown adipose tissue, RNA was pooled from four separate mice and processed by the UCLA microarray core facility using Gene-Chip Mouse Gene 1.0 ST Arrays (Affymetrix, Santa Clara, CA). The results were analyzed using Genespring GX (Agilent, Santa Clara).
Protein Analysis
Cells were homogenized using RIPA buffer plus proteinase inhibitor and sonicated for 10 s using Bioruptor. Brown adipose tissue was homogenized using RIPA buffer plus proteinase inhibitor using dounce (Perkin Elmer) and placed on nutator for 30 min. Cell or tissue homogenate was spun at 12,000g for 10 min at 4 C, supernatant was saved and protein was quantified using a BCA protein assay (Piercenet). Proteins were diluted using Nupage loading dye and heated at 70 C for 20 min and run on 4–12% Bis-Tris Nupage Gels (Invitrogen). Proteins were transferred to hybond ECL(GE Healthcare) and blotted using TLE3 antibody (Proteintech), Hmg1 (Abcam), UCP1 (Abcam), PPARγ (Cell Signaling), or V5 antibody (Invitrogen).
Coimmunoprecipitation
293 cells were transfected using BioT (Bioland Scientific) transfection reagent in 10 cm dish when cells were 80% confluent. Cells were transfected with expression plasmids of V5-Prdm16, Flag-TLE3, Flag-PPARg2 or Myc-CEBPβ (gift from Stephen R. Farmer) as indicated in the legend. 2-days post transfection cells were lysed with RIPA buffer plus proteinase inhibitor (Roche) as described above. Lysate was incubated with V5 (Abcam) followed by protein A beads. Lysate was allowed to incubate overnight at 4C. Agarose beads were washed with RIPA buffer and with PBS for the final wash. Proteins were eluted with Nupage loading dye (Invitrogen) and were immunoblotted using M2 Flag (Sigma Aldrich), V5(Invitrogen), or Myc (Covance Research Products) antibodies as described above.
Chromatin Immunoprecipitation
ChIP studies were completed as described previously (Villanueva et al., 2011). Briefly, 10T1/2 cells were washed with PBS and fixed with 1% formaldehyde for 10 min. Lysed cells were sonicated with Bioruptor using 30 cycles of 30 s ON/OFF. Chromatin was spun at 12,000 g for 10 min and supernatant was precleared with ProteinA or ProteinG agarose beads. A portion of precleared chromatin was saved for input or incubated with TLE3 (Protein Tech), PPARγ (Santa Cruz) or Flag (Sigma Aldrich) antibodies at 4C overnight. ProteinA agarose beads were incubate for 4 h, spun and washed. Crosslinking was reversed and DNA was purified using columns (Qiagen). DNA enrichment was quantified by real-time PCR using SybrGreen (Diagenode) and ABI 7900. Genomic regions to be tested were determined by mining genomewide PPARγ ChIP seq. experiments available in GEO Datasets (NCBI). Primers were designed using the Universal Probe Library (Roche) and are listed in Table S1. Relative occupancy was quantified by using a standard curve and normalizing samples to input.
Animal Studies
Transgenic mice (C57BL6J inbred strain) expressing TLE3 from the −5.4kb aP2 enhancer have been described (Villanueva et al., 2011). For cold exposure experiments, control littermates and aP2-TLE3 Tg mice were housed individually without food and bedding immediately before the start of experiments. Mice were allowed free access to water and placed at 4 °C while core body temperature was monitored every hour using a rectal probe. A conditional knock-out allele for Tle3 was generated using a sequence-replacement “targeted trapping/conditional-ready” gene-targeting vector. The targeting vector was electroporated into strain 129/OlaHsd ES cells and G418-resistant targeted clones identified by long-range PCR at both the 5′ and 3′ ends using primers 5′-1033F (tgt gtc tgt gtg gga tga act ttcc) and 5′-4432R (acc gta atg gga tag gtt acg ttg g) for the 5′ end, and primers 3′-71F (gtg gag agg cta ttc ggc tat gact) and 3′-1370R (atc cac tct tcc tga ctg tgg ctt c) for the 3′ end. Nineteen targeted clones (out of 168 G418-resistant clones screened) were identified. Two targeted ES cell clones (#38, #50) were injected into C57BL/6 blastocysts to generate chimeric mice. High-percentage male chimeras were obtained, and resulting chimeras were crossed to C57BL/6J mice to establish germline transmission. Heterozygous mice carrying the conditional-ready knockout allele were mated with mice carrying a Flp recombinase deleter transgene. Flp recombinase excised the gene-trapping cassette within intron 2 of Tle3, producing a conditional knockout allele containing a lox71 site in intron 2 and a loxP site in intron 3. Genotyping was completed by amplification of genomic region flanking loxp sites using the following primer pairs, forward gct ccc ttc ttc agc ttc ct and reverse, gct cca aga ggg att ttt at. TLE3F/F mice were crossed with adiponectin-Cre mice obtained from Evan Rosen. Mice were challenged with cold exposure as described above. Energy expenditure, food intake, and ambulatory activity were determined by using Oxymax Lab Animal Monitoring System: CLAMS (Columbus Instruments) as described in Villanueva et al, 2011. Mice were treated with 1 mg/kg/day of CL-316,243 for 3-days.
