Abstract
Horizontal gene transfer is important in the evolution of bacterial and archaeal genomes. An interesting genetic exchange process is carried out by diverse phage-like gene transfer agents (GTAs) that are found in a wide range of prokaryotes. Although GTAs resemble phages, they lack the hallmark capabilities that define typical phages, and they package random pieces of the producing cell’s genome. In this Review, we discuss the defining characteristics of the GTAs that have been identified to date, along with potential functions for these agents and the possible evolutionary forces that act on the genes involved in their production.
The prevalence of horizontal gene transfer (HGT), in which DNA is exchanged between closely or distantly related lineages, has altered the way we think about evolution1,2, having profound effects on our views of the evolutionary relationships among living organisms. It has caused a shift from a bifurcating ‘tree of life’ view to a ‘net of life’ or ‘web of life’ view3–5, in which “highways of gene sharing” (REF. 6) represent major connections, with genes (or DNA segments) viewed as “public goods, available for all organisms to integrate into their genomes” (REF. 7). HGT occurs within and between all three domains of life, and it has been estimated that 0.05–80% of genes in bacterial and archaeal genomes have been affected by HGT, depending on the analytical method used and the genome analysed8–10. Although HGT is widespread, it is not random: patterns of preferential gene exchange among specific groups of organisms that share ancestry or habitat have been observed6,9,11,12. Such patterns are probably a consequence of existing barriers to HGT, which have been reviewed in detail elsewhere13,14.
Although we have known about some HGT mechanisms for decades, novel processes that expand the existing repertoire of gene exchange methods are continually being identified. Transformation was the first mechanism of gene exchange to be discovered15, wherein DNA is taken up directly from the environment (reviewed elsewhere16,17). In transduction, DNA transfer is mediated by viral particles (BOX 1). In conjugation, which was discovered in the 1940s18, DNA is transferred from a donor to a recipient cell during cell-to-cell contact; conjugation is used in the transfer of plasmids, transposons, integrons, and integrative and conjugative elements (ICEs)19–21. There are other modes of gene exchange that do not fit well into any of these three canonical mechanisms, including temporary cell fusion followed by chromosomal recombination and plasmid exchange22, intercellular connection through nanotubes23, and the release of membrane vesicles containing chromosomal, plasmid and phage DNA that can then merge with nearby cells24. In addition, a novel mechanism of gene transfer that has features of both membrane vesicle trafficking and viral infection was recently reported and dubbed serial transduction; in this mechanism, membrane vesicle-like particles from sea water containing large fragments of DNA were capable of transducing cells, and the recipient cells were then able to produce similar membrane vesicles25.
Box 1. Transduction: phages and transmission of cellular DNA.
During phage reproduction, particles are occasionally produced that contain DNA from the cellular genome as opposed to, or in addition to, the phage genome. This DNA can then be transferred to a subsequent host cell in a process named transduction92. There are two classes of transduction: specialized and generalized. In specialized transduction, a particular phage packages only a specific region of the host genome. This is exemplified by phage λ, which occasionally packages the gal (galactose metabolism) or bio (biotin metabolism) genes93,94 that are located adjacent to its integration site in the Escherichia coli chromosome. In generalized transduction, a phage packages a nonspecific portion of the host genome, as exemplified by E. coli phage P1 (REFS 95,96). Transposable phages, such as E. coli phage Mu97, package pieces of host DNA at the ends of the phage genome. For phage Mu, this results in ~100 bp of host DNA at one end of the phage genome and ~2 kb at the other end98, whereas for a Mu-like phage in Rhodobacter capsulatus, RcapMu, the particles contain ~30 bp of host DNA at one end and ~3 kb at the other99. The presence of such transposing phages in viral samples used for metagenomics would result in cellular genes being detected, as is common86.
Transduction may affect virulence in bacteria100–102. For example, in Vibrio cholerae the cholera toxin, a key virulence determinant, is encoded in the genome of the temperate phage ΦCTX103. As another example, Staphylococcus aureus pathogenicity islands (SaPIs) excise from the chromosome and replicate following induction by an infecting phage, and the SaPI DNA is packaged in modified phage particles and transferred to other S. aureus cells104,105 and even to species from other genera106.
Transduction has been documented in natural settings, including freshwater107, marine108, plant-associated109, rumen110, milk106 and wastewater111,112 environments. The wastewater studies are particularly revealing about the ability of phages to carry host genes, as these studies identify diverse bacterial 16S ribosomal DNA sequences in the viral component of the waste water111,112. Some phages are thought to carry cellular genes because of a selective advantage that is accrued by the phage host, as exemplified by the photosynthesis genes that are carried by cyanophages113. Many viral metagenomes contain a large proportion of sequences with homologues that are classified as cellular in GenBank86, but it is uncertain what proportion of these are homologues of prophage genes in bacterial genomes rather than homologues of cellular genes that are packaged by the phages. Because a variety of gene transfer agents have been identified in diverse prokaryotes, it is possible that some of the cellular DNA found in the viral fraction of natural environments is present in gene transfer agent particles28,76,77,114. At the present time, however, the reason for the occurrence of such a large number of cellular genes in viral metagenomes is not known.
Although some of these mechanisms might owe their existence to functions other than HGT, and have been recruited for HGT secondarily, the benefits of HGT probably contribute to their maintenance. Here, we review one gene exchange process, that mediated by gene transfer agents (GTAs), and discuss the possible origins and roles of these entities in the evolution of bacteria and archaea.
GTAs
A GTA is a phage-like entity with the following characteristics: it contains a random piece of the genome of the producing cell, and the amount of DNA that it contains is insufficient to encode the protein components of the particle itself. Therefore, a GTA particle does not necessarily contain any GTA-encoding genes, and it cannot transfer a complete set of structural GTA genes to a recipient cell. This is distinct from a generalized transducing phage, for which usually only an occasional particle contains host genes, and the fragments of packaged DNA are the size of the phage genome.
The production of GTAs by a cell is outlined and contrasted with the production of transducing phage particles in FIG. 1. The production of GTAs is not the result of a phage infection. Rather, the genes encoding the phage-like structure of the GTA particles are contained within the genome of the cell that produces the GTAs. The resultant GTA particles contain random pieces of the producing cell’s genome that are smaller than would be required to encode the GTA particle itself. Therefore, only the occasional particle will contain GTA-encoding genes, and even then they will not contain enough DNA to transfer the ability to produce a GTA to another cell. All known GTAs have tailed-phage structures and are presumably released into the environment by lysis of the producing cell. Released GTA particles may transfer DNA from the producing cell to a recipient cell. By contrast, transducing phage particles are produced during the replication of a phage inside a host cell, and only the occasional phage particle contains host DNA in place of phage DNA (FIG. 1). The amount of DNA in the transducing particles is equivalent to the normal phage genome size.
Figure 1. Comparison of gene transfer agent and transducing phage production.
a | The genes encoding the gene transfer agent (GTA) particles are located on the host chromosome, and their expression leads to the production of GTA particles (black). GTA genes have never been found to excise from the genome as part of GTA production. Random DNA segments from the producing cell are packaged in the particles (blue particle heads), and only the occasional particle contains GTA genes (red particle head). For all genetically characterized GTAs, the amount of DNA packaged is insufficient to encode the phage-like structure (as indicated by the small heads). It is presumed that GTAs require lysis (dashed line) for release from cells. b | In the production of transducing phages, phage or prophage genes within the host genome are expressed, resulting in the production of phage particles (black) and replication of the phage genome (not shown). Packaging of the complete phage genome then occurs (orange phage heads), with occasional packaging of non-phage DNA (blue phage head). Note that this is an over simplification and some transducing phage particles can contain both phage and cellular genomic DNA. Tailed phage structures require lysis to be released from cells.
Four genetically unrelated GTAs have been identified to date: RcGTA in the alphaproteobacterium Rhodobacter capsulatus (then called Rhodopseudomonas capsulata), Dd1 in the deltaproteobacterium Desulfovibrio desulfuricans, VSH-1 (virus of Serpulina hyodysenteriae) in the spirochaete Brachyspira hyodysenteriae (previously known as Serpulina hyodysenteriae), and voltae transfer agent (VTA) in the archaeon Methanococcus voltae. There are also some apparently functional GTAs that are related to RcGTA in other alphaproteobacteria. The various GTAs are discussed in detail below, along with several phage-like agents that display some properties of GTAs (BOX 2).
Box 2. Other gene transfer agent-like, but non-gene-transfering, elements.
In addition to the gene transfer agent (GTA) particles that have been identified in bacteria and archaea, there are several other phage-like elements that package small fragments of cellular genomic DNA, but these elements have not been documented to transfer DNA to recipient cells.
Phage-like particles, named bacteriophage-like particles (BLPs), are produced by multiple species of Bartonella, a genus in the Rhizobiales order (within the class Alphaproteobacteria) that appears to lack Rhodobacter capsulatus GTA (RcGTA)-like gene homologues28,115. The first BLP observed was associated with Bartonella bacilliformis116, and the first BLP to be characterized was from Bartonella henselae (then known as Rochalimaea henselae)117. The B. henselae particles contain seemingly random 14 kb pieces of the host genome, on the basis of the gel banding pattern after restriction endonuclease digestion, sequence analysis of cloned DNA from the particles and Southern blot hybridization117. Analysis of the DNA from the B. bacilliformis BLPs indicated that the genomic origin of the packaged DNA is biased to certain regions118. A more recent study used whole-genome microarrays to characterize DNA packaging by BLPs produced by Bartonella grahamii115. The 14 kb DNA fragments from the BLPs contained sequences from throughout the genome but were biased towards a high-plasticity region that is amplified within cells by run-off replication initiating at the BLP-encoding sequences115. Therefore, in respect to DNA packaging, BLPs have properties that are intermediate between a GTA and a specialized transducing phage.
