Abstract
Background
Alternative splicing (AS) is a major mechanism for modulating gene expression of an organism, allowing the synthesis of several structurally and functionally distinct mRNAs and protein isoforms from a unique gene. Related to AS is the Transcription Induced Chimerism (TIC) or Tandem Chimerism, by which chimeric RNAs between adjacent genes can be found, increasing combinatorial complexity of the proteome. The Ly6g5b gene presents particular behaviours in its expression, involving an intron retention event and being capable to form RNA chimera transcripts with the upstream gene Csnk2b. We wanted to characterise these events more deeply in four tissues in six different mammals and analyse their protein products.
Results
While canonical Csnk2b isoform was widely expressed, Ly6g5b canonical isoform was less ubiquitous, although the Ly6g5b first intron retained transcript was present in all the tissues and species analysed. Csnk2b-Ly6g5b chimeras were present in all the samples analysed, but with restricted expression patterns. Some of these chimeric transcripts maintained correct structural domains from Csnk2b and Ly6g5b. Moreover, we found Csnk2b, Ly6g5b, and Csnk2b-Ly6g5b transcripts that present exon skipping, alternative 5' and 3' splice site and intron retention events. These would generate truncated or aberrant proteins whose role remains unknown. Some chimeric transcripts would encode CSNK2B proteins with an altered C-terminus, which could affect its biological function broadening its substrate specificity. Over-expression of human CSNK2B, LY6G5B, and CSNK2B-LY6G5B proteins, show different patterns of post-translational modifications and cell distribution.
Conclusions
Ly6g5b intron retention and Csnk2b-Ly6g5b transcript chimerism are broadly distributed in tissues of different mammals.
Background
To date, a high number of eukaryotic genomes have been sequenced. Surprisingly, it is interesting to observe that Homo sapiens and Caenorhabditis elegans genomes contain a similar number of protein-coding genes (~21.000), according to the Ensembl (http://www.ensembl.org/) database. Initially, these findings disconcerted researchers who thought the number of genes should be correlated with developmental and physiological complexity, and made them realize that other mechanisms should be involved in this evolutionary variety. Alternative splicing (AS) is a major mechanism for modulating gene expression of an organism, and enables a single gene to increase its expression capacity, allowing the synthesis of several structurally and functionally distinct mRNAs and protein isoforms from a unique gene (For reviews see [1-3]). This mechanism, which was initially described in viruses [4-6], is now known to affect 95% of all human genes [7] and has been proposed as a primary driver of the evolution of phenotypic complexity in mammals [8-10]. The human Major Histocompatibility Complex (MHC) is located on chromosome 6, and is ~4 Mb in length. It is composed by three regions, the class I and class II regions flanking the central class III region. The class III region is ~0.9 Mb in length and contains 62–64 genes and 2–3 pseudogenes, depending on the haplotype [11,12]. Previously, our group precisely defined the AS patterns of a five gene cluster from the Lymphocyte antigen-6 (LY-6) superfamily [13] and characterised the expression of the corresponding proteins [14] in human and mouse. LY-6 superfamily members are cysteine-rich, generally GPI-anchored, cell surface proteins, which have definite or putative immune-related roles [15]. Among these LY-6 MHC class III region genes, Ly6g5b showed a particular behaviour in the regulation of its expression [13,16], involving an intron retention event in human and mouse, the rarest form of alternative splicing found in metazoan species [17]. The intron retained is the first one after the initial exon and interrupts the open reading frame (ORF) just after the signal peptide introducing a premature stop codon (PSC). The presence of a PSC at this position should cause this intron-retained transcript to undergo Nonsense Mediated Decay (NMD) [18,19]. However, this transcript seemed to escape NMD and was more abundant than the correctly spliced mRNA [13,16]. In addition, findings in our laboratory showed the presence of LY6G5B gene exons in transcripts derived from the upstream gene CSNK2B[16], which encodes the Casein Kinase II β subunit, a ubiquitous protein kinase which regulates metabolic pathways, signal transduction, transcription, translation, and replication [20,21]. The Transcription Induced Chimerism (TIC) or Tandem Chimerism, is a phenomenon whose mechanism still remains largely unknown, although it is being promoted as a novel way to increase combinatorial complexity of the proteome [22,23]. At least 4%-5% of the tandem gene pairs in the human genome can be eventually transcribed into a single RNA sequence encoding a putative chimeric protein [22] but recent bioinformatic analyses, partially supported by experimental data, show that this phenomenon could be quite frequent [24,25]. Recently, it has been showed that these chimeras significantly exploit signal peptides and transmembrane domains, which could alter the cellular localisation of cognate proteins, and that chimeric RNAs are more tissue specific than non-chimeric transcripts [26]. Thus, this novel mechanism directly related to AS could have an important role in evolution divergence. In this regard, the majority of these studies give relevance to this phenomenon in Homo sapiens[24], but detailed comparative analyses among different species are, to our knowledge, not described. Here, we have deeply characterised the expression of Ly6g5b and Csnk2b transcripts independently and of the Csnk2b-Ly6g5b chimeric transcripts in four defined tissues among six different mammals. We conclude that Ly6g5b intron retention and Csnk2b-Ly6g5b chimerism is present in the tissues of the analysed mammalian group. In addition, we have made a comparative analysis of human CSNK2B, LY6G5B, and CSNK2B-LY6G5B protein expressions.