Cellular Metabolism
For determination of fatty acid oxidation and uptake, interscapular BAT was excised and immediately placed in warm serum free DMEM containing 1% fatty-acid free BSA and 0.5μM palmitate, with or without nor-adrenaline (10μM). Tissues were allowed to incubate for 1hr at 37 degrees before the addition of 1 μCi/mL of 14C-palmitate (Perkin Elmer) followed by 2 h incubation at 37 degrees. Tissue and media were analyzed for fatty acid oxidation and uptake as previously described (Ribas et al., 2011). Cellular metabolic rates were measured using a XF24 Analyzer (Seahorse Bioscience). Immediately before the measurement, cells were washed with unbuffered DMEM as described (Wu et al., 2007). Plates were placed into the XF24 instrument for measurement of oxygen consumption (OCR) and extracellular acidification (ECAR) rates. Mixing, waiting and measurement times were 4, 2, and 2 min, respectively.
Supplementary Material
Physical activity (A) and food intake (B) in Cre-negative and Cre-positive TLE3F/F mice (N=6–10/group) determined by housing in Oxymax metabolic cages.
Research Highlights.
TLE3 is a transcriptional cofactor that activates white fat-selective gene expression
TLE3 and Prdm16 reciprocally regulate thermogenic gene expression
Recruitment of TLE3 and Prdm16 to adipocyte promoters is mutually exclusive
Modulation of TLE3 expression alters thermogenesis in white and brown fat
Acknowledgments
We thank members of our laboratories for discussions and technical support. P.T. is an investigator of the Howard Hughes Medical Institute. This work was supported by NIH grants HL-090553, S10RR026744 and DK063491 and by a UCLA Jonsson Comprehensive Cancer Center Fellowship.
Footnotes
The authors have no financial interests related to this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Atit R, Sgaier SK, Mohamed OA, Taketo MM, Dufort D, Joyner AL, Niswander L, Conlon RA. Beta-catenin activation is necessary and sufficient to specify the dorsal dermal fate in the mouse. Dev Biol. 2006;296:164–176. doi: 10.1016/j.ydbio.2006.04.449. [DOI] [PubMed] [Google Scholar]
- Barak Y, Nelson MC, Ong ES, Jones YZ, Ruiz-Lozano P, Chien KR, Koder A, Evans RM. PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol Cell. 1999;4:585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- Bouillaud F, Ricquier D, Thibault J, Weissenbach J. Molecular approach to thermogenesis in brown adipose tissue: cDNA cloning of the mitochondrial uncoupling protein. Proc Natl Acad Sci U S A. 1985;82:445–448. doi: 10.1073/pnas.82.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- Chao L, Marcus-Samuels B, Mason MM, Moitra J, Vinson C, Arioglu E, Gavrilova O, Reitman ML. Adipose tissue is required for the antidiabetic, but not for the hypolipidemic, effect of thiazolidinediones. J Clin Invest. 2000;106:1221–1228. doi: 10.1172/JCI11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman RA, Bell RM. Selective changes in enzymes of the sn-glycerol 3-phosphate and dihydroxyacetone-phosphate pathways of triacylglycerol biosynthesis during differentiation of 3T3-L1 preadipocytes. J Biol Chem. 1980;255:7681–7687. [PubMed] [Google Scholar]
- Cook KS, Min HY, Johnson D, Chaplinsky RJ, Flier JS, Hunt CR, Spiegelman BM. Adipsin: a circulating serine protease homolog secreted by adipose tissue and sciatic nerve. Science. 1987;237:402–405. doi: 10.1126/science.3299705. [DOI] [PubMed] [Google Scholar]
- Cousin B, Cinti S, Morroni M, Raimbault S, Ricquier D, Penicaud L, Casteilla L. Occurrence of brown adipocytes in rat white adipose tissue: molecular and morphological characterization. J Cell Sci. 1992;103 ( Pt 4):931–942. doi: 10.1242/jcs.103.4.931. [DOI] [PubMed] [Google Scholar]
- Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng YH, Doria A, Kolodny GM, Kahn CR. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi J, Wang X, Yu S, Kershaw EE, Chiu PC, Dushay J, Estall JL, Klein U, Maratos-Flier E, Rosen ED. Transcriptional control of adipose lipid handling by IRF4. Cell Metab. 2011;13:249–259. doi: 10.1016/j.cmet.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halaas JL, Gajiwala KS, Maffei M, Cohen SL, Chait BT, Rabinowitz D, Lallone RL, Burley SK, Friedman JM. Weight-reducing effects of the plasma protein encoded by the obese gene. Science. 1995;269:543–546. doi: 10.1126/science.7624777. [DOI] [PubMed] [Google Scholar]