Another phage-like particle with some similarities to GTAs is the defective phage PBSX of Bacillus subtilis119,120. The 28 kb PBSX gene cluster in the B. subtilis genome contains phage head, tail, lysis and lysogeny genes but no replication functions84,121. The particles appear to contain random ~13 kb pieces of the B. subtilis genome, but the activity of these particles is very different to that of GTAs or the Bartonella BLPs. These elements kill B. subtilis cells that do not carry the genes encoding the particles. This might be considered analogous to antibiotic or bacteriocin production — that is, a means of inhibiting competing cells in the environment — although in this case the producing cells presumably lyse to release the particles. Tailed phage particles have never been found to be released from cells without lysis, and it is difficult to imagine such a structure escaping from a bacterial or archaeal cell in a non-lytic event. The PBSX phenomenon is somewhat similar to the situation for GTAs, as the cell releasing the particles must die but the genes required for particle production have nonetheless been maintained in the host species.
The R. capsulatus GTA
The first GTA to be discovered was RcGTA, which was found in the purple photosynthetic bacterium R. capsulatus26. Co-culture of strains with different antibiotic resistance phenotypes yielded doubly resistant strains, and the mode of genetic exchange was determined to be similar to transduction in that it did not require cell–cell contact and was resistant to DNase26. However, an estimation of the size of the agent (based on the sedimentation rate) showed that it was unusually small compared with known transducing phages, and this led to the moniker gene transfer agent26. This was subsequently abbreviated to GTA27, and then became RcGTA when sequence analysis revealed that GTAs are present in other taxonomic groups28.
A detailed biochemical characterization found that RcGTA particles contain linear double-stranded DNA of a similar size to the ~5 kb linearized genome of simian virus 40 (SV40)29. An alternative to a small transducing phage-like particle could not be ruled out26,27,29 until the creation of an overproducer mutant strain enabled purification of sufficient material for the phage-like nature of RcGTA particles to be visualized by electron microscopy30 (FIG. 2a). RcGTA particles resemble a tailed phage, with a head of ~30 nm in diameter and a tail of ~50 nm in length30. The overproducer mutant also allowed the RcGTA-packaged DNA to be visualized by gel electrophoresis, yielding an ~4 kb band that produces a smear on the gel after treatment with a restriction endonuclease and has renaturation kinetics that are similar to those of R. capsulatus genomic DNA30. Hybridization of the DNA from RcGTA particles to an R. capsulatus whole-genome microarray confirmed that all of the genes in the R. capsulatus genome can be packaged within the particles, with no effect of genome position on packaging frequency31.
Figure 2. Electron micrographs of gene transfer agent particles.
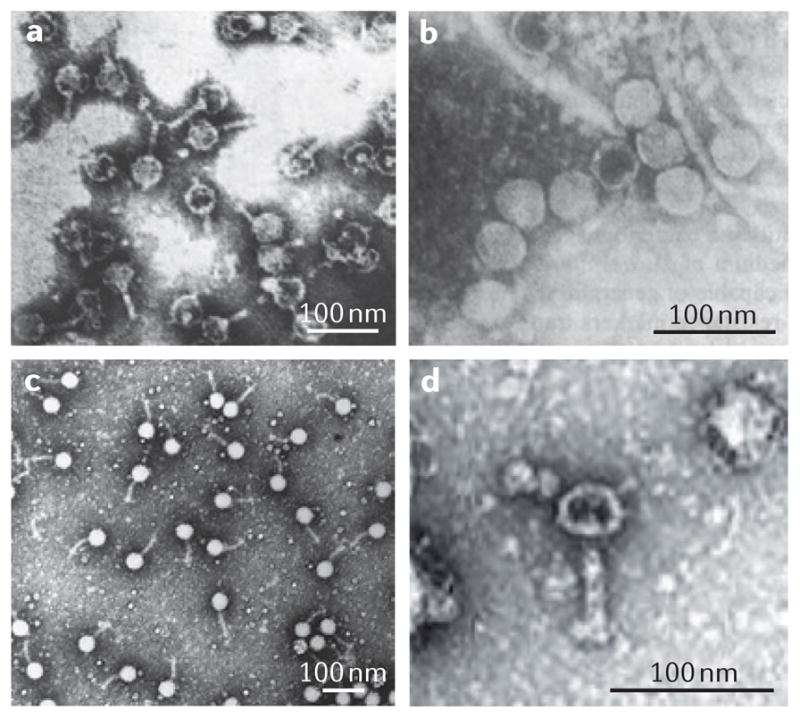
The estimated sizes of the particles are given in TABLE 1. a | Rhodobacter capsulatus gene transfer agents (RcGTAs)30. b | Dd1 particles from Desulfovibrio desulfuricans48. c | Virus of Serpulina hyodysenteriae (VSH 1) particles in Brachyspira hyodysenteriae50. d | A voltae transfer agent (VTA) particle in Methanococcus voltae67. Parts a,c and d are reproduced, with permission, from (respectively) REF. 30 © (1979) Elsevier; REF. 50 © (1997) American Society for Microbiology; and REF. 67© (1999) Society for General Microbiology. Part b is modified, with permission, from REF. 48 © (1987) US National Academy of Sciences.
The genes encoding the RcGTA particle were discovered 26 years after the first report of GTA-mediated genetic recombination in R. capsulatus32. The particle is encoded in an ~14 kb cluster of 15–17 apparently co-transcribed genes (FIG. 3a), and it is possible to make a plausible evolutionary connection between most of these genes and genuine phage genes28,32–34. Each RcGTA particle, which contains ~4 kb of DNA, carries only ~30% of the minimal number of base pairs needed to encode its structure. Proteomic analysis of RcGTA particles identified proteins that are not encoded in the RcGTA gene cluster35, indicating that additional RcGTA genes are located elsewhere in the R. capsulatus genome. Genetic experiments indicate that two of these genes, which are located ~670 kb away from the RcGTA cluster (FIG. 3a), encode proteins that are not essential for RcGTA activity but increase the ability of RcGTA particles to bind to R. capsulatus cells and transfer DNA, and are therefore likely to encode tail fibre proteins (A.S.L., unpublished observations). A putative endolysin, which is located ~1.2 Mb away from the RcGTA gene cluster, was recently found to be required for release of RcGTA particles from cells31.
Figure 3. Gene transfer agent-encoding gene clusters.
The general organizations of genes encoding the two characterized gene transfer agents (GTAs). Encoded protein functions are indicated above, according to the literature28,32,57 and our own analyses (A.S.L., unpublished observations), and ORF locus tags are indicated below. GTA encoding regions are in blue, and non-GTA-encoding regions are in green. Genes that have not yet been verified as having a role in GTA production are shown in white57. Arrows indicate the direction of transcription. a | The Rhodobacter capsulatus GTA (RcGTA) gene cluster in R. capsulatus. The archetypal RcGTA ORFs are labelled 1–15 (REF. 37). b | The virus of Serpulina hyodysenteriae (VSH 1) gene cluster in Brachyspira hyodysenteriae. McpB, methyl-accepting chemotaxis protein.
Shortly after the discovery of RcGTA, experiments on 33 independent R. capsulatus isolates showed that 16 of these isolates produce RcGTA and 25 are capable of receiving genetic information via RcGTA36; however, tests of numerous other species for interspecies transfer via RcGTA were unsuccessful36. These findings indicate that RcGTA is fairly specific in its host range. Four of the isolates that are incapable of transferring DNA36 were found to contain RcGTA genes on the basis of a Southern blot37, although two of these RcGTA gene clusters yielded banding patterns that were different to those of the other isolates after digestion with a restriction enzyme, and therefore contain sequence differences compared with the other clusters. This means that even the R. capsulatus isolates that are incapable of gene transfer contain RcGTA genes, in agreement with the ubiquitous presence of RcGTA genes in members of the order Rhodobacterales.
Early work showed that the production of RcGTA is strongly dependent on the phase of host cell growth, with increased production in stationary phase27. This results from increased transcription of the RcGTA genes in the late phases of growth32,38 owing to modulation by a quorum sensing system39,40. A cellular phosphorelay involving the R. capsulatus response regulator CtrA is also involved, as loss of this response regulator causes a complete loss of transcription of RcGTA structural genes32,38. However, the exact mechanism of CtrA-mediated regulation remains to be determined41. The control of RcGTA gene expression by cellular systems is consistent with the absence of RcGTA induction by treatment with the DNA-damaging agents mitomycin C or ultraviolet light; such induction would indicate that transcription of the RcGTA cluster is controlled by a phage repressor protein, which is not the case.