Results
Csnk2b transcript analysis
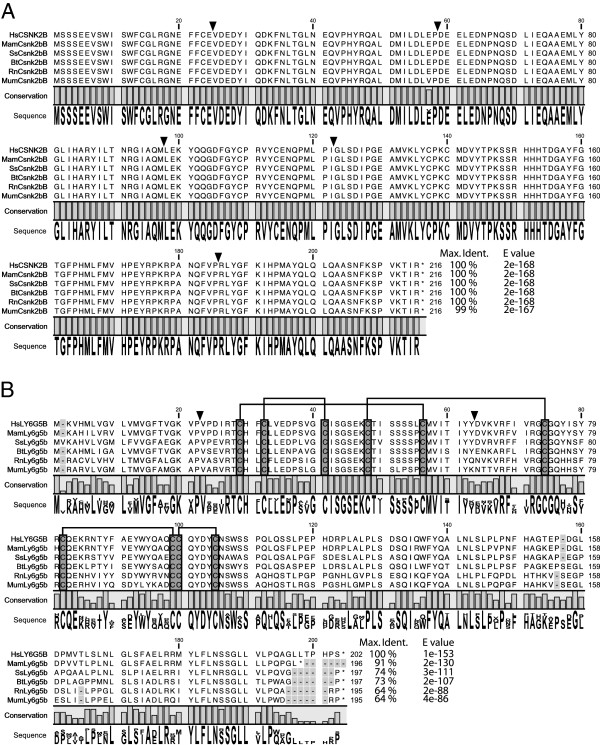
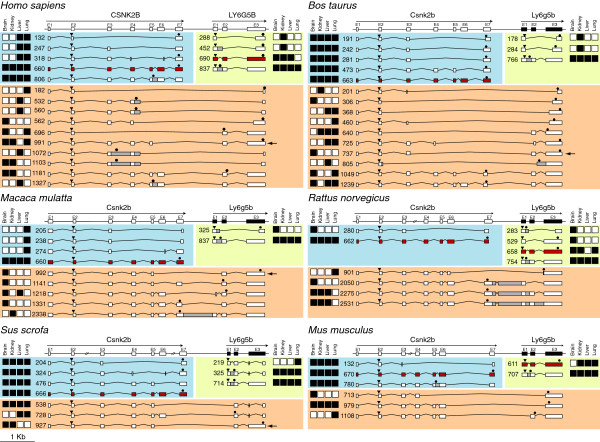
Canonical Csnk2kb ORF orthologue sequences from Homo sapiens (NM_001320), Macaca mulata (XM_001112478), Sus scrofa (XM_001928731), Bos taurus (NM_001046454), Rattus norvegicus (NM_031021) and Mus musculus (NM_009975) were analysed in order to find common features among them. Comparative analysis showed a total conservation rate in protein sequence among these six species, except for Mus musculus which presents a unique change in position 57 (V→ E) (Figure 1A). Through RT-PCR analysis, we found five different transcripts for Csnk2b in Homo sapiens, four in Macaca mulata, four in Sus scrofa, five in Bos taurus, two in Rattus norvegicus and three in Mus musculus (Figure 2 and Additional file 1). Only the canonical transcript sequences were on databases, except for Bos taurus for which BtCsnk2b-473 was also present (see Additional file 2: Table S1). We could detect the presence of the canonical Csnk2b transcript in each tested species and tissues (Figure 2), and it was also the isoform expressed at the highest level (data not shown). In addition, Csnk2b expression also generated other transcripts (Figure 2) expressed at lower levels (data not shown), but with a remarkable specificity among the analysed tissues in these six species (Figure 2). Some of them presented quite restricted expression patterns, such as the Macaca mulata ones that are only expressed in lung, or the Rattus norvegicus one, expressed only in brain. By contrast, the variants from Sus scrofa, Bos taurus and Mus musculus are broadly expressed. In Homo sapiens, the isoform which retains intron 5 is widely expressed; however the other three are only expressed in liver and lung. Through AS these CSNK2B transcripts present exon skipping, alternative 5’ and 3’ splice site and intron retention events (Figure 2) in the different species, which would generate severely truncated or aberrant proteins by using the canonical start codon.
Figure 1.
Homo sapiens (Hs), Macaca mulata (Mam), Sus scrofa (Ss), Bos taurus (Bt), Rattus norvegicus (Rn) and Mus musculus (Mum) ORF alignments. A) Csnk2b: N-terminal region α1-α5 (5-104), juxta-dimer interface region including zinc-finger (105-147) and α6 (163-175), C-terminal region (178-205) including interaction with CSNK2α (175-193) and dimerization (190-205) regions [27], B) Ly6g5b: The conserved cysteines, characteristic for Ly-6 domain, are shown highlighted in grey and their connectivity with interconnected lines. Exon-exon junctions are indicated by inverted triangles. The canonical (or theoretical canonical) sequence for each specie is only shown. The percentage of identity of each sequence respect to the human one is shown as well as the corresponding E values.
Figure 2.
Schematic representation of all Csnk2b and Ly6g5b transcripts detected through nested RT-PCR on each analysed tissue for Homo sapiens, Macaca mulata, Sus scrofa, Bos Taurus, Rattus norvegicus and Mus musculus. White boxes represent Csnk2b, Ly6g5b and chimera Csnk2b-Ly6g5b exons. Grey boxes represent intron sequences retained on each transcript. Red boxes indicate canonical isoforms. ORFs are delimited by triangles (ATG codon) and hexagons (Stop codon). Squares at the left or right, represent the presence (black) or absence (white) of expression of each transcript on the indicated tissue. Black arrows indicate Ly6g6b-Csnk2b chimeras that carry structural domains from Csnk2b and Ly6g5b, respectively. The corresponding nucleotide and protein sequences and the accession numbers (EMBL: HE864415-HE864490) are shown in Additional file 1.
Ly6g5b transcript analysis
Canonical Ly6g5b ORF orthologue sequences from Homo sapiens (NM_021221), Macaca mulata (XR_014070), Sus scrofa (XM_001926307), Bos taurus (XM_585827), Rattus norvegicus (NM_001001934) and Mus musculus (NM_148939) were compared (Figure 1B). Although some differences in amino acid sequence can be detected, a LY-6 protein domain conservation in these Ly6g5b orthologues is clearly present (Figure 1B). This domain is composed of ~80 amino acids and is characterised by a conserved pattern of eight to ten cysteine residues that have a defined disulfide-bounding pattern [14]. Through RT-PCR analysis, we found four different transcripts for Ly6g5b in Homo sapiens, two in Macaca mulata, three in Sus scrofa, three in Bos taurus, four in Rattus norvegicus and two in Mus musculus (Figure 2 and Additional file 1). The majority of these transcripts were not available on databases; even the canonical sequences (see Additional file 2: Table S1). Curiously, the presence of the canonical Ly6g5b transcript was only detected in three of the analysed species (Homo sapiens, Rattus norvegicus and Mus musculus). Similarly to what happened for Csnk2b, different transcript variants are generated for Ly6g5b through AS. These transcripts present a remarkable specificity among the analysed tissues and species, and assuming that they could be translated into proteins starting from the first canonical start codon, only truncated or aberrant proteins could be generated by them. Nevertheless, there is an interesting feature that should be stressed, the retention of the first intron in Ly6g5b transcripts, giving rise to a particular isoform (exon 1, intron 1, exon 2 and exon 3) that is present in all the tissues and analysed species (Figure 2), indicating conservation, and presenting the highest expression levels (data not shown). This isoform contains a PSC after the canonical start codon, in the middle of the retained intron, and therefore should be degraded through control mechanisms like NMD [18,19]. However this seems not to be the case, as we have previously described in human [16].
Csnk2b-Ly6g5b Chimeric transcript analysis
Through RT-PCR analysis, we found ten different Csnk2b-Ly6g5b chimeric transcripts in Homo sapiens, five in Macaca mulata, three in Sus scrofa, ten in Bos taurus, four in Rattus norvegicus and three in Mus musculus (Figure 2 and Additional file 1). Only human Csnk2b - Ly6g5b -1181 was found on databases (see Additional file 2: Table S1). As it happened with Csnk2b and Ly6g5b independent transcripts described above, AS seems to play an important role in generating these chimeras, and results in a set of transcripts that greatly vary in terms of composition and size. Indeed, exon skipping, intron retention and intergenic region retention events are present in these transcripts. The majority of the described chimeras (26/35) have a common characteristic: the total lack of the last exon (exon 7) of the upstream gene (Csnk2b) as well as the first exon of the downstream gene (Ly6g5b). There are also four chimeric transcripts that partially lack Csnk2b last exon (exon 7) (two in Macaca mulata and two in Bos taurus), one that partially maintains Ly6g5b first exon (in Macaca mulata) and four that retain the intergenic regions (three in Rattus norvegicus and one in Macaca mulata). Although chimeras function is still unknown, some authors defend that this kind of fusion might generate bi-functional proteins which would have the properties of both original proteins [23,26]. Assuming this, we determined the number of chimeric transcripts which conserved the ORF of both Csnk2b and Ly6g5b genes. We found such transcripts in Homo sapiens, Macaca mulata, Sus scrofa and Bos taurus, of which only HsCsnk2b - Ly6g5b -991, MamCsnk2b-Ly6g5b-992 and SsCsnk2b-Ly6g5b-927 (Figures 2, 3 and Additional file 1) maintain the N-terminal functional domains (alpha helices 1 to 6) from Csnk2b (see Figures 1A and 3) [27], such as the acidic loop (aa 55-64) and nuclear localization sequence (aa 9-14 or α1), as well as the LY-6 structural domain (see Figures 1B and 3) [14], allowing the possibility to create potentially bi-functional proteins [26,28,29]. These particular transcripts which contain the same exon-intron structure are expressed in different tissues, but commonly in brain. Bos taurus BtCsnk2b-Ly6g5b-737 maintains only exons 1 to 3 of Csnk2b and then would not encode its entire N-terminal domain [27]. We did not find this type of “bi-functional” chimeric transcripts in Rattus norvegicus or Mus musculus (Figure 2).