- Jacobsson A, Stadler U, Glotzer MA, Kozak LP. Mitochondrial uncoupling protein from mouse brown fat. Molecular cloning, genetic mapping, and mRNA expression. J Biol Chem. 1985;260:16250–16254. [PubMed] [Google Scholar]
- Kajimura S, Seale P, Kubota K, Lunsford E, Frangioni JV, Gygi SP, Spiegelman BM. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. 2009;460:1154–1158. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S, Seale P, Tomaru T, Erdjument-Bromage H, Cooper MP, Ruas JL, Chin S, Tempst P, Lazar MA, Spiegelman BM. Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes Dev. 2008;22:1397–1409. doi: 10.1101/gad.1666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura M, Jensen DF, Wancewicz EV, Joy LL, Khoo JC, Steinberg D. Hormone-sensitive lipase in differentiated 3T3-L1 cells and its activation by cyclic AMP-dependent protein kinase. Proc Natl Acad Sci U S A. 1981;78:732–736. doi: 10.1073/pnas.78.2.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lean ME, James WP, Jennings G, Trayhurn P. Brown adipose tissue uncoupling protein content in human infants, children and adults. Clin Sci (Lond) 1986;71:291–297. doi: 10.1042/cs0710291. [DOI] [PubMed] [Google Scholar]
- Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J Clin Invest. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthias A, Ohlson KB, Fredriksson JM, Jacobsson A, Nedergaard J, Cannon B. Thermogenic responses in brown fat cells are fully UCP1-dependent. UCP2 or UCP3 do not substitute for UCP1 in adrenergically or fatty scid-induced thermogenesis. J Biol Chem. 2000;275:25073–25081. doi: 10.1074/jbc.M000547200. [DOI] [PubMed] [Google Scholar]
- Mirbolooki MR, Constantinescu CC, Pan ML, Mukherjee J. Quantitative assessment of brown adipose tissue metabolic activity and volume using 18F-FDG PET/CT and Beta3-adrenergic receptor activation. EJNMMI Res. 2012;1:1–11. doi: 10.1186/2191-219X-1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno H, Shinoda K, Spiegelman BM, Kajimura S. PPARgamma agonists induce a white-to-brown fat conversion through stabilization of PRDM16 protein. Cell Metab. 15:395–404. doi: 10.1016/j.cmet.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno H, Shinoda K, Spiegelman BM, Kajimura S. PPARgamma agonists induce a white-to-brown fat conversion through stabilization of PRDM16 protein. Cell Metab. 2012;15:395–404. doi: 10.1016/j.cmet.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Rauch JC, Hayward JS. Topography and vascularization of brown fat in a small nonhibernator (deer mouse, Peromyscus maniculatus) Can J Zool. 1969;47:1301–1314. doi: 10.1139/z69-203. [DOI] [PubMed] [Google Scholar]
- Reitman ML, Arioglu E, Gavrilova O, Taylor SI. Lipoatrophy revisited. Trends Endocrinol Metab. 2000;11:410–416. doi: 10.1016/s1043-2760(00)00309-x. [DOI] [PubMed] [Google Scholar]
- Reue K, Peterfy M. Mouse models of lipodystrophy. Curr Atheroscler Rep. 2000;2:390–396. doi: 10.1007/s11883-000-0077-1. [DOI] [PubMed] [Google Scholar]
- Ribas V, Drew BG, Le JA, Soleymani T, Daraei P, Sitz D, Mohammad L, Henstridge DC, Febbraio MA, Hewitt SC, Korach KS, Bensinger SJ, Hevener AL. Myeloid-specific estrogen receptor alpha deficiency impairs metabolic homeostasis and accelerates atherosclerotic lesion development. Proc Natl Acad Sci U S A. 2011;108:16457–16462. doi: 10.1073/pnas.1104533108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Cuenca S, Monjo M, Frontera M, Gianotti M, Proenza AM, Roca P. Sex steroid receptor expression profile in brown adipose tissue. Effects of hormonal status. Cell Physiol Biochem. 2007;20:877–886. doi: 10.1159/000110448. [DOI] [PubMed] [Google Scholar]