GTAs in other alphaproteobacteria
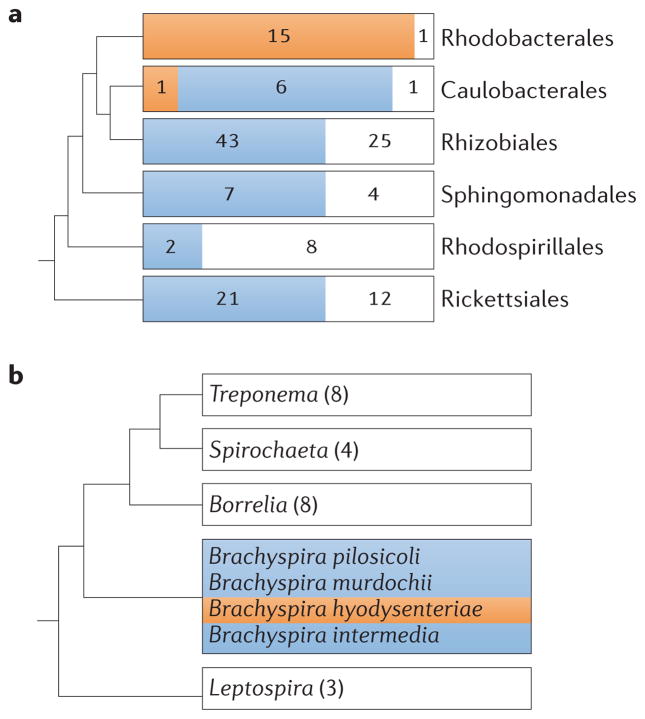
Soon after the RcGTA gene cluster was identified, it became apparent that RcGTA-like genes are widespread in alphaproteobacteria. An RcGTA-like gene cluster was first identified in another purple photosynthetic bacterium, Rhodopseudomonas palustris37, followed by clusters in Rhodobacter sphaeroides, Caulobacter crescentus, Agrobacterium tumefaciens and Brucella melitensis42, and then other species as additional complete genome sequences became available28,43,44. To date, RcGTA-like genes have been found in all taxonomic orders that are represented by complete genome sequences in the class Alphaproteobacteria (FIG. 4a), which suggests that a GTA gene cluster was present in the last common ancestor of this class and subsequently lost in some lineages (discussed below).
Figure 4. Distribution of gene transfer agent genes in alphaproteobacteria and spirochaetes.
a | The presence of gene transfer agent (GTA) genes within the alphaproteo-bacterial orders is shown. The number of genomes with a complete set of the archetypal 15 genes (orange), with at least one homologue of any of these 15 genes (blue) or lacking any detectable homologues (white) is indicated. We cannot exclude the possibility that, in genomes with only a few GTA gene homologues, the genes represent true prophages and not GTA genes. However, in several closely examined genomes, the evidence is in favour of true GTA gene homologues28,43. The BLASTP (protein basic local alignment search tool) similarity searches were carried out on 146 alphaproteobacterial genomes listed as complete in the GenBank database in October 2011, using Rhodobacter capsulatus GTA (RcGTA) ORFs 1–15 (see FIG. 3) as queries and retaining matches with E values of <10−4. The phylogenetic relationships between orders are based on previous analysis of alphaproteobacteria75. b | The distribution of virus of Serpulina hyodysenteriae (VSH 1) genes within the order Spirochaetales is represented on a phylogenetic tree. VSH 1 is produced by Brachyspira hyodysenteriae (orange). Homologues of the genes encoding VSH 1 are found only within other members of the Brachyspira genus (blue), but the organization of these genes is not syntenic to that in B. hyodysenteriae62. White indicates genera in the order that do not contain known VSH 1 gene homologues. This simplified tree is based on a 16S ribosomal RNA gene phylogeny of strains with completed genome sequences, rooted with sequences from alpha-proteobacteria. The number of complete genome sequences within each genus available for analysis is indicated.
Beyond conservation of RcGTA-like gene clusters, there is evidence of GTA production in several members of the Rhodobacterales. Ruegeria pomeroyi (previously known as Silicibacter pomeroyi) contains a complete RcGTA-like gene cluster and is capable of gene transfer that is attributed to this cluster43. Tail-less particles varying in size from 50 nm to 70 nm have been observed in this species43, and the R. pomeroyi genome seems to be free of other prophage sequences43. Two other marine Rhodobacterales species, Roseovarius nubinhibens and Ruegeria mobilis, have also been shown to produce particles classified as GTAs45. When these particles were added to natural microbial communities, the transfer rates of antibiotic resistance markers were remarkable — as much as 106-fold higher than previous estimates of transformation and transduction rates in natural environments45. Equally striking was the finding that the gene transfer events occurred between bacteria in different phyla, and it appears that none of the recipients recovered was an alphaproteobacterium. In addition, the rates of transfer to heterologous recipients were higher than those observed for transfer to the original, marker-less GTA donor in laboratory culture experiments45. The R. nubinhibens GTAs were reported to be tail-less particles with an ~50 nm diameter, in contrast to R. mobilis particles, which have heads of ~25 nm in diameter and tails of ~40 nm in length45. As was the case in the work on R. pomeroyi GTAs43, no biochemical or genetic analysis was carried out for the R. nubinhibens GTA particles, nor was there a demonstration that GTA genes are required for the observed effects. Such experiments are needed to validate the proposed role of GTAs in the extraordinarily high frequencies of gene transfer reported.
The presence of GTA capsid proteins has been confirmed in multiple lineages within the order Rhodobacterales, from a range of geographically separated marine environments46. The possibility of widespread conservation of GTA production in this particular group of bacteria is especially interesting because roseobacters (within the order Rhodobacterales) appear to account for >25% of the total bacterial and archaeal community in some marine environments47. Therefore, GTAs could make up a sizable portion of the viral community in places where the abundance of Rhodobacterales members is high.
The D. desulfuricans GTA
The GTA of D. desulfuricans, Dd1, resembles a tailed phage48, in this case a podovirus (a capsid with a very short tail; FIG. 2b). Dd1 transfers multiple types of marker, and purification of the DNA from Dd1 particles showed it to be a 13.6 kb double-stranded DNA that produces a smear after treatment with a restriction endonuclease. Like RcGTA, Dd1 is not induced after treatment with the DNA-damaging agent mitomycin C.
There are several possible prophage regions in the genome of D. desulfuricans subsp. desulfuricans str. ATCC 27774, the strain that produces Dd1. One of these regions, ORFs Ddes_0706–Ddes_0726 (encompassing ~17.8 kb), contains several genes that are predicted to encode homologues of structural phage proteins, and of podoviral proteins in particular. This is the only prophage region in this genome in which we can identify multiple clear homologues of podoviral proteins, and it is possible that this cluster encodes Dd1.
The B. hyodysenteriae GTA
B. hyodysenteriae GTA particles have a head diameter of 45 nm and a tail length of 64 nm49 (FIG. 2c), and contain 7.5 kb of linear double-stranded DNA50. The element is named VSH-1, and the packaged DNA is a random collection of fragments from the genome of the producing cell50. VSH-1 mediates the transfer of many markers between B. hyodysenteriae cells50–53. Although mitomycin C is required to observe the particles49,50, DNA fragments matching those packaged by VSH-1 have been found in B. hyodysenteriae that has not been treated with mitomycin C54,55, and co-culture experiments have confirmed that VSH-1 can be produced in the absence of an exogenous inducer52,53. Two important differences between VSH-1 and other GTAs are the fact that VSH-1 production can be induced with mitomycin C and the fact that detectable cell lysis is associated with particle release49 (discussed further below).
The genes in the B. hyodysenteriae genome that are needed for production of VSH-1 (REFS 56,57) (FIG. 3b) include a gene that is similar to a putative phage endolysin and is located next to the region encoding the VSH-1 structure. The protein encoded by this gene degrades B. hyodysenteriae peptidoglycan56, and a putative holin-encoding gene has also been identified close by56. One of the proteins associated with VSH-1 did not correspond to any of the genes that were originally mapped56, and subsequent work identified a separate cluster of three contiguous VSH-1 tail genes located a short distance away57 (FIG. 3b).
Transcription studies and an analysis of putative regulatory sequences indicate that the two VSH-1 gene clusters are co-regulated57. The larger ‘head–tail–lysis’ gene cluster appears to be transcribed from a single promoter, because transcripts spanning neighbouring sequences have been identified for all of the genes in this cluster, although the mRNA sizes indicate that post-initiation processes affect the final mRNA structures58. Quantitative measurements of gene copy number after the induction of VSH-1 production do not reveal any bias for packaging of VSH-1 genes relative to non-VSH-1 genes58, although VSH-1-specific transcription increases by >200-fold after induction57.
Particles that are similar in size and shape to VSH-1 have been observed in other studies of B. hyodysenteriae and other species of Brachyspira49,59–61. VSH-1 gene distribution, using the current VSH-1-encoding region56,57 (FIG. 3b), appears to be limited to members of the Brachyspira genus (FIG. 4b), with gene organization varying among species62.
B. hyodysenteriae is a spirochaete pathogen that causes swine dysentery. It has been suggested that VSH-1 plays a part in the genomic and population structures and the pathogenic properties of Brachyspira spp.63,64, and VSH-1 does transfer virulence and antibiotic resistance genes between cells50–53. VSH-1 is induced by treatment of Brachyspira spp. with hydrogen peroxide58 and the antibiotics carbadox and metronidazole53 (antibiotics that are commonly used in the swine industry53). VSH-1-like GTAs are likely to be produced by spirochaetes that infect humans49,59,60,65, indicating that these elements could affect the virulence of human pathogens.
The M. voltae GTA
GTAs are not restricted to the bacterial domain, as VTA was discovered as a mediator of genetic exchange in the archaeon M. voltae66. VTA particles contain 4.4 kb of double-stranded DNA that appears to be randomly packaged from the genome of the producing cell, although there is some evidence for partial enrichment of one genomic region66. The particles have a tailed phage structure, with a head of 40 nm in diameter and a tail of 61 nm in length67 (FIG. 2d). Tailed phages are uncommon in archaea68. One region of the M. voltae genome, over a span of ~12 genes (ORFs Mvol_0401–Mvol_0414), contains several matches to bacteriophage sequences, and siphoviruses in particular. This region was previously named Mvol-Pro1 and hypothesized to encode the VTA structure69.
