Abstract
To identify the trans-acting factors involved in autonomously replicating sequence (ARS) function, we initiated a screen for Saccharomyces cerevisiae mutants capable of stabilizing a plasmid that contains a defective ARS element. The amm (altered minichromosome maintenance) mutations recovered in this screen defined at least four complementation groups. amm1, a mutation that has been studied in detail, gave rise to a 17-fold stabilization of one defective ARS1 plasmid over the level seen in wild-type cells. The mutation also affected the stability of at least one plasmid bearing a wild-type ARS element. amm1 is an allele of the previously identified TUP1 gene and exhibited the same pleiotropic phenotypes as other tup1 mutants. Plasmid maintenance was also affected in strains bearing a TUP1 gene disruption. Like the amm1 mutant, the tup1 disruption mutant exhibited ARS-specific plasmid stabilization; however, the ARS specificities of these two mutants differed. The recovery of second-site mutations that suppressed many of the tup1 phenotypes but not the increased plasmid maintenance demonstrates that the plasmid stability phenotype of tup1 mutants is not a consequence of the other defects caused by tup1.
Full text
PDF


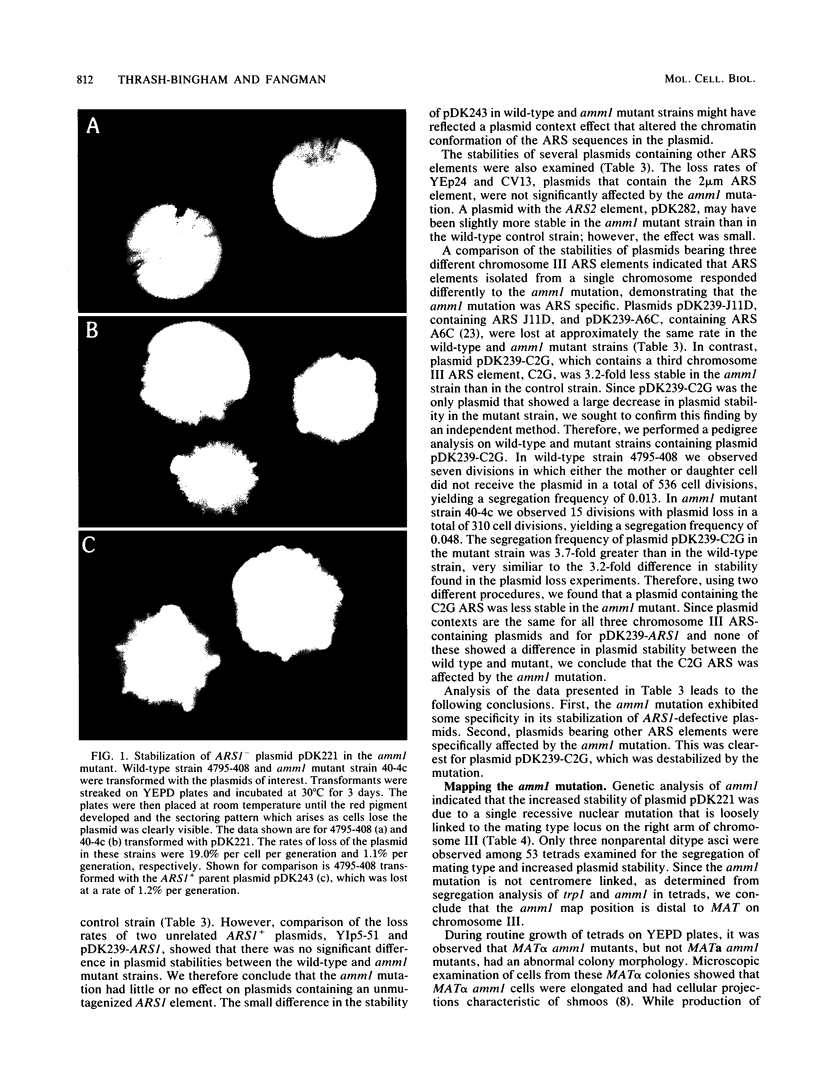
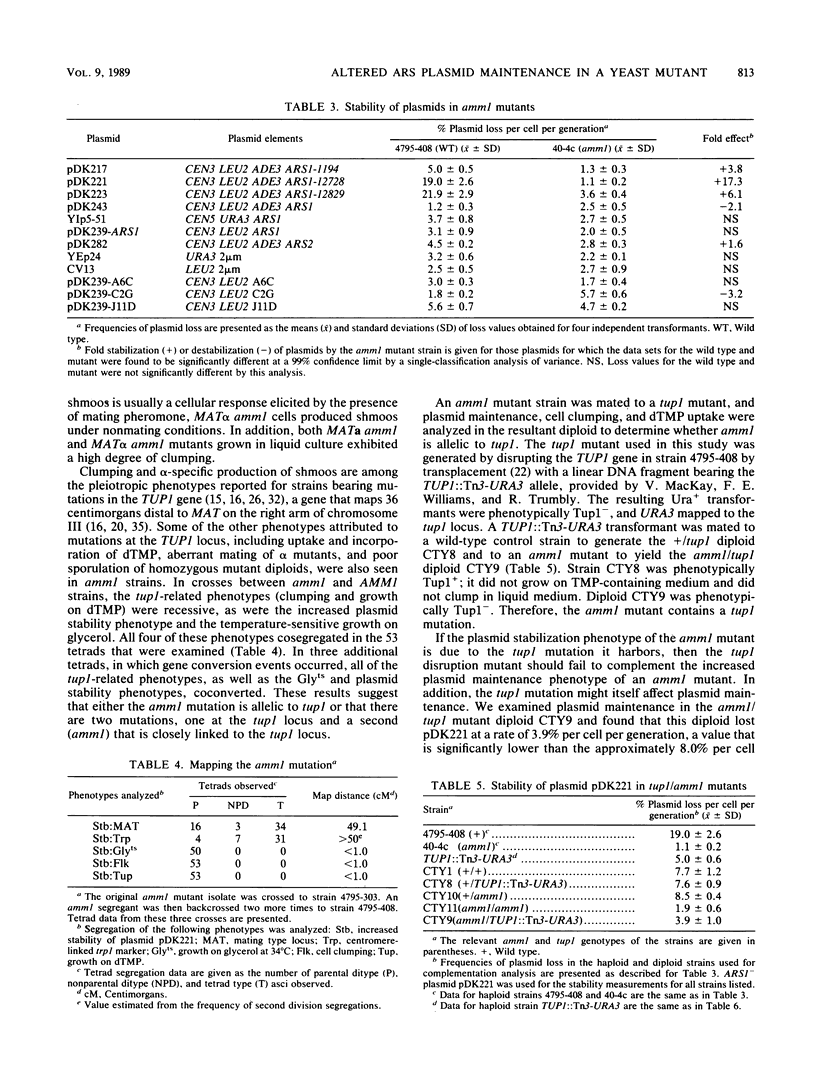


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brewer B. J., Fangman W. L. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987 Nov 6;51(3):463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- Broach J. R. Construction of high copy yeast vectors using 2-microns circle sequences. Methods Enzymol. 1983;101:307–325. doi: 10.1016/0076-6879(83)01024-1. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Campbell J. L. Eukaryotic DNA replication. Annu Rev Biochem. 1986;55:733–771. doi: 10.1146/annurev.bi.55.070186.003505. [DOI] [PubMed] [Google Scholar]
- Clarke L., Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980 Oct 9;287(5782):504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- Dotto G. P., Zinder N. D. Increased intracellular concentration of an initiator protein markedly reduces the minimal sequence required for initiation of DNA synthesis. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1336–1340. doi: 10.1073/pnas.81.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto G. P., Zinder N. D. Reduction of the minimal sequence for initiation of DNA synthesis by qualitative or quantitative changes of an initiator protein. Nature. 1984 Sep 20;311(5983):279–280. doi: 10.1038/311279a0. [DOI] [PubMed] [Google Scholar]