- Rosen ED, Sarraf P, Troy AE, Bradwin G, Moore K, Milstone DS, Spiegelman BM, Mortensen RM. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol Cell. 1999;4:611–617. doi: 10.1016/s1097-2765(00)80211-7. [DOI] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, Tempst P, Rudnicki MA, Beier DR, Spiegelman BM. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Conroe HM, Estall J, Kajimura S, Frontini A, Ishibashi J, Cohen P, Cinti S, Spiegelman BM. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J Clin Invest. 2011;121:96–105. doi: 10.1172/JCI44271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, Tavernier G, Langin D, Spiegelman BM. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura I, Hammer RE, Ikemoto S, Brown MS, Goldstein JL. Leptin reverses insulin resistance and diabetes mellitus in mice with congenital lipodystrophy. Nature. 1999;401:73–76. doi: 10.1038/43448. [DOI] [PubMed] [Google Scholar]
- Siersbaek MS, Loft A, Aagaard MM, Nielsen R, Schmidt SF, Petrovic N, Nedergaard J, Mandrup S. Genome-Wide Profiling of Peroxisome Proliferator-Activated Receptor gamma in Primary Epididymal, Inguinal, and Brown Adipocytes Reveals Depot-Selective Binding Correlated with Gene Expression. Molecular and cellular biology. 2012;32:3452–3463. doi: 10.1128/MCB.00526-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RE, Roberts JC. Thermogenesis of Brown Adipose Tissue in Cold-Acclimated Rats. Am J Physiol. 1964;206:143–148. doi: 10.1152/ajplegacy.1964.206.1.143. [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Hu E, Graves RA, Budavari AI, Spiegelman BM. mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev. 1994a;8:1224–1234. doi: 10.1101/gad.8.10.1224. [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994b;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARgamma. Annu Rev Biochem. 2008;77:289–312. doi: 10.1146/annurev.biochem.77.061307.091829. [DOI] [PubMed] [Google Scholar]
- Uldry M, Yang W, St-Pierre J, Lin J, Seale P, Spiegelman BM. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006;3:333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JM, Kemerink GJ, Bouvy ND, Schrauwen P, Teule GJ. Cold-activated brown adipose tissue in healthy men. N Engl J Med. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- Villanueva CJ, Waki H, Godio C, Nielsen R, Chou WL, Vargas L, Wroblewski K, Schmedt C, Chao LC, Boyadjian R, Mandrup S, Hevener A, Saez E, Tontonoz P. TLE3 is a dual-function transcriptional coregulator of adipogenesis. Cell Metab. 2011;13:413–427. doi: 10.1016/j.cmet.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen KA, Lidell ME, Orava J, Heglind M, Westergren R, Niemi T, Taittonen M, Laine J, Savisto NJ, Enerback S, Nuutila P. Functional brown adipose tissue in healthy adults. N Engl J Med. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- Wu J, Bostrom P, Sparks LM, Ye L, Choi JH, Giang AH, Khandekar M, Virtanen KA, Nuutila P, Schaart G, Huang K, Tu H, van Marken Lichtenbelt WD, Hoeks J, Enerback S, Schrauwen P, Spiegelman BM. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Neilson A, Swift AL, Moran R, Tamagnine J, Parslow D, Armistead S, Lemire K, Orrell J, Teich J, Chomicz S, Ferrick DA. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am J Physiol Cell Physiol. 2007;292:C125–C136. doi: 10.1152/ajpcell.00247.2006. [DOI] [PubMed] [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- Yamauchi T, Kamon J, Waki H, Terauchi Y, Kubota N, Hara K, Mori Y, Ide T, Murakami K, Tsuboyama-Kasaoka N, Ezaki O, Akanuma Y, Gavrilova O, Vinson C, Reitman ML, Kagechika H, Shudo K, Yoda M, Nakano Y, Tobe K, Nagai R, Kimura S, Tomita M, Froguel P, Kadowaki T. The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat Med. 2001;7:941–946. doi: 10.1038/90984. [DOI] [PubMed] [Google Scholar]
- Young P, Arch JR, Ashwell M. Brown adipose tissue in the parametrial fat pad of the mouse. FEBS Lett. 1984;167:10–14. doi: 10.1016/0014-5793(84)80822-4. [DOI] [PubMed] [Google Scholar]
- Zechner R, Kienesberger PC, Haemmerle G, Zimmermann R, Lass A. Adipose triglyceride lipase and the lipolytic catabolism of cellular fat stores. J Lipid Res. 2009;50:3–21. doi: 10.1194/jlr.R800031-JLR200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Physical activity (A) and food intake (B) in Cre-negative and Cre-positive TLE3F/F mice (N=6–10/group) determined by housing in Oxymax metabolic cages.