Transfer of packaged DNA into recipient cells
Because most GTAs are tailed phage-like particles, it is likely that GTAs bind to recipient cells via specific tail–receptor interactions, but the receptor has not been identified for any of the GTAs. There are several possible fates for the packaged DNA after release into the cytoplasm, including degradation by cellular nucleases, which would block gene transfer. To be maintained in the cell and in its progeny, the DNA needs to be incorporated into the host cell genome by recombination, as discussed below.
DNA recombinase A (RecA)-type homologous recombination is thought to be needed for a recipient cell to integrate DNA from an RcGTA particle70. Furthermore, when RcGTA is used to replace a chromosomal allele with a marked disruption of a cloned gene, the marker is invariably integrated at the expected location. One study demonstrated that eight of 33 tested R. capsulatus strains were incapable of receiving a gene via RcGTA36. This study assessed the transfer of rifampicin resistance, which occurs through mutations in rpoB (encoding DNA-directed RNA polymerase subunit-β), a gene that should not vary greatly among strains. It is therefore unlikely that the failure of transfer was caused by inadequate sequence similarity between the donor and recipient genes. Instead, the results may reflect strain-to-strain variation in the restriction–modification systems that degrade incoming DNA.
The frequency of homologous recombination between incoming DNA and a chromosome decreases as identity between the two DNA regions decreases. For illegitimate recombination, in which the incoming DNA is not homologous to the recipient DNA, the frequency is expected to be orders of magnitude lower than the frequency for homologous sequences, especially for linear fragments as opposed to circular molecules, as reviewed elsewhere14. In silico analyses of gene transfer patterns indicate that HGT is much more frequent between closely related organisms than between distantly related ones9,11,71. This is consistent with the idea that GTAs have played an important part in creating such HGT patterns, as GTA-mediated HGT requires specific interactions between the host and the GTA particle for successful attachment and injection of the DNA72, as well as DNA sequence similarity to facilitate incorporation of a transferred allele by homologous recombination14.
Origins of GTAs
There are indisputable connections between the known GTAs and tailed phages, including structural similarities with established phage morphotypes (siphoviruses and podoviruses) for all four GTAs, and conservation of gene organization and sequence similarity for phages and the two genetically characterized GTAs (RcGTA and VSH-1). Broadly outlined, three evolutionary scenarios can explain the shared evolutionary connections between GTAs and phages: GTAs descended from phages multiple independent times; GTAs came first and subsequently gave rise to ‘selfish’ tailed phages; or extinct, or undiscovered, elements led independently to GTAs and phages. In the following sections, we discuss the evidence relating to past evolutionary events that led to GTAs.
Relationships between GTAs and phages
The RcGTA and VSH-1 gene clusters have overall synteny with the gene organization for double-stranded-DNA tailed phages28,32,57, although there appear to have been rearrangements resulting in non-contiguous tail and tail fibre genes (FIG. 3). Unlike RcGTA genes in the order Rhodobacterales, VSH-1 genes are present in only one genus of the order Spirochaetales (the genus Brachyspira) (FIG. 4), and the order of VSH-1 genes in different Brachyspira spp. is not conserved62. This might reflect a more recent origin of VSH-1 in this genus relative to RcGTA in alphaproteobacteria. Indeed, assuming that GTAs arose from phages, the induction of VSH-1 by DNA-damaging agents and the large-scale lysis of host cells on VSH-1 induction indicate that this GTA is still under a phage-like control, as opposed to the apparently purely cellular control of RcGTA production. Although the composition and arrangement of VTA and Dd1 gene clusters has not been confirmed, we note that the putative sets of genes (from inspection of the genome sequences; see above) resemble variants of phage head–tail gene clusters.
Several RcGTA genes show clear evolutionary relationships to genuine phage sequences, as was initially most obvious for the DNA packaging and head assembly region (FIG. 3, genes 2–5)28,32. More recently, phages ΦJL001 and RDJLΦ1, which infect marine Rhizobiales str. JL001 (REF. 34) and Roseobacter denitrificans33, respectively, were found to have genes with significant similarity to four RcGTA genes that are predicted to encode tail proteins (FIG. 3, genes 12–15). In alphaproteobacteria and betaproteobacteria, prophages were identified that also contain sequences which are homologous to these four RcGTA genes, and the prophage genes have greater sequence identity to each other than to the RcGTA genes33. Unfortunately, few genome sequences are available for phages that infect alphaproteobacteria (of the ~650 phage sequences present in GenBank in December 2011, only seven are for alphaproteobacterium-infecting phages), but perhaps as more genome sequences are determined for these phages there will be new insights into the origins of RcGTA-like elements.
The connections between RcGTA genes and phage (or prophage) genes indicate that these elements have a long and tangled evolutionary history73. Initial speculation on the origins of RcGTA30 led to the suggestion that this GTA represents a defective phage or arose from a phage progenitor30. However, the notion that GTAs predate phages is not substantiated by the current evidence.
For all four documented GTAs, it appears that the DNA packaged by the particles is smaller in size than the particle- encoding structural genes (TABLE 1). Only Dd1, which has a podoviral structure, packages DNA (13.6 kb) comparable to the known size range packaged by phages with the same structure (for example, Mycoplasma phage P1, a podovirus, has a genome of ~11.7 kb74). If GTAs evolved from phages, this would indicate that mutations occurred which resulted in smaller packaging capacities than those of the hypothetical ancestral phages.
Table 1.
Properties of gene transfer agents and related elements
| Species | Element | Carries out gene transfer? | Particle structure | Head diameter | Tail length | Size of packaged DNA | Encoding DNA | References |
|---|---|---|---|---|---|---|---|---|
| Rhodobacter capsulatus | RcGTA | Yes | Siphovirus | 30 nm | 50 nm | 4.4 kb | 14.1 kb, 1.2 kb | 29.30,32 (A.S.L., unpublished observations) |
| Desulfovibrio desulfuricans | Dd1 | Yes | Podovirus | 43 nm | 7 nm | 13.6 kb | 17.8 kb* | 48 |
| Brachyspira hyodysenteriae | VSH 1 | Yes | Siphovirus | 45 nm | 64 nm | 7.5 kb | 16.3 kb, 3.6 kb | 49,50,56,57 |
| Methanococcus voltae | VTA | Yes | Siphovirus | 40 nm | 61 nm | 4.4 kb | 14.2 kb* | 66,67,69 |
| Bartonella spp. | BLP | No | Unclassified | 40 nm | No tail observed | 14 kb | 32 kb | 115,117 |
| Bacillus spp. | PBSX | No | Myovirus | 41 nm | 190 nm | 13 kb | 28 kb | 119,121 |
BLP, bacteriophage like particle; GTA, gene transfer agent; RcGTA, Rhodobacter capsulatus GTA; VSH 1, virus of Serpulina hyodysenteriae. *The region proposed as possibly encoding the GTA has not been experimentally demonstrated.
On the antiquity of GTAs
Homologues of the genes encoding the RcGTA structure are found throughout alphaproteobacteria as isolated genes, partial gene clusters and complete RcGTA- like gene clusters28,43,44 (FIG. 4a). Two analyses have shown that GTA genes form clusters which correspond to the established taxonomic alphaproteobacterial orders28,43 (some discrepancies observed in these analyses reflect incorrect taxonomic assignments; for example, the genera Oceanicaulis and Maricaulis were recently reclassified75). The simplest explanation of this distribution of GTA genes is that a single RcGTA-like element was present in the last common ancestor of the alphaproteobacteria, and that there was subsequent loss in some lineages28. An alternative scenario would require multiple HGT events or multiple instances of independent GTA gain and loss. Recent GTA-mediated propagation of GTAs between the alphaproteobacterial orders is unlikely owing to the limited host range of well-characterized GTAs and their inability to package a complete GTA gene cluster. It is conceivable that a GTA ancestor had a greater packaging capacity and, therefore, that GTA-mediated transfer of GTA clusters occurred in the past. GTA propagation via other HGT mechanisms is not out of the question, and biased gene transfer11 cannot be ruled out as the cause of the observed relationships.
An undiscovered widespread distribution for GTAs?
For the four substantiated GTAs, none of the GTA genes (or putative GTA genes, in the case of Dd1 and VTA) is recognizably similar to any unrelated GTA gene, on the basis of BLAST (basic local alignment search tool) searches. Also, RcGTA, VSH-1 and VTA have sipho-viral structures, whereas Dd1 has a podoviral structure (TABLE 1). Therefore, it appears that these four GTAs arose independently. However, the large amount of cellular and viral metagenomic sequence without recognizable similarity to any known genes is indicative of our limited ability to uncover structural elements of viral genomes using similarity searches alone. It is possible that these GTAs in four distantly related groups of prokaryotes (alphaproteobacteria, deltaproteobacteria, spirochaetes and euryarchaeotes) represent only the tip of the iceberg of GTA diversity76–78.
Functions of GTAs
The GTAs discussed in this Review were discovered because of their gene transfer activity. So, is their production beneficial for the host organisms? Are GTAs maintained mainly for DNA repair, or do they confer additional advantages, such as facilitating the spread of novel traits through a population or providing DNA for nutrition? It has been speculated that, under some conditions, a partial diploid state resulting from GTA-dependent transduction enhances cell survival78. Although such questions remain unanswered, we discuss some of them below and review intriguing recent suggestions regarding the role of GTAs in evolution.
Maintenance of GTAs: gene or kin selection?
The well-documented GTAs all have tailed-phage structures, and there is no plausible mechanism other than lysis to explain their release from producing cells. Although the cell releasing the GTA particles may die, the recipient cell (or cells) could benefit from an allele acquired by GTA transfer. From an evolutionary standpoint, this is compatible with a microbial social behaviour79,80. Are GTAs public goods and a means of altruistic cooperation between related microorganisms, such that the host range specificity and quorum sensing-mediated regulation of GTAs result in kin discrimination? There are known examples of evolutionarily stable cooperation between related microorganisms via the production of public goods81. Intercellular homologous recombination in a population of cells, facilitated by GTAs, could be beneficial in certain circumstances82. Although this possibility requires further exploration, it may explain the maintenance of costly (that is, deadly for the donor cell) GTA production: selection on a population level would impose purifying selection on the GTA genes in individual genomes (see below).
An alternative possibility is that a special type of gene-level selection, known as selective conversion83, acts on GTA gene clusters independently of organism- or population-level selection. The mathematical model that was developed to study DNA secretion and transformation83, another costly form of genetic exchange, predicts that there are special conditions under which such a mode of selection is possible. Whether a similar model fits GTA maintenance and production remains to be tested.
Evidence for purifying selection acting on GTA genes
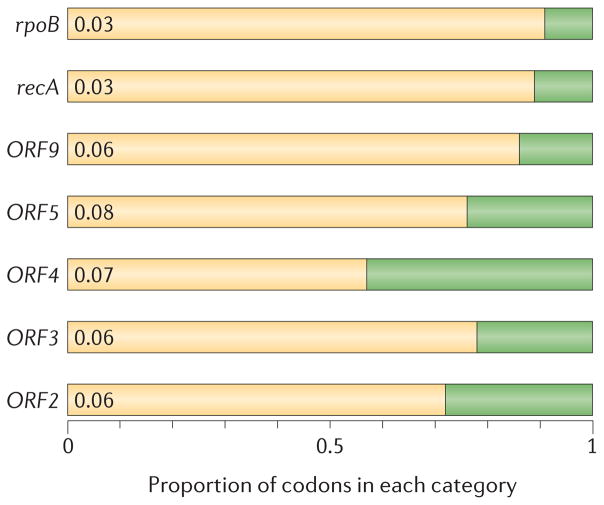
Bacterial and archaeal genomes evolve under a strong deletion bias that sees unnecessary genes being inactivated through mutation and then lost, as exemplified by rapidly degrading regions containing prophages84. If GTAs provide a selective advantage, the GTA genes would be under selection against mutations that cause amino acid changes, to prevent inactivation of function. For members of the order Rhodobacterales, in which RcGTA-like clusters comprise the archetypal ~15 genes in most completely sequenced genomes, analysis of the ratio of non-synonymous-to-synonymous amino acid substitutions (dN/dS, or ω) shows that the majority of codons in the GTA genes are indeed under purifying selection (as indicated by an ω value of <1 in FIG. 5). Although the selection pressure is not as strong as for two housekeeping genes, recA and rpoB, which both have an ω value of 0.03 in most of their codons, other functionally conserved genes in bacterial genomes have ω values that are comparable to those of the RcGTA genes85. Therefore, it appears that there is selective pressure for the functional conservation of these GTA genes in members of the order Rhodobacterales.
Figure 5. Evidence of purifying selection in selected gene transfer agent genes in the Rhodobacterales order of the class Alphaproteobacteria.
The ratio of non synonymous to synonymous (dN/dS, or ω) codon substitution rates can be used as an indication of the selective pressure acting on a protein-coding gene. If a gene (or gene segment) is under purifying selection, non-synonymous mutations (that is, mutations resulting in a change of the encoded amino acid) will be selected against, resulting in ω < 1. In neutrally evolving proteins, synonymous and non-synonymous mutations will be fixed at approximately the same rate, resulting in ω = 1. Because different regions of a protein-coding gene may be under different selective constraints, codons can be divided into two categories: those with ω < 1 (yellow; the estimated ω is indicated on the bar) and those with ω fixed to 1 (green); the estimated proportion of codons in each category is given on the horizontal axis. Using this model (known as the M1a model122), we estimated ω values for 15 GTA genes in members of the order Rhodobacterales (on the basis of alignments of homologues in 48 completed and draft genomes). The majority of the codons in all GTA genes are under purifying selection (only some genes are shown; ORF numbers are as given in FIG. 3). The M1a model fits significantly better than either a model with ω fixed to 1 across all codons or a model (M0) in which all codons have the same ω value (data not shown). Predictions for the housekeeping genes rpoB (encoding DNA-dependent RNA polymerase subunit-β) and recA (encoding DNA recombinase A), which are expected to be under strong purifying selection, are shown for comparison.
GTAs and ‘cloud’ genes
Among the numerous surprises from metagenomics is the enormous diversity of genes in the virosphere that are considered to be non-viral86. In viral metagenomes, 70% of genes do not have recognizable homologues in GenBank86, and this vast unknown sequence space has been dubbed dark matter, a term that was originally applied to phages87. A recent analysis of viral and cellular metagenomic data showed that the viral metagenome contains an apparently random collection of cellular genes76. Metabolic and information-processing cellular genes exist in phage genomes86 but are a minor component of the viral pan-genome. In addition to genes with known functions, a substantial fraction (~29%) of the typical bacterial genome consists of poorly conserved ‘accessory’, or ‘cloud’, genes88. These observations led to the hypothesis that the viral metagenome is dominated not by functional viruses but by virus-like entities (such as GTAs) that contain cellular genome fragments and, as further speculation, that GTAs have a bias towards packaging cloud genes. For the currently verified GTAs, no packaging bias has been observed (as discussed above), and we would therefore expect cloud genes to appear in GTA particles at the frequency of their presence in a donor genome. Whether a GTA metagenome consists predominantly of cloud genes owing to post-release selection pressure or to other unknown biases, or whether the skewed gene distribution in viral metagenomes is due to technical biases, is a conundrum.
GTAs and the origins of eukaryotes
One important milestone of eukaryogenesis was the symbiosis of a proto-eukaryotic cell with an alphaproteobacterium to give rise to mitochondria. However, molecular analyses of eukaryotic genomes reveal that many nuclear genes have a complex ancestry and some have possible origins in a variety of bacterial and archaeal groups89,90. These puzzling discoveries led to an intriguing hypothesis91 based on suggestions that the GTA system in alpha-proteobacteria is ancient28, and may therefore have been active at the time of this alphaproteobacterium– proto-eukaryote symbiosis, and is also capable of delivering alphaproteobacterial genes to a wide range of recipients45. Armed with these assumptions, it was speculated that rampant GTA transfer activities contributed to the complex evolutionary histories of nuclear genes91 because GTA-mediated transfer introduced alphaproteobacterial genes into a variety of hosts, including the proto-eukaryotic cell itself, both before and after the symbiosis. If the assumptions of this hypothesis hold true, ancient GTA-mediated transfers may make deciphering the order of events in past evolutionary phenomena, such as eukaryogenesis, even more daunting.
Concluding remarks
Gene transfer between cells is a powerful evolutionary force. One interesting mode of gene transfer is carried out by GTAs, which are phage-like elements that package small segments of the genome of a GTA-producing cell and transmit these genes throughout the environment. For at least one GTA, there is compelling evidence that the production of the GTA is a specifically maintained and controlled cellular function. GTAs could also be a previously unrecognized important component of the global viral community.
Much remains to be elucidated about even the best understood GTAs. For RcGTA of R. capsulatus, several regulatory genes and induction signals remain unknown; it is not clear whether we have identified all of the genes that are needed to assemble the complete particle; the nature of attachment to a recipient cell is a mystery; the genuine host range, restriction barriers aside, is unclear; although plausible evolutionary pathways and forces have been suggested, much about this is conjecture; and we have no clear idea how abundant RcGTA-like particles are in natural environments. These questions also apply to other GTAs, and we look forward to exciting discoveries in the field of GTA research in the near future.
Acknowledgments
This work is supported by funding from the National Science and Engineering Research Council of Canada and the Newfoundland and Labrador Research and Development Corporation to A.S.L.; by West Virginia University (Morgantown, USA) startup funds to O.Z.; and by the Canadian Institutes of Health Research to J.T.B. The authors thank R. T. Papke and J. P. Gogarten for stimulating discussions, and F. Eiserling, B. Marrs, T. Stanton and J. Wall for permission to use their electron microscopy images in figure 2. The authors regret their inability to cite all relevant research literature owing to space constraints.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
FURTHER INFORMATION
Andrew S. Lang’s homepage: http://www.mun.ca/biology/aslang/index.php
Olga Zhaxybayeva’s homepage: http://carrot.mcb.uconn.edu/~olgazh/
J. Thomas Beatty’s homepage: http://www.microbiology.ubc.ca/Beatty
GenBank: http://www.ncbi.nlm.nih.gov/genbank/
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Gogarten MB, Gogarten JP, Olendzenski L, editors. Horizontal Gene Transfer: Genomes in Flux. Humana Press; 2009. [Google Scholar]
- 2.Zhaxybayeva O, Doolittle WF. Lateral gene transfer: a primer. Curr Biol. 2011;21:R242–R246. doi: 10.1016/j.cub.2011.01.045. [DOI] [PubMed] [Google Scholar]
- 3.Dagan T, Martin W. Getting a better picture of microbial evolution en route to a network of genomes. Phil Trans R Soc B. 2009;364:2187–2196. doi: 10.1098/rstb.2009.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olendzenski L, Gogarten JP. Evolution of genes and organisms: the tree/web of life in light of horizontal gene transfer. Ann NY Acad Sci. 2009;1178:137–145. doi: 10.1111/j.1749-6632.2009.04998.x. [DOI] [PubMed] [Google Scholar]
- 5.Puigbo P, Wolf YI, Koonin EV. The tree and net components of prokaryote evolution. Genome Biol Evol. 2010;2:745–756. doi: 10.1093/gbe/evq062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beiko RG, Harlow TJ, Ragan MA. Highways of gene sharing in prokaryotes. Proc Natl Acad Sci USA. 2005;102:14332–14337. doi: 10.1073/pnas.0504068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McInerney JO, Pisani D, Bapteste E, O’Connell MJ. The public goods hypothesis for the evolution of life on earth. Biol Direct. 2011;6:41. doi: 10.1186/1745-6150-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lukjancenko O, Wassenaar TM, Ussery DW. Comparison of 61 sequenced Escherichia coli genomes. Microb Ecol. 2010;60:708–720. doi: 10.1007/s00248-010-9717-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhaxybayeva O, Gogarten JP, Charlebois RL, Doolittle WF, Papke RT. Phylogenetic analyses of cyanobacterial genomes: quantification of horizontal gene transfer events. Genome Res. 2006;16:1099–1108. doi: 10.1101/gr.5322306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakamura Y, Itoh T, Matsuda H, Gojobori T. Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nature Genet. 2004;36:760–766. doi: 10.1038/ng1381. [DOI] [PubMed] [Google Scholar]
- 11.Andam CP, Williams D, Gogarten JP. Biased gene transfer mimics patterns created through shared ancestry. Proc Natl Acad Sci USA. 2010;107:10679–10684. doi: 10.1073/pnas.1001418107. This paper highlights the challenge of distinguishing the patterns of HGT and of vertical inheritance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kloesges T, Popa O, Martin W, Dagan T. Networks of gene sharing among 329 proteobacterial genomes reveal differences in lateral gene transfer frequency at different phylogenetic depths. Mol Biol Evol. 2011;28:1057–1074. doi: 10.1093/molbev/msq297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Popa O, Dagan T. Trends and barriers to lateral gene transfer in prokaryotes. Curr Opin Microbiol. 2011;14:615–623. doi: 10.1016/j.mib.2011.07.027. [DOI] [PubMed] [Google Scholar]
- 14.Thomas CM, Nielsen KM. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nature Rev Microbiol. 2005;3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 15.Griffith F. The significance of pneumococcal types. J Hyg (Lond) 1928;27:113–159. doi: 10.1017/s0022172400031879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen I, Dubnau D. DNA uptake during bacterial transformation. Nature Rev Microbiol. 2004;2:241–249. doi: 10.1038/nrmicro844. [DOI] [PubMed] [Google Scholar]
- 17.Claverys JP, Martin B, Polard P. The genetic transformation machinery: composition, localization, and mechanism. FEMS Microbiol Rev. 2009;33:643–656. doi: 10.1111/j.1574-6976.2009.00164.x. [DOI] [PubMed] [Google Scholar]
- 18.Lederberg J, Tatum EL. Gene recombination in Escherichia coli. Nature. 1946;158:558. doi: 10.1038/158558a0. The discovery of conjugation, which ushered in the field of bacterial and archaeal genetics. [DOI] [PubMed] [Google Scholar]
- 19.Chen I, Christie PJ, Dubnau D. The ins and outs of DNA transfer in bacteria. Science. 2005;310:1456–1460. doi: 10.1126/science.1114021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mazel D. Integrons: agents of bacterial evolution. Nature Rev Microbiol. 2006;4:608–620. doi: 10.1038/nrmicro1462. [DOI] [PubMed] [Google Scholar]
- 21.Wozniak RA, Waldor MK. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nature Rev Microbiol. 2010;8:552–563. doi: 10.1038/nrmicro2382. [DOI] [PubMed] [Google Scholar]
- 22.Rosenshine I, Tchelet R, Mevarech M. The mechanism of DNA transfer in the mating system of an archaebacterium. Science. 1989;245:1387–1389. doi: 10.1126/science.2818746. [DOI] [PubMed] [Google Scholar]
- 23.Dubey GP, Ben-Yehuda S. Intercellular nanotubes mediate bacterial communication. Cell. 2011;144:590–600. doi: 10.1016/j.cell.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 24.Mashburn-Warren LM, Whiteley M. Special delivery: vesicle trafficking in prokaryotes. Mol Microbiol. 2006;61:839–846. doi: 10.1111/j.1365-2958.2006.05272.x. [DOI] [PubMed] [Google Scholar]
- 25.Chiura HX, Kogure K, Hagemann S, Ellinger A, Velimirov B. Evidence for particle-induced horizontal gene transfer and serial transduction between bacteria. FEMS Microbiol Ecol. 2011;76:576–591. doi: 10.1111/j.1574-6941.2011.01077.x. A description of membrane vesicle-like transducing viral particles with a broad host range. [DOI] [PubMed] [Google Scholar]
- 26.Marrs BL. Genetic recombination in Rhodopseudomonas capsulata. Proc Natl Acad Sci USA. 1974;71:971–973. doi: 10.1073/pnas.71.3.971. This paper describes the first GTA, now known as RcGTA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Solioz M, Yen HC, Marrs B. Release and uptake of gene transfer agent by Rhodopseudomonas capsulata. J Bacteriol. 1975;123:651–657. doi: 10.1128/jb.123.2.651-657.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lang AS, Beatty JT. Importance of widespread gene transfer agent genes in α-proteobacteria. Trends Microbiol. 2007;15:54–62. doi: 10.1016/j.tim.2006.12.001. An article discussing the remarkable distribution of GTA genes in alphaproteobacteria and the possible ramifications for widespread GTA production. [DOI] [PubMed] [Google Scholar]
- 29.Solioz M, Marrs B. The gene transfer agent of Rhodopseudomonas capsulata: purification and characterization of its nucleic acid. Arch Biochem Biophys. 1977;181:300–307. doi: 10.1016/0003-9861(77)90508-2. [DOI] [PubMed] [Google Scholar]
- 30.Yen H-C, Hu NT, Marrs BL. Characterization of the gene transfer agent made by an overproducer mutant of Rhodopseudomonas capsulata. J Mol Biol. 1979;131:157–168. doi: 10.1016/0022-2836(79)90071-8. The isolation of a GTA overproducer mutant allows visualization of the particles and packaged DNA. The methods used were important for allowing subsequent cloning and identification of structural and regulatory GTA genes in R. capsulatus. [DOI] [PubMed] [Google Scholar]
- 31.Hynes AP, Mercer RG, Watton DE, Buckley CB, Lang AS. DNA packaging bias and differential expression of gene transfer agent genes within a population during production and release of the Rhodobacter capsulatus gene transfer agent, RcGTA. Mol Microbiol. 2012 May 28; doi: 10.1111/j.1365-2958.2012.08113.x. [DOI] [PubMed] [Google Scholar]
- 32.Lang AS, Beatty JT. Genetic analysis of a bacterial genetic exchange element: the gene transfer agent of Rhodobacter capsulatus. Proc Natl Acad Sci USA. 2000;97:859–864. doi: 10.1073/pnas.97.2.859. This paper describes the RcGTA gene cluster and regulatory genes in R. capsulatus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang SK, Zhang YU, Chen F, Jiao NZ. Complete genome sequence of a marine roseophage provides evidence into the evolution of gene transfer agents in alphaproteobacteria. Virol J. 2011;8:124. doi: 10.1186/1743-422X-8-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lohr JE, Chen F, Hill RT. Genomic analysis of bacteriophage ΦJL001: Insights into its interaction with a sponge-associated alpha-proteobacterium. Appl Environ Microbiol. 2005;71:1598–1609. doi: 10.1128/AEM.71.3.1598-1609.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen F, et al. Proteomic analysis and identification of the structural and regulatory proteins of the Rhodobacter capsulatus gene transfer agent. J Proteome Res. 2008;8:967–973. doi: 10.1021/pr8006045. The identification of protein components of RcGTA particles, some of which are encoded outside of the RcGTA gene cluster. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wall JD, Weaver PF, Gest H. Gene transfer agents, bacteriophages, and bacteriocins of Rhodopseudomonas capsulata. Arch Microbiol. 1975;105:217–224. doi: 10.1007/BF00447140. [DOI] [PubMed] [Google Scholar]
- 37.Lang AS, Beatty JT. The gene transfer agent of Rhodobacter capsulatus and “constitutive transduction” in prokaryotes. Arch Microbiol. 2001;175:241–249. doi: 10.1007/s002030100260. [DOI] [PubMed] [Google Scholar]
- 38.Mercer RG, et al. Loss of the response regulator CtrA causes pleiotropic effects on gene expression but does not affect growth phase regulation in Rhodobacter capsulatus. J Bacteriol. 2010;192:2701–2710. doi: 10.1128/JB.00160-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leung MM, Brimacombe CA, Spiegelman GB, Beatty JT. The GtaR protein negatively regulates transcription of the gtaRI operon and modulates gene transfer agent (RcGTA) expression in Rhodobacter capsulatus. Mol Microbiol. 2012;83:759–774. doi: 10.1111/j.1365-2958.2011.07963.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schaefer AL, Taylor TA, Beatty JT, Greenberg EP. Long-chain acyl-homoserine lactone quorum-sensing regulation of Rhodobacter capsulatus gene transfer agent production. J Bacteriol. 2002;184:6515–6521. doi: 10.1128/JB.184.23.6515-6521.2002. The discovery of quorum sensing-mediated regulation of RcGTA production. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mercer RG, et al. Regulatory systems controlling motility and gene transfer agent production and release in Rhodobacter capsulatus. FEMS Microbiol Lett. 2012;331:53–62. doi: 10.1111/j.1574-6968.2012.02553.x. [DOI] [PubMed] [Google Scholar]
- 42.Lang AS, Taylor TA, Beatty JT. Evolutionary implications of phylogenetic analyses of the gene transfer agent (GTA) of Rhodobacter capsulatus. J Mol Evol. 2002;55:534–543. doi: 10.1007/s00239-002-2348-7. [DOI] [PubMed] [Google Scholar]
- 43.Biers EJ, et al. Occurrence and expression of gene transfer agent genes in marine bacterioplankton. Appl Environ Microbiol. 2008;74:2933–2939. doi: 10.1128/AEM.02129-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paul JH. Prophages in marine bacteria: dangerous molecular time bombs or the key to survival in the seas? ISME J. 2008;2:579–589. doi: 10.1038/ismej.2008.35. This paper raises cogent questions about phage- and GTA-mediated transduction of genes in ocean waters. [DOI] [PubMed] [Google Scholar]
- 45.McDaniel LD, et al. High frequency of horizontal gene transfer in the oceans. Science. 2010;330:50. doi: 10.1126/science.1192243. This article reports remarkable GTA-attributed gene transfer rates in natural environments. [DOI] [PubMed] [Google Scholar]
- 46.Fu Y, et al. High diversity of Rhodobacterales in the subarctic North Atlantic Ocean and gene transfer agent protein expression in isolated strains. Aquat Microb Ecol. 2010;59:283–293. [Google Scholar]
- 47.Buchan A, Gonzalez JM, Moran MA. Overview of the marine Roseobacter lineage. Appl Environ Microbiol. 2005;71:5665–5677. doi: 10.1128/AEM.71.10.5665-5677.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rapp BJ, Wall JD. Genetic transfer in Desulfovibrio desulfuricans. Proc Natl Acad Sci USA. 1987;84:9128–9130. doi: 10.1073/pnas.84.24.9128. A report detailing the second GTA to be discovered. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Humphrey SB, Stanton TB, Jensen NS. Mitomycin C induction of bacteriophages from Serpulina hyodysenteriae and Serpulina innocens. FEMS Microbiol Lett. 1995;134:97–101. doi: 10.1111/j.1574-6968.1995.tb07921.x. [DOI] [PubMed] [Google Scholar]
- 50.Humphrey SB, Stanton TB, Jensen NS, Zuerner RL. Purification and characterization of VSH-1, a generalized transducing bacteriophage of Serpulina hyodysenteriae. J Bacteriol. 1997;179:323–329. doi: 10.1128/jb.179.2.323-329.1997. A paper describing the spirochaete GTA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li C, et al. The spirochete FlaA periplasmic flagellar sheath protein impacts flagellar helicity. J Bacteriol. 2000;182:6698–6706. doi: 10.1128/jb.182.23.6698-6706.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stanton TB, Matson EG, Humphrey SB. Brachyspira (Serpulina) hyodysenteriae gyrB mutants resistance. and interstrain transfer of coumermycin A1. Appl Environ Microbiol. 2001;67:2037–2043. doi: 10.1128/AEM.67.5.2037-2043.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stanton TB, Humphrey SB, Sharma VK, Zuerner RL. Collateral effects of antibiotics: carbadox and metronidazole induce VSH-1 and facilitate gene transfer among Brachyspira hyodysenteriae strains. Appl Environ Microbiol. 2008;74:2950–2956. doi: 10.1128/AEM.00189-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Combs BG, Hampson DJ, Harders SJ. Typing of Australian isolates of Treponema hyodysenteriae by serology and by DNA restriction endonuclease analysis. Vet Microbiol. 1992;31:273–285. doi: 10.1016/0378-1135(92)90085-8. [DOI] [PubMed] [Google Scholar]
- 55.Turner AK, Sellwood R. Extracellular DNA from Serpulina hyodysenteriae consists of 6.5 kbp random fragments of chromosomal DNA. FEMS Microbiol Lett. 1997;150:75–80. doi: 10.1111/j.1574-6968.1997.tb10352.x. [DOI] [PubMed] [Google Scholar]
- 56.Matson EG, Thompson MG, Humphrey SB, Zuerner RL, Stanton TB. Identification of genes of VSH-1, a prophage-like gene transfer agent of Brachyspira hyodysenteriae. J Bacteriol. 2005;187:5885–5892. doi: 10.1128/JB.187.17.5885-5892.2005. The identification of VSH-1 genes in the B. hyodysenteriae genome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stanton TB, Humphrey SB, Bayles DO, Zuerner RL. Identification of a divided genome for VSH-1, the prophage-like gene transfer agent of Brachyspira hyodysenteriae. J Bacteriol. 2009;191:1719–1721. doi: 10.1128/JB.01359-08. The finding that VSH-1 genes are not contiguous in the genome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matson EG, Zuerner RL, Stanton TB. Induction and transcription of VSH-1, a prophage-like gene transfer agent of Brachyspira hyodysenteriae. Anaerobe. 2007;13:89–97. doi: 10.1016/j.anaerobe.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 59.Calderaro A, et al. Bacteriophages induced from weakly beta-haemolytic human intestinal spirochaetes by mitomycin C. J Basic Microbiol. 1998;38:323–335. [PubMed] [Google Scholar]
- 60.Calderaro A, et al. Search for bacteriophages spontaneously occurring in cultures of haemolytic intestinal spirochaetes of human and animal origin. J Basic Microbiol. 1998;38:313–322. [PubMed] [Google Scholar]
- 61.Ritchie A, Robinson I, Joens L, Kinyon J. A bacteriophage for Treponema hyodysenteriae. Vet Rec. 1978;103:34–35. doi: 10.1136/vr.103.2.34. [DOI] [PubMed] [Google Scholar]
- 62.Motro Y, et al. Identification of genes associated with prophage-like gene transfer agents in the pathogenic intestinal spirochaetes Brachyspira hyodysenteriae, Brachyspira pilosicoli and Brachyspira intermedia. Vet Microbiol. 2009;134:340–345. doi: 10.1016/j.vetmic.2008.09.051. This report describes the distribution of VSH-1 genes in species of spirochaete other than Serpulina hyodysenteriae. [DOI] [PubMed] [Google Scholar]
- 63.Trott DJ, Oxberry SL, Hampson DJ. Evidence for Serpulina hyodysenteriae being recombinant, with an epidemic population structure. Microbiology. 1997;143:3357–3365. doi: 10.1099/00221287-143-10-3357. [DOI] [PubMed] [Google Scholar]
- 64.Zuerner RL, et al. Genetic variation in Brachyspira: chromosomal rearrangements and sequence drift distinguish B. pilosicoli from B. hyodysenteriae. Anaerobe. 2004;10:229–237. doi: 10.1016/j.anaerobe.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 65.Stanton TB, Thompson MG, Humphrey SB, Zuerner RL. Detection of bacteriophage VSH-1 svp38 gene in Brachyspira spirochetes. FEMS Microbiol Lett. 2003;224:225–229. doi: 10.1016/S0378-1097(03)00438-5. [DOI] [PubMed] [Google Scholar]
- 66.Bertani G. Transduction-like gene transfer in the methanogen Methanococcus voltae. J Bacteriol. 1999;181:2992–3002. doi: 10.1128/jb.181.10.2992-3002.1999. This paper details the discovery of VTA, the first archaeal GTA to be found. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eiserling F, Pushkin A, Gingery M, Bertani G. Bacteriophage-like particles associated with the gene transfer agent of Methanococcus voltae PS. J Gen Virol. 1999;80:3305–3308. doi: 10.1099/0022-1317-80-12-3305. This work elucidates the structure of VTA. [DOI] [PubMed] [Google Scholar]
- 68.Pina M, Bize A, Forterre P, Prangishvili D. The archeoviruses. FEMS Microbiol Rev. 2011;35:1035–1054. doi: 10.1111/j.1574-6976.2011.00280.x. [DOI] [PubMed] [Google Scholar]
- 69.Krupovič M, Forterre P, Bamford DH. Comparative analysis of the mosaic genomes of tailed archaeal viruses and proviruses suggests common themes for virion architecture and assembly with tailed viruses of bacteria. J Mol Biol. 2010;397:144–160. doi: 10.1016/j.jmb.2010.01.037. [DOI] [PubMed] [Google Scholar]
- 70.Genthner FJ, Wall JD. Isolation of a recombination-deficient mutant of Rhodopseudomonas capsulata. J Bacteriol. 1984;160:971–975. doi: 10.1128/jb.160.3.971-975.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Popa O, Hazkani-Covo E, Landan G, Martin W, Dagan T. Directed networks reveal genomic barriers and DNA repair bypasses to lateral gene transfer among prokaryotes. Genome Res. 2011;21:599–609. doi: 10.1101/gr.115592.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bertin A, de Frutos M, Letellier L. Bacteriophage-host interactions leading to genome internalization. Curr Opin Microbiol. 2011;14:492–496. doi: 10.1016/j.mib.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 73.Hendrix RW, Smith MCM, Burns RN, Ford ME, Hatfull GF. Evolutionary relationships among diverse bacteriophages and prophages: all the world’s a phage. Proc Natl Acad Sci USA. 1999;96:2192–2197. doi: 10.1073/pnas.96.5.2192. A study that elegantly illustrates the movement of genes in the phage (and prophage) pan-genome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tu AHT, Voelker LL, Shen X, Dybvig K. Complete nucleotide sequence of the mycoplasma virus P1 genome. Plasmid. 2001;45:122–126. doi: 10.1006/plas.2000.1501. [DOI] [PubMed] [Google Scholar]
- 75.Williams KP, Sobral BW, Dickerman AW. A robust species tree for the alphaproteobacteria. J Bacteriol. 2007;189:4578–4586. doi: 10.1128/JB.00269-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kristensen DM, Mushegian AR, Dolja VV, Koonin EV. New dimensions of the virus world discovered through metagenomics. Trends Microbiol. 2010;18:11–19. doi: 10.1016/j.tim.2009.11.003. This article suggests a role for GTAs in generating the unexpected cellular-gene composition of viral metagenomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lang AS, Beatty JT. In: Extracellular Nucleic Acids. Kikuchi Y, Rykova EY, editors. Springer; 2010. pp. 15–24. [Google Scholar]
- 78.Stanton TB. Prophage-like gene transfer agents — novel mechanisms of gene exchange for Methanococcus, Desulfovibrio, Brachyspira, and Rhodobacter species. Anaerobe. 2007;13:43–49. doi: 10.1016/j.anaerobe.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 79.Diggle SP. Microbial communication and virulence: lessons from evolutionary theory. Microbiology. 2010;156:3503–3512. doi: 10.1099/mic.0.045179-0. [DOI] [PubMed] [Google Scholar]
- 80.West SA, Griffin AS, Gardner A, Diggle SP. Social evolution theory for microorganisms. Nature Rev Microbiol. 2006;4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- 81.West SA, Diggle SP, Buckling A, Gardner A, Griffin AS. The social lives of microbes. Ann Rev Ecol Evol Syst. 2007;38:53–77. [Google Scholar]
- 82.Vos M. Why do bacteria engage in homologous recombination? Trends Microbiol. 2009;17:226–232. doi: 10.1016/j.tim.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 83.Draghi JA, Turner PE. DNA secretion and gene-level selection in bacteria. Microbiology. 2006;152:2683–2688. doi: 10.1099/mic.0.29013-0. This investigation leads to the development of a model that rationalizes the maintenance of costly activities and may be applicable to GTA production. [DOI] [PubMed] [Google Scholar]
- 84.Canchaya C, Proux C, Fournous G, Bruttin A, Brussow H. Prophage genomics. Microbiol Mol Biol Rev. 2003;67:238–276. doi: 10.1128/MMBR.67.2.238-276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chattopadhyay S, et al. High frequency of hotspot mutations in core genes of Escherichia coli due to short-term positive selection. Proc Natl Acad Sci USA. 2009;106:12412–12417. doi: 10.1073/pnas.0906217106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rosario K, Breitbart M. Exploring the viral world through metagenomics. Curr Opin Virol. 2011;1:289–297. doi: 10.1016/j.coviro.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 87.Filée J, Tétart F, Suttle CA, Krisch HM. Marine T4-type bacteriophages, a ubiquitous component of the dark matter of the biosphere. Proc Natl Acad Sci USA. 2005;102:12471–12476. doi: 10.1073/pnas.0503404102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lapierre P, Gogarten JP. Estimating the size of the bacterial pan-genome. Trends Genet. 2009;25:107–110. doi: 10.1016/j.tig.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 89.Esser C, et al. A genome phylogeny for mitochondria among α-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol Biol Evol. 2004;21:1643–1660. doi: 10.1093/molbev/msh160. [DOI] [PubMed] [Google Scholar]
- 90.Rivera MC, Jain R, Moore JE, Lake JA. Genomic evidence for two functionally distinct gene classes. Proc Natl Acad Sci USA. 1998;95:6239–6244. doi: 10.1073/pnas.95.11.6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Richards TA, Archibald JM. Cell evolution: gene transfer agents and the origin of mitochondria. Curr Biol. 2011;21:R112–R114. doi: 10.1016/j.cub.2010.12.036. [DOI] [PubMed] [Google Scholar]
- 92.Zinder ND, Lederberg J. Genetic exchange in Salmonella. J Bacteriol. 1952;64:679–699. doi: 10.1128/jb.64.5.679-699.1952. The disovery of transduction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Del Campillo-Campbell A, Kayajanian G, Campbell A, Adhya S. Biotin-requiring mutants of Escherichia coli K-12. J Bateriol. 1967;94:2065–2066. doi: 10.1128/jb.94.6.2065-2066.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Morse ML, Lederberg EM, Lederberg J. Transduction in Escherichia coli K-12. Genetics. 1956;41:142–156. doi: 10.1093/genetics/41.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ikeda H, Tomizawa J-i. Transducing fragments in generalized transduction by phage P1: I Molecular origin of the fragments. J Mol Biol. 1965;14:85–109. doi: 10.1016/s0022-2836(65)80232-7. [DOI] [PubMed] [Google Scholar]
- 96.Lennox ES. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955;1:190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- 97.Howe MM. Transduction by bacteriophage MU-1. Virology. 1973;55:103–117. doi: 10.1016/s0042-6822(73)81012-8. [DOI] [PubMed] [Google Scholar]
- 98.Bukhari AI, Froshauer S, Botchan M. Ends of bacteriophage Mu DNA. Nature. 1976;264:580–583. doi: 10.1038/264580a0. [DOI] [PubMed] [Google Scholar]
- 99.Fogg PCM, Hynes AP, Digby E, Lang AS, Beatty JT. Characterization of a newly discovered Mu-like bacteriophage, RcapMu, in Rhodobacter capsulatus strain SB1003. Virology. 2011;421:211–221. doi: 10.1016/j.virol.2011.09.028. [DOI] [PubMed] [Google Scholar]
- 100.Brussow H, Canchaya C, Hardt WD. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev. 2004;68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cheetham BF, Katz ME. A role for bacteriophages in the evolution and transfer of bacterial virulence determinants. Mol Microbiol. 1995;18:201–208. doi: 10.1111/j.1365-2958.1995.mmi_18020201.x. [DOI] [PubMed] [Google Scholar]
- 102.Wagner PL, Waldor MK. Bacteriophage control of bacterial virulence. Infect Immun. 2002;70:3985–3993. doi: 10.1128/IAI.70.8.3985-3993.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Waldor MK, Mekalanos JJ. Lysogenic conversion by filamentous phage encoding a cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 104.Novick RP, Christie GE, Penadés JR. The phage-related chromosomal islands of Gram-positive bacteria. Nature Rev Microbiol. 2010;8:541–551. doi: 10.1038/nrmicro2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tormo MA, et al. Staphylococcus aureus pathogenicity island DNA is packaged in particles composed of phage proteins. J Bacteriol. 2008;190:2434–2440. doi: 10.1128/JB.01349-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen J, Novick RP. Phage-mediated intergeneric transfer of toxin genes. Science. 2009;323:139–141. doi: 10.1126/science.1164783. [DOI] [PubMed] [Google Scholar]
- 107.Kenzaka T, Tani K, Nasu M. High-frequency phage-mediated gene transfer in freshwater environments determined at single-cell level. ISME J. 2010;4:648–659. doi: 10.1038/ismej.2009.145. [DOI] [PubMed] [Google Scholar]
- 108.Jiang SC, Paul JH. Gene transfer by transduction in the marine environment. Appl Environ Microbiol. 1998;64:2780–2787. doi: 10.1128/aem.64.8.2780-2787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kidambi SP, Ripp S, Miller RV. Evidence for phage-mediated gene transfer among Pseudomonas aeruginosa strains on the phylloplane. Appl Environ Microbiol. 1994;60:496–500. doi: 10.1128/aem.60.2.496-500.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cornick NA, Helgerson AF, Mai V, Ritchie JM, Acheson DWK. In vivo transduction of an Stx-encoding phage in ruminants. Appl Environ Microbiol. 2006;72:5086–5088. doi: 10.1128/AEM.00157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Del Casale A, Flanagan PV, Larkin MJ, Allen CCR, Kulakov LA. Extent and variation of phage-borne bacterial 16S rRNA gene sequences in wastewater environments. Appl Environ Microbiol. 2011;77:5529–5532. doi: 10.1128/AEM.00457-11. This work identifies taxa that are responsible for producing transducing phages in the environment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Del Casale A, Flanagan PV, Larkin MJ, Allen CCR, Kulakov LA. Analysis of transduction in wastewater bacterial populations by targeting the phage-derived 16S rRNA gene sequences. FEMS Microbiol Ecol. 2011;76:100–108. doi: 10.1111/j.1574-6941.2010.01034.x. [DOI] [PubMed] [Google Scholar]
- 113.Lindell D, Jaffe JD, Johnson ZI, Church GM, Chisholm SW. Photosynthesis genes in marine viruses yield proteins during host infection. Nature. 2005;438:86–89. doi: 10.1038/nature04111. [DOI] [PubMed] [Google Scholar]
- 114.Rohwer F, Vega Thurber R. Viruses manipulate the marine environment. Nature. 2009;459:207–212. doi: 10.1038/nature08060. [DOI] [PubMed] [