- Duntze W., MacKay V., Manney T. R. Saccharomyces cerevisiae: a diffusible sex factor. Science. 1970 Jun 19;168(3938):1472–1473. doi: 10.1126/science.168.3938.1472. [DOI] [PubMed] [Google Scholar]
- Greenfield L., Simpson L., Kaplan D. Conversion of closed circular DNA molecules to single-nicked molecules by digestion with DNAase I in the presence of ethidium bromide. Biochim Biophys Acta. 1975 Oct 15;407(3):365–375. doi: 10.1016/0005-2787(75)90104-5. [DOI] [PubMed] [Google Scholar]
- Huberman J. A., Spotila L. D., Nawotka K. A., el-Assouli S. M., Davis L. R. The in vivo replication origin of the yeast 2 microns plasmid. Cell. 1987 Nov 6;51(3):473–481. doi: 10.1016/0092-8674(87)90643-x. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearsey S. E., Edwards J. Mutations that increase the mitotic stability of minichromosomes in yeast: characterization of RAR1. Mol Gen Genet. 1987 Dec;210(3):509–517. doi: 10.1007/BF00327205. [DOI] [PubMed] [Google Scholar]
- Koshland D., Kent J. C., Hartwell L. H. Genetic analysis of the mitotic transmission of minichromosomes. Cell. 1985 Feb;40(2):393–403. doi: 10.1016/0092-8674(85)90153-9. [DOI] [PubMed] [Google Scholar]
- Lemontt J. F., Fugit D. R., Mackay V. L. Pleiotropic Mutations at the TUP1 Locus That Affect the Expression of Mating-Type-Dependent Functions in SACCHAROMYCES CEREVISIAE. Genetics. 1980 Apr;94(4):899–920. doi: 10.1093/genetics/94.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemontt J. F. Pathways of ultraviolet mutability in Saccharomyces cerevisiae. III. Genetic analysis and properties of mutants resitant to ultraviolet-induced forward mutation. Mutat Res. 1977 May;43(2):179–204. doi: 10.1016/0027-5107(77)90003-3. [DOI] [PubMed] [Google Scholar]
- Maine G. T., Sinha P., Tye B. K. Mutants of S. cerevisiae defective in the maintenance of minichromosomes. Genetics. 1984 Mar;106(3):365–385. doi: 10.1093/genetics/106.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maine G. T., Surosky R. T., Tye B. K. Isolation and characterization of the centromere from chromosome V (CEN5) of Saccharomyces cerevisiae. Mol Cell Biol. 1984 Jan;4(1):86–91. doi: 10.1128/mcb.4.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer R. K., Schild D. Genetic map of Saccharomyces cerevisiae, edition 9. Microbiol Rev. 1985 Sep;49(3):181–213. doi: 10.1128/mr.49.3.181-213.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newlon C. S., Petes T. D., Hereford L. M., Fangman W. L. Replication of yeast chromosomal DNA. Nature. 1974 Jan 4;247(5435):32–35. doi: 10.1038/247032a0. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palzkill T. G., Oliver S. G., Newlon C. S. DNA sequence analysis of ARS elements from chromosome III of Saccharomyces cerevisiae: identification of a new conserved sequence. Nucleic Acids Res. 1986 Aug 11;14(15):6247–6264. doi: 10.1093/nar/14.15.6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes T. D., Williamson D. H. Fiber autoradiography of replicating yeast DNA. Exp Cell Res. 1975 Oct 1;95(1):103–110. doi: 10.1016/0014-4827(75)90614-x. [DOI] [PubMed] [Google Scholar]