Figure 3.
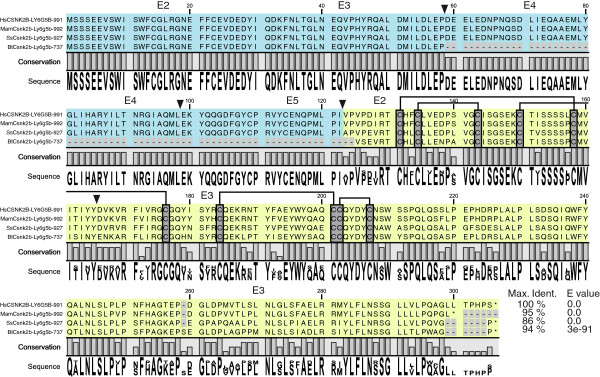
Homo sapiens (Hs), Macaca mulata (Mam), Sus scrofa (Ss) and Bos Taurus (Bt) Csnk2b- Ly6g5b chimeras ORF alignment. Blue and light green colour indicates Csnk2b and Ly6g5b domains, respectively. Exon-exon junctions are indicated by inverted triangles. The conserved cysteines, characteristic for Ly-6 domain, are shown highlighted in grey and their connectivity with interconnected lines. The percentage of identity of each sequence respect to the human one is shown as well as the corresponding E values.
It is interesting to note that several (23/35) chimeric transcripts could encode Csnk2b truncated proteins (7/35) or with modifications in their C-terminus (16/35). The CSNK2B C-terminal part is involved in homodimerization and binding to CSNK2A subunit (see Figure 1) [27]. Some of these detected chimeric transcripts are generated by replacing the canonical sequence of exon 7 by sequences encoded by total or partial exon 2 or 3 of Ly6g5b but not corresponding to LY-6 amino-acid sequences due to changes in the reading frames. These are chimeras HsCsnk2b-Ly6g5b-1181, MamCsnk2b-Ly6g5b-1218, SsCsnk2b-Ly6g5b-728, MumCsnk2b-Ly6g5b-979 and MumCsnk2b-Ly6g5b-1108, with variable tissue distribution, except MumCsnk2b-Ly6g5b-979 that is expressed in the four tissues. Other transcripts are altered on Csnk2b exon 6 or 7, lacking the zinc-finger domain, α6 and C-terminal regions of CSNK2B (see Figure 1), such as MamCsnk2b-Ly6g5b-1141 (only expressed in brain) and BtCsnk2b-Ly6g5b-1049 (expressed in brain and lung) which maintain the same exon-intron structure (lack of Csnk2b exon 6 and Ly6g6b exon 1), and SsCsnk2b-Ly6g5b-538 (expressed in the four tissues) (see Figure 2). In addition, there are some chimeric transcripts that would encode a complete CSNK2B protein considering that they contain all exons (1-7) of Csnk2b including the stop codon and in which the Ly6g5b nucleotide sequences will act as 3´ UTRs. These are MamCsnk2b-Ly6g5b-1331, MamCsnk2b-Ly6g5b-2338, BtCsnk2b-Ly6g5b-1239, RnCsnk2b-Ly6g5b-2050, RnCsnk2b-Ly6g5b-2275 and RnCsnk2b-Ly6g5b-2531. They present variable tissue distribution and exon-intron structure (see Figure 2).
Csnk2b, Ly6g5b and Csnk2b -Ly6g5b Chimera protein analysis
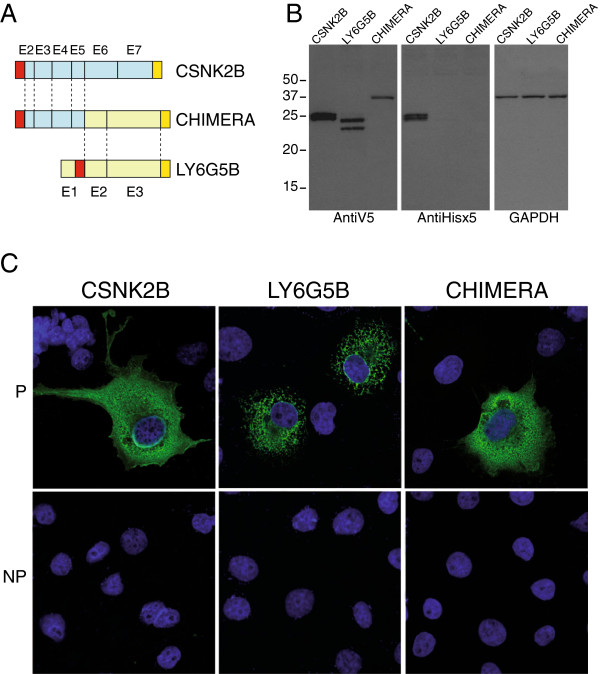
In order to analyse post-translational modifications and sub-cellular localisation of human CSNK2B, LY6G5B and CSNK2B-LY6G5B proteins, we over-expressed them in the COS7 cell line (Figure 4), using a double tag strategy. Thus, CSNK2B, LY6G5B and CSNK2B-LY6G5B proteins were C-terminally tagged by adding a Hisx6 tag. On the other hand, N-terminal V5 epitope was added upstream the first ATG to CSNK2B and CSNK2B-LY6G5B proteins, but due to the presence of a signal peptide in LY6G5B [13] and in order to tag the mature LY6G5B protein (Figure 4A) the V5 epitope tag was inserted after the signal peptide. Western blot analysis using anti-V5 or anti-PentaHis antibodies showed interesting results. Anti-V5 antibodies showed two close intense bands for CSNK2B protein of the estimated size (Figure 4B). These two bands could correspond to post-translational modifications of CSNK2B such as phosphorilation [30]. For LY6G5B, also two clear bands were also present, in agreement with previous results [14]. Interestingly, anti-V5 antibodies western-blot analysis showed a single discrete band of the predicted size for CSNK2B-LY6G5B protein, indicating lack of post-translational modifications.
Figure 4.
Human CSNK2B, LY6G5B and CSNK2B-LY6G5B chimeric protein characterisation. Schematic representation of A) V5 and Hisx6 double tag strategy. V5 (red box) and Hisx6 (yellow box) epitopes are shown. Exon numbers are indicated. B) Western-blot analysis showing in vitro expression of CSNK2B, LY6G5B and CSNK2B-LY6G5B chimera double tagged proteins. GAPDH expression is shown as internal control. C) Immunofluorescence experiments in permeable (P) and non permeable (NP) conditions showing CSNK2B, LY6G5B and CSNK2B-LY6G5B chimeric protein cellular distribution.
On the other hand, anti-PentaHis antibodies showed similar pattern for CSNK2B protein to that showed by anti-V5 antibodies, but no signal of LY6G5B protein or CSNK2B-LY6G5B protein was detected (Figure 4B). This lack of detection on LY6G5B could be due to a C-terminal processing cleaving also the Tag. This cleavage signal sequence would also be present in CSNK2B-LY6G5B protein and for that also prone to be processed.
Cellular CSNK2B protein distribution has been described before in cytoplasm, nuclei, and other organelles [21]. Our results, through immunoflourescent confocal microscopy experiments by using anti-V5 antibodies under permeabilised conditions, showed mainly cytoplasmic cellular CSNK2B protein distribution, in agreement with previous data [21]. Under the same conditions, LY6G5B showed a protein distribution clearly related with ER pattern, and not extracellular staining, as previously described [14]. Here, for the first time, we show CSNK2B-LY6G5B protein distribution, which is quite similar to the one presented by CSNK2B and which clearly differs from the one of the LY6G5B protein.
Although the LY6G5B protein belongs to a GPI-anchored protein family, it has not been found to be located on the outside of the cellular membrane [14]. In addition, it is known that CSNK2B can be exported to the external side of the cellular membrane [31], and CSNK2B-LY6G5B presents CSNK2B domains needed for its exportation to cell surface and/or its excretion, as well as a mature Ly-6 domain (Figure 3). To know whether CSNK2B-LY6G5B could be on the cell surface we carried out two experimental strategies. The first one consisted of immunoflourescent confocal microscopy experiments under non-permeabilised conditions. Our results showed the absence of CSNK2B, LY6G5B as well as CSNK2B-LY6G5B chimeric proteins in the cell surface (Figure 4C), in COS 7 cells. The second one was to test CSNK2B, LY6G5B and/or CSNK2B-LY6G5B proteins presence in supernatant by western-blot experiment. It was not possible to observe expression of these proteins in the supernatant, either when loaded directly on a gel or when TCA-precipitated (data not shown).
Discussion
After the challenge supposed by the human genome sequencing, as well as of other organisms with medical or commercial interest, the determination of the transcriptome of these species is a prerequisite for fully understanding not only their molecular biology, but also for translating this information to technical applications in medicine, pharmacology and biotechnology. In this sense, high throughput technologies supported by systematic cDNA libraries sequencing has been the main approach used for transcriptome characterisation. Thus, sequencing of random transcript cDNA clones that results in short partial sequences, known as expressed sequence tags [32] (EST) or, more recently, methods for full length isolation and sequencing of random clones from cDNA libraries have been used [33,34]. In addition, in the last few years massive sequencing technologies have also been developed for transcriptome analysis [7,24,35]. All these high throughput techniques present the advantage to generate a considerable volume of information in a relative short time, but these techniques seem to be inefficient for discovering relative rare transcripts. This affirmation is supported by recent data that suggest the existence of a wealth of transcripts which had, so far, escaped detection through systematic sequencing of cDNA libraries [36]. In a recent published work [26] combining EST and RNAseq data, it has been showed that chimeras are lowly expressed transcripts. Thus, here we present that nested RT-PCR shows to be an efficient tool to discover a number of transcripts expressed from a concrete locus, not only those highly expressed but also those expressed at lower levels. In fact, 62 of 76 transcripts detected in our experiments had never been described before. This supposes 81.5% of novel sequences, showing how powerful this technique is in order to detect transcripts from a concrete locus. In this respect, our results show that current gene and transcript annotation sets might cover only a small fraction of the total transcriptional output of the human and other organism described genomes, and in this sense, a major enforce should be taken in order to detect more transcripts of a particular organism.
A detailed human and mouse Ly6g5b transcript analysis has been previously described by our group [13,16]. However, here we have extended this comparative analysis among different mammalian species, showing that Ly6g5b first intron retention is a ubiquitous and important characteristic conserved in mammals. Its apparent capacity to escape NMD together with its conservation in the mammalian group studied points to an important role for this non-coding RNA, which should be investigated. So, we could conclude that Ly6g5b gene presents a double expression pattern. The first one, quite similar among tissues and species, is constituted by Ly6g5b transcripts with first intron retention event. The second one seems to be tissue and species specific and is constituted by canonical Ly6g5b transcripts with a complete Ly-6 ORF as well as aberrant and non conserved transcripts, with no common pattern distribution.
For Csnk2b gene expression we can also describe a double expression pattern. The first one constituted by the canonical isoform, which is quite similar among tissues and species, and the second one which seems to be tissue and species specific and is constituted by aberrant and non conserved transcripts, with no common pattern distribution.
CSNK2B-LY6G5B human chimerism was initially described by Calvanesse et al (2008) by using different human cell lines [16]. However, here we show the first comparative analysis of this TIC event using RNA from different mammalian tissues and species. Our results show that, far to be a human characteristic, Csnk2b-Ly6g5b chimerism is widely conserved in mammals. Its conservation among all the species analysed in this study shows how Csnk2b-Ly6g5b chimerism is not a trivial event. The majority of them lack the last exon of the upstream gene as well as the first exon of the downstream gene which is consistent with previous reported data [22,23]. This eliminates the stop codon as well as the molecular targets present in the 3’ UTR region of the transcripts of the upstream gene. We also agree with these authors that run-off is the most likely mechanism involved in the origin of TIC, since some chimeric transcripts detected in our study maintain the intergenic region. Other mechanisms proposed for generating chimeric transcripts, like trans-splicing, are not likely to maintain these intergenic regions.
In addition to the canonical Csnk2b, Ly6g5 and Csnk2b-Ly6g5b transcripts with a coherent ORF, other transcripts detected in our study present exon skipping as well as intron retention events which allow to generate, assuming that all these transcripts could be translated into protein by using canonical start codon, truncated or aberrant proteins. Others are non-coding RNAs. The observation that there are tens of thousands of non-coding RNA (ncRNA) expressed in mammals, and that most of the genome is transcribed, confronts and contradicts the traditional protein-centric view of genetic information and genome organisation [37], [38]. Thus, there are two opposing alternatives either the bulk of the transcription which does not yield mRNAs is ‘transcriptional noise’ and/or the residue of evolutionary baggage retained or accumulated within genes, or this transcription comprises another level of expression and transaction of RNA information that is important to the evolution and developmental ontogeny of the higher organisms [39]. If one assumes that all this is transcriptional noise and that all these transcripts are the result of transcriptional machinery mistakes while it is working, they should not be distributed in a specific manner.
Some authors defend that chimerism might generate bi-functional proteins having properties from both original proteins [23]. Through different analyses of our results, we could identify chimeras which maintain the ORFs of Csnk2b and Ly6g5b susceptible to form bi-functional chimeric proteins in Homo sapiens, Macaca mulata, and Sus scrofa. The fact that these were not found in cow, rat and mouse does not indicate functional chimera absence in these species, since they could be present in other tissues not analysed in this study. These hypothetical new chimeric proteins all carry N-terminus domains from Csnk2b involved in structural aspects that are required for Csnk2b exportation to the cellular surface [21] and/or its regulation [27], as well as Ly-6 domain amino acid sequence [13,14] at their C-terminus. However, the chimera “bi-functional” protein will affect the juxta-dimer interface region [27] containing the zinc-finger involved in the homo-dimerisation, as well as all the C-terminal domain involved in the interaction with the CSNK2A subunits and the crucial last 20 amino-acids also involved in the homo-dimerisation. In addition, the Ly-6 domain, with 10 Cysteins, could not be folded as such due to the intracellular localisation. These two facts indicate that the “bi-functional” chimera would not be formed, and would only have one function: the one of CSNK2B, although possibly binding to other kinase different to CSNK2A due to the alterations produced on the C-terminal domain commented above. Other chimeras would only be affected from exon 7, containing then the juxta-dimer interface region but differing at the very end C-terminal region and not containing the Ly-6 domain sequence due to frameshifts, and probably only affecting the binding to CSNK2A. It has been proposed that CSNK2B might bind other kinases such as Ras-1 and Mos to modify their catalytic affinity in a CK2-independent fashion. The alternative C-terminal ends, generated by the chimeric transcripts, could increment the binding repertoire of CSNK2B to other kinases or non-kinase proteins converting CSNK2B in even a wider “wild-card” regulator subunit than previously proposed [27].
In addition, we have found chimeric transcripts that would encode a complete CSNK2B protein, but with different 3´UTR due to the Ly6g5b sequence, in Macaca mulata, Bos taurus and Rattus norvegicus. These transcripts could have different mRNA localisations or stabilities which could have altered protein functional implications [40,41]. It has been described that some 3´UTR contain “localization elements” or “zip codes” which target mRNAs to specific subcellular sites.
Conclusions
Alternative splicing has an important role in Csnk2b and Ly6g5b gene expression. Ly6g5b intron retention and Csnk2b-Ly6g5b chimera transcripts are present in many tissues of different mammals. The data and analysis we have performed should serve as a valuable resource for further characterizing the possible functional role of TIC and the mechanisms that affect it.
Methods
Computational analysis
Csnk2b and Ly6g5b orthologue ORF sequences Homo sapiens, Macaca mulata, Sus scrofa, Bos taurus, Rattus norvegicus and Mus musculus were obtained from the National Centre for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/BLAST/) [42]. Multiple alignments of sequences were performed with ClustalW2 software (http://www.ebi.ac.uk/Tools/msa/clustalw2/) [43,44]. Protein sequences analyses were carried out using Pfam (http://www.sanger.ac.uk/resources/databases/pfam.html) [45], SMART (http://smart.embl-heidelberg.de/) and InterProScan (http://www.ebi.ac.uk/InterProScan/) [46-49] databases. The sequencing results were evaluated using the BLAST algorithm at the NCBI web page and the MultAlin alignment software (http://bioinfo.genotoul.fr/multalin/multalin.html) [50]. Primers were designed using the Primer3 software (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi).
mRNA extraction and retrotranscription
Homo sapiens, Macaca mulata, Sus scrofa, Bos taurus, Rattus norvegicus and Mus musculus brain, kidney, liver and lung tissue total RNAs were obtained from BioChain® (USA) http://www.biochain.com through one of their Europe distributor "AMS" http://www.amsbio.com (UK). One μg of total RNA from each tissue was used for oligo-dT primed cDNA synthesis which was performed using the ImProm-II(TM) Reverse Transcription System (Promega) in a 20 μl reaction volume following the manufacturer's instructions.
Nested RT-PCR
In order to detect Csnk2b, Ly6g5b and Csnk2b-Ly6g5b transcripts, first and second rounds of nested RT-PCR were performed with the primers indicated on Table 1 by using GoTaq® Green Master Mix (Promega) in a 20 μl reaction volume. For the first round of PCR 1 μl of cDNA was used in each reaction. Amplification conditions were: 95°C for 5 min followed by 40 cycles of 95°C for 30 s, 60°C for 30 s and 72°C for 90 s followed by 72°C for 10 min. For the second round of PCR, 1 μl from the first round product diluted 1/25 was used in each PCR reaction. Amplification conditions were the same as before but only 30 cycles were used. PCR products were separated by size through electrophoresis, purified from gel (Wizard® SV Gel and PCR Clean-Up System, Promega) and finally cloned into pGEM-T Easy Vector (Promega). Sequencing of PCR products and plasmids were carried out by the sequencing service of the Instituto de Investigaciones Biomédicas “Alberto Sols” (Madrid, Spain. http://www.iib.uam.es).
Table 1.
Nested-PCR primers
| |
|
CSNK2B |
LY6G5B |
||
|---|---|---|---|---|---|
| First Round | Second Round | First Round | Second Round | ||
|
H.sapiens |
F |
HsCSNK2BE1f01 |
Hs-MamCSNK2BE1-2f01 |
HsLY6G5BE1f01 |
HsLY6G5BE1f02 |
| CGTTCCCTGGAAGTAGCAA |
GCTGACGTGAAGATGAGCAG |
GAGCATGGTCACAGGAAGGT |
CATCTCCCCAGAATTCCAAA |
||
|
R |
Hs-SsCSNK2BE7r02 |
Hs-Mam-RnCSNK2BE7r01 |
HsLY6G5BE3r02 |
Hs-MamLY6G5BE3r01 |
|
| CCCACCACAATAACGACTCC |
TCAGCGAATCGTCTTGACTG |
CGGAGGCCTAAGAAATCACA |
TGTTTTCAGAGAGGGCAGTG |
||
|
M.mulata |
F |
MamCsnk2bE1iso1f01 |
Hs-MamCsnk2bE1-2f01 |
MamLy6g5bE1f01 |
MamLy6g5bE1f02 |
| ACCCTCCCCAATTTCCACT |
GCTGACGTGAAGATGAGCAG |
TCACAGGAAGGTGGGGTTT |
CGTCTCCCCAGAATTCCATA |
||
|
R |
Mam-BtCsnk2bE7r02 |
Hs-Mam-RnCsnk2bE7r01 |
MamLy6g5bE3r02 |
Hs-MamLy6g5bE3r01 |
|
| TCCCACCACAATAATGACTCC |
TCAGCGAATCGTCTTGACTG |
CAACAGAGGGAGGCCTAAGA |
TGTTTTCAGAGAGGGCAGTG |
||
|
S.scrofa |
F |
SsCsnk2bE1f01 |
SsCsnk2bE1-2f01 |
SsLy6g5bE1f01 |
SsLy6g5bE1f02 |
| TCCTTGGAAGCAGAAACTCC |
CGCTGAAGTGAAGATGAGCA |
CTCAGGGATCACCCCTCTC |
TCACGTGCTCGTAGGTATGC |
||
|
R |
Hs-SsCsnk2bE7r02 |
SsCsnk2bE7r01 |
SsLy6g5bE3r02 |
SsLy6g5bE3r01 |
|
| CCCACCACAATAACGACTCC |
GGGAATCAGCGAATTGTCTT |
GAATGCTGGGGTCTTAGGG |
CCGGGAAGGATGAGATGTTA |
||
|
B.taurus |
F |
BtCsnk2bE1f01 |
BtCsnk2bE1-2f01 |
BtLy6g5bE1f01 |
BtLy6g5bE1f02 |
| GAAGCAGAAACTCCCCTTCC |
CCGACGTGAAGATGAGCAG |
GTCAGAACCACCCTGCAGTT |
CTCTCCCCAGAAGTCCATGA |
||
|
R |
Mam-BtCsnk2bE7r02 |
BtCsnk2bE7r01 |
BtLy6g5bE3r02 |
BtLy6g5bE3r01 |
|
| TCCCACCACAATAATGACTCC |
GGAATCAGCGAATCGTCTTC |
GGGGTGTCAGGAGTGAGATG |
AATCTATCTGCGACGGGAAA |
||
|
R.norvegicus |
F |
Rn-MumCsnk2bE1f01 |
Rn-MumCsnk2bE1-2f01 |
RnLy6g5bBE1f01 |
RnLy6g5bE1f02 |
| GTTCCTTGGAAGCACAGCTC |
CCGCGGACATAAAGATGAGT |
AGTGTGATCCCAGGAAGGTG |
CTACTCCACGGGAGTTGCTC |
||
|
R |
RnCsnk2bE7r02 |
Hs-Mam-RnCsnk2bE7r01 |
RnLy6g5bE3r02 |
RnLy6g5bE3r01 |
|
| GTGTCACAGGCAGAGGAGGT |
TCAGCGAATCGTCTTGACTG |
CCATGGAGAGCAGAAGGAAG |
CAGAGCAGAATCTGGGAAGG |
||
| M.musculus |
F |
Rn-MumCsnk2bE1f01 |
Rn-MumCsnk2bE1-2f01 |
MumLy6g5bE1f01 |
MumLy6g5bE1f02 |
| GTTCCTTGGAAGCACAGCTC |
CCGCGGACATAAAGATGAGT |
ACTGCCTGTCAAACCCATTC |
TCCCCCAAATTCCATAATGA |
||
| R | MumCsnk2bE7r02 |
MumCsnk2bE7r01 |
MumLy6g5bE3r02 |
MumLy6g5bE3r01 |
|
| AGCAGAGGAATGGTGGTGTC | GTGGGCAATCAGCGAATAGT | GAGTGTTCACAGACCGCAGA | GGGGAACACATCAGGGTCTA | ||
Name and sequence (5’-3’ orientation). F, forward primer; R, reverse primer.
Expression constructs
The full-length coding sequence of human CSNK2B, LY6G5B and CSNK2B-LY6G5B were amplified from the previously cloned pGEM-T Easy vectors with the specific primers showed on Table 2 by using Pfu DNA Polymerase (Promega). Amplification conditions were: 95°C for 5 min followed by 35 cycles of 95°C for 30 s, 60°C for 30 s and 74°C for 3 min followed by 74°C for 10 min. PCR products were cloned in BamHI/AgeI cloning sites present in pcDNA3.1/V5-His-TOPO plasmid (Invitrogen), removing V5 epitope and in frame with the His tag creating CSNK2B-His, CSNK2B-LY6G5B-His and exon 2-exon 3 LY6G5B-His plasmids. To create a V5Tag-pcDNA3.1/Zeo vector we designed long primers containing the V5 epitope sequence (Table 2). These primers were hybridised (100°C for 4 min followed by 1 min cycles decreasing 0,5°C per cycle from 100°C to 4°C) to create the dsV5 Tag sequence, which was cloned in NotI/EcoRV cloning sites present in pcDNA3.1/Zeo(-) plasmid (Invitrogen) ending with the V5-Tag-pcDNA3.1/Zeo vector. Then, ATG-Stop CSNK2B-His and CSNK2B-LY6G5B-His and exon 2-exon 3 LY6G5B-His coding sequences were amplified with primers showed on Table 2 by using Pfu DNA Polymerase (Promega) under the same PCR conditions as above and cloned in BamHI/HindIII cloning sites present in the new V5-Tag-pcDNA3.1/Zeo vector in frame with the N-terminal V5 epitope creating V5-CSNK2B-His, V5-CSNK2B-LY6G5B-His and V5-exon 2-exon 3 LY6G5B-His. Finally, LY6G5B exon 1 was amplified from LY6G5B-His plasmid using the primers showed on Table 2 and Pfu DNA Polymerase (Promega) under the same PCR conditions described above and cloned in XbaI/NotI cloning sites present in V5-exon 2-exon 3 LY6G5B-His plasmid to finally create an exon1-V5-exon 2-exon 3 LY6G5B-His vector. Sequencing of PCR products and plasmids were carried out by the sequencing service of the Instituto de Investigaciones Biomédicas “Alberto Sols” (Madrid, Spain. http://www.iib.uam.es).
Table 2.
Expression constructs primers
|
Long primers for V5 epitope cloning in pcDNA3.1/Zeo(-) | |
| Name |
Sequence |
| MCV5_F01 |
GGGGCGGCCGCATGGGAAAGCCGATCCCAAACCCTCTATTAGGTCTGGACTCCACCGGATCCTGAGATATCGGG |
| MCV5_R01 |
CCCGATATCTCAGGATCCGGTGGAGTCCAGACCTAATAGAGGGTTTGGGATCGGCTTTCCCATGCGGCCGCCCC |
|
Primers for CSNK2B, LY6G5B and CSNK2B-LY6G5B Chimera ORFs cloning in pcDNA3.1/V5-His-TOPO | |
| Name |
Sequence |
| HsCSNK2BORF_F01 |
CCCGGATCCATGAGCAGCTCAGAGG |
| HsCSNK2BORF_R01 |
CCCACCGGTGCGAATCGTCTTGAC |
| HsLY6G5BORF_F01 |
CCCGGATCCATGAAGGTCCATATGC |
| HsLY6G5BORF_R01 |
CCCACCGGTGGAAGGGTGAGGTGTC |
|
Primers for final construct in pcDNA3.1/Zeo(-) | |
| Name |
Sequence |
| HsCSNK2BORF_F01 |
CCCGGATCCATGAGCAGCTCAGAGG |
| HsLY6G5BORFE2_F02 |
CCCGGATCCGTTCCTGTTCCCGACATC |
| Hisx6HindIIIR_01 |
CCCAAGCTTTCAATGGTGATGGTGATGATG |
| HsLY6G5BPS_F01 |
GGGTCTAGAATGAAGGTCCATATGC |
| HsLY6G5BPS_R01 | CCCGCGGCCGCTCTTTCCTACTGTGAA |
Restriction enzyme target sequences are in bold and italics.
Transfections and western blot analyses
Transfections of COS-7 cells were carried in 24-well plates, with 0.5 μg of plasmid per well, by using TransIT® COS Transfection kit (Mirus-BioNova) following the manufacturer´s instructions. For Western Blot analyses, two days after transfection, cells were harvested in Laemmli´s SDS sample buffer. Samples were resolved on SDS 12% (w/v) polyacrylamide gels and the proteins were transferred onto nitrocellulose membranes (Protran). After blocking the membrane 30 minutes in PBST (PBS-0,05% (v/v) Tween) containing 5% (w/v) skimmed milk powder (blocking solution), the blot was first incubated for 90 min either in a 1/2000 dilution of mouse anti-Hisx5 (Qiagen), a 1/10000 dilution of mouse anti-V5 (Sigma) or a 1/5000 dilution of mouse anti-GAPDH monoclonal antibodies in blocking solution, washed three times for 10 min with PBST, and then incubated for 30 min in a 1:5000 dilution of peroxidase-conjugated anti-mouse IgG antibody (Sigma) in PBST. Membrane was washed twice for 10 min with PBST followed by a final wash in PBS for 10 min. Bound antibodies were detected by ECL Plus Western Blotting (Amershan). Secreted proteins were TCA-precipitated from the culture media after the addition of 1 volume of TCA to 4 volumes of media, incubated 10 min at 4°C, washed with 200 μl cold acetone, resuspended in Laemmli´s SDS sample buffer, and analysed by SDS-PAGE followed by Western blotting, as described above.
Immunofluorescences
Transfections were performed as described above. Two days after transfection, cells were fixed for 15 min at room temperature with 4% (v/v) paraformaldehyde in PBS. Following fixation, cells were washed three times for 15 min with PBS, treated with ammonium chloride 50 mM for 30 min and permeabilised for 20 min with PBS containing 0.1% (v/v) Triton X-100 (wash solution). Cells were blocked for 30 min in wash solution with 5% (w/v) BSA (Bovine Serum Albumin, blocking solution) and then incubated for 1 h with 1:400 dilution of mouse anti-V5 monoclonal antibody (Sigma) in blocking solution. After that, cells were washed three times for 15 min with wash solution and then incubated with a secondary antibody coupled to Alexa 488 (anti mouse IgG, Invitrogen) in a 1:500 dilution for 1 h in blocking solution. Cells were washed three times for 15 min with wash solution and nucleic acids were stained with To-Pro3 in a 1:500 dilution in blocking solution. Finally, cells were washed three times for 15 min with wash solution, mounted with Prolong Gold Antifade Reagent (Molecular Probes) and photographed with a Leika confocal microscope. Non-permeabilised immunofluorescence experiments were carried out under the same conditions described above but avoiding Triton X-100 addition.
Abbreviations
RT-PCR: Reverse transcription polymerase chain reaction; CDS: Coding sequence; AS: Alternative splicing; MHC: Major Histocompatibility Complex; ORF: Open reading frame; PSC: Premature stop codon; NMD: Nonsense mediated decay; TIC: Transcription induced chimerism; EST: Expressed sequence tag; cDNA: Complementary DNA; mRNA: Messenger RNA; ncRNA: Non-coding RNA; TCA: Trichloroacetic acid; ER: Endoplasmic reticulum
Competing interest
The authors declare that they have no competing interests.
Authors’ contributions
FH-T and AR designed and carried out the experiments, and analysed and interpreted the data. FH-T drafted the manuscript. BA conceived the study, participated in its design and data interpretation, and completed the manuscript. All authors read and approved the final manuscript.
Supplementary Material
List of all the nucleotide and protein sequences of the mRNAs described on this article with their corresponding names and accession numbers.
Table that contains all the different spliced isoforms detected in all the species studied in this work indicating whether they were previously described (deposited on databases) and, in the case they were found on databases the access number of the corresponding sequence or of the ESTs are indicated.
Contributor Information
Francisco Hernández-Torres, Email: fraheto@ujaen.es.
Alberto Rastrojo, Email: arastrojo@cbm.uam.es.
Begoña Aguado, Email: baguado@cbm.uam.es.
Acknowledgements
We are grateful to Fernando Carrasco from the Genomics Service at CBMSO for his technical expertise and advice. This work was supported by grants from the Ministerio de Educación y Ciencia (BFU2005-03683), Ministerio de Ciencia e Innovación (BFU 2008-03126, BFU2009-09117), Comunidad de Madrid (GR/SAL/0670/2004, and 200620 M078) and Fundación Ramón Areces. F Hernández-Torres was funded by Genoma España and A Rastrojo by a postgraduate fellowship (FPU) from the Ministerio de Educación y Ciencia. B Aguado held a Programa Ramón y Cajal (MEC) contract and an Amarouto (Comunidad Madrid-Fundación Severo Ochoa) contract. We acknowledge support of the publication fee by the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI). The CBMSO receives an institutional grant from Fundación Ramón Areces.
References
- Villate O, Rastrojo A, Lopez-Diez R, Hernandez-Torres F, Aguado B. Differential splicing, disease and drug targets. Infect Disord Drug Targets. 2008;8(4):241–251. doi: 10.2174/187152608786734188. [DOI] [PubMed] [Google Scholar]
- Irimia M, Blencowe BJ. Alternative splicing: decoding an expansive regulatory layer. Curr Opin Cell Biol. 2012;24(3):323–332. doi: 10.1016/j.ceb.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Kalsotra A, Cooper TA. Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet. 2011;12(10):715–729. doi: 10.1038/nrg3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow LT, Gelinas RE, Broker TR, Roberts RJ. An amazing sequence arrangement at the 5' ends of adenovirus 2 messenger RNA. Cell. 1977;12(1):1–8. doi: 10.1016/0092-8674(77)90180-5. [DOI] [PubMed] [Google Scholar]
- Gelinas RE, Roberts RJ. One predominant 5'-undecanucleotide in adenovirus 2 late messenger RNAs. Cell. 1977;11(3):533–544. doi: 10.1016/0092-8674(77)90071-X. [DOI] [PubMed] [Google Scholar]
- Berget SM, Moore C, Sharp PA. Spliced segments at the 5' terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci USA. 1977;74(8):3171–3175. doi: 10.1073/pnas.74.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF. Schroth GP. Burge CB: Alternative isoform regulation in human tissue transcriptomes. Nature; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302(5653):2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA. The sequence of the human genome. Science. 2001;291(5507):1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Xie T, Rowen L, Aguado B, Ahearn ME, Madan A, Qin S, Campbell RD, Hood L. Analysis of the gene-dense major histocompatibility complex class III region and its comparison to mouse. Genome Res. 2003;13(12):2621–2636. doi: 10.1101/gr.1736803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The MHC sequencing consortium. Complete sequence and gene map of a human major histocompatibility complex. Nature. 1999;401(6756):921–923. doi: 10.1038/44853. [DOI] [PubMed] [Google Scholar]
- Mallya M, Campbell RD, Aguado B. Transcriptional analysis of a novel cluster of LY-6 family members in the human and mouse major histocompatibility complex: five genes with many splice forms. Genomics. 2002;80(1):113–123. doi: 10.1006/geno.2002.6794. [DOI] [PubMed] [Google Scholar]
- Mallya M, Campbell RD, Aguado B. Characterization of the five novel Ly-6 superfamily members encoded in the MHC, and detection of cells expressing their potential ligands. Protein Sci. 2006;15(10):2244–2256. doi: 10.1110/ps.062242606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroncek DF, Caruccio L, Bettinotti M. CD177: A member of the Ly-6 gene superfamily involved with neutrophil proliferation and polycythemia vera. J Transl Med. 2004;2(1):8. doi: 10.1186/1479-5876-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvanese V, Mallya M, Campbell RD, Aguado B. Regulation of expression of two LY-6 family genes by intron retention and transcription induced chimerism. BMC Mol Biol. 2008;9:81. doi: 10.1186/1471-2199-9-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Magen A, Ast G. Different levels of alternative splicing among eukaryotes. Nucleic Acids Res. 2007;35(1):125–131. doi: 10.1093/nar/gkm529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F, Maquat LE. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr Opin Cell Biol. 2005;17(3):309–315. doi: 10.1016/j.ceb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Conti E, Izaurralde E. Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr Opin Cell Biol. 2005;17(3):316–325. doi: 10.1016/j.ceb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Jakobi R, Voss H, Pyerin W. Human phosvitin/casein kinase type II. Molecular cloning and sequencing of full-length cDNA encoding subunit beta. Eur J Biochem. 1989;183(1):227–233. doi: 10.1111/j.1432-1033.1989.tb14917.x. [DOI] [PubMed] [Google Scholar]
- Rodriguez FA, Contreras C, Bolanos-Garcia V, Allende JE. Protein kinase CK2 as an ectokinase: the role of the regulatory CK2beta subunit. Proc Natl Acad Sci USA. 2008;105(15):5693–5698. doi: 10.1073/pnas.0802065105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G, Reymond A, Dabbouseh N, Dermitzakis ET, Castelo R, Thomson TM, Antonarakis SE, Guigo R. Tandem chimerism as a means to increase protein complexity in the human genome. Genome Res. 2006;16(1):37–44. doi: 10.1101/gr.4145906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiva P, Toporik A, Edelheit S, Peretz Y, Diber A, Shemesh R, Novik A, Sorek R. Transcription-mediated gene fusion in the human genome. Genome Res. 2006;16(1):30–36. doi: 10.1101/gr.4137606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacu S, Yuan W, Kan Z, Bhatt D, Rivers CS, Stinson J, Peters BA, Modrusan Z, Jung K, Seshagiri S. Deep RNA sequencing analysis of readthrough gene fusions in human prostate adenocarcinoma and reference samples. BMC Med Genomics. 2011;4:11. doi: 10.1186/1755-8794-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoeud F, Kapranov P, Ucla C, Frankish A, Castelo R, Drenkow J, Lagarde J, Alioto T, Manzano C, Chrast J. Prominent use of distal 5' transcription start sites and discovery of a large number of additional exons in ENCODE regions. Genome Res. 2007;17(6):746–759. doi: 10.1101/gr.5660607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel-Morgenstern M, Lacroix V, Ezkurdia I, Levin Y, Gabashvili A, Prilusky J, Del Pozo A, Tress M, Johnson R, Guigo R. Chimeras taking shape: Potential functions of proteins encoded by chimeric RNA transcripts. Genome Res. 2012;22(7):1231–1242. doi: 10.1101/gr.130062.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolanos-Garcia VM, Fernandez-Recio J, Allende JE, Blundell TL. Identifying interaction motifs in CK2beta–a ubiquitous kinase regulatory subunit. Trends Biochem Sci. 2006;31(12):654–661. doi: 10.1016/j.tibs.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Kumar-Sinha C, Kalyana-Sundaram S, Chinnaiyan AM. SLC45A3-ELK4 chimera in prostate cancer: spotlight on cis-splicing. Cancer discovery. 2012;2(7):582–585. doi: 10.1158/2159-8290.CD-12-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gong M, Yuan H, Park HG, Frierson HF, Li H. Chimeric transcript generated by cis-splicing of adjacent genes regulates prostate cancer cell proliferation. Cancer discovery. 2012;2(7):598–607. doi: 10.1158/2159-8290.CD-12-0042. [DOI] [PubMed] [Google Scholar]
- Ackerman P, Glover CV, Osheroff N. Stimulation of casein kinase II by epidermal growth factor: relationship between the physiological activity of the kinase and the phosphorylation state of its beta subunit. Proc Natl Acad Sci USA. 1990;87(2):821–825. doi: 10.1073/pnas.87.2.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez F, Allende CC, Allende JE. Protein kinase casein kinase 2 holoenzyme produced ectopically in human cells can be exported to the external side of the cellular membrane. Proc Natl Acad Sci USA. 2005;102(13):4718–4723. doi: 10.1073/pnas.0501074102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams MD, Soares MB, Kerlavage AR, Fields C, Venter JC. Rapid cDNA sequencing (expressed sequence tags) from a directionally cloned human infant brain cDNA library. Nat Genet. 1993;4(4):373–380. doi: 10.1038/ng0893-373. [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C. The transcriptional landscape of the mammalian genome. Science. 2005;309(5740):1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Kawai J, Shinagawa A, Shibata K, Yoshino M, Itoh M, Ishii Y, Arakawa T, Hara A, Fukunishi Y, Konno H. Functional annotation of a full-length mouse cDNA collection. Nature. 2001;409(6821):685–690. doi: 10.1038/35055500. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Gerhardt DJ, Dinger ME, Crawford J, Trapnell C, Jeddeloh JA, Mattick JS, Rinn JL. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol. 2012;30(1):99–104. doi: 10.1038/nbt.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Kapranov P, Foissac S, Lagarde J, Reymond A, Ucla C, Wyss C, Drenkow J, Dumais E, Murray RR. Efficient targeted transcript discovery via array-based normalization of RACE libraries. Nat Methods. 2008;5(7):629–635. doi: 10.1038/nmeth.1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker JR, Bickmore WA, Barroso I, Pritchard JK, Gilad Y, Segal E. Genomics: ENCODE explained. Nature. 2012;489(7414):52–55. doi: 10.1038/489052a. [DOI] [PubMed] [Google Scholar]
- Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res. 2011;39(Database issue):D146–D151. doi: 10.1093/nar/gkq1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(Spec No 1):R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- Holt CE, Bullock SL. Subcellular mRNA localization in animal cells and why it matters. Science. 2009;326(5957):1212–1216. doi: 10.1126/science.1176488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136(4):719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38(Web Server issue):W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K. The Pfam protein families database. Nucleic Acids Res. 2010;38(Database issue):D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher P, Karplus K, Moeri N, Hofmann K. A flexible motif search technique based on generalized profiles. Comput Chem. 1996;20(1):3–23. doi: 10.1016/S0097-8485(96)80003-9. [DOI] [PubMed] [Google Scholar]
- Scordis P, Flower DR, Attwood TK. FingerPRINTScan: intelligent searching of the PRINTS motif database. Bioinformatics. 1999;15(10):799–806. doi: 10.1093/bioinformatics/15.10.799. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14(9):755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16(22):10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of all the nucleotide and protein sequences of the mRNAs described on this article with their corresponding names and accession numbers.
Table that contains all the different spliced isoforms detected in all the species studied in this work indicating whether they were previously described (deposited on databases) and, in the case they were found on databases the access number of the corresponding sequence or of the ESTs are indicated.