- ROMAN H. Studies of gene mutation in Saccharomyces. Cold Spring Harb Symp Quant Biol. 1956;21:175–185. doi: 10.1101/sqb.1956.021.01.015. [DOI] [PubMed] [Google Scholar]
- Schamhart D. H., Ten Berge A. M., Van De Poll K. W. Isolation of a catabolite repression mutant of yeast as a revertant of a strain that is maltose negative in the respiratory-deficient state. J Bacteriol. 1975 Mar;121(3):747–752. doi: 10.1128/jb.121.3.747-752.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J., Carlson M. Molecular analysis of SSN6, a gene functionally related to the SNF1 protein kinase of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Oct;7(10):3637–3645. doi: 10.1128/mcb.7.10.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortle D. R., Margolskee R. F., Nathans D. Mutational analysis of the simian virus 40 replicon: pseudorevertants of mutants with a defective replication origin. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6128–6131. doi: 10.1073/pnas.76.12.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha P., Chang V., Tye B. K. A mutant that affects the function of autonomously replicating sequences in yeast. J Mol Biol. 1986 Dec 20;192(4):805–814. doi: 10.1016/0022-2836(86)90030-6. [DOI] [PubMed] [Google Scholar]
- Snyder M., Buchman A. R., Davis R. W. Bent DNA at a yeast autonomously replicating sequence. Nature. 1986 Nov 6;324(6092):87–89. doi: 10.1038/324087a0. [DOI] [PubMed] [Google Scholar]
- Stark H. C., Fugit D., Mowshowitz D. B. Pleiotropic properties of a yeast mutant insensitive to catabolite repression. Genetics. 1980 Apr;94(4):921–928. doi: 10.1093/genetics/94.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcomb D. T., Struhl K., Davis R. W. Isolation and characterisation of a yeast chromosomal replicator. Nature. 1979 Nov 1;282(5734):39–43. doi: 10.1038/282039a0. [DOI] [PubMed] [Google Scholar]
- Trumbly R. J. Isolation of Saccharomyces cerevisiae mutants constitutive for invertase synthesis. J Bacteriol. 1986 Jun;166(3):1123–1127. doi: 10.1128/jb.166.3.1123-1127.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B. Mutants of Saccharomyces cerevisiae that incorporate deoxythymidine-5'-monophosphate into deoxyribonucleic acid in vivo. J Bacteriol. 1974 Jan;117(1):252–260. doi: 10.1128/jb.117.1.252-260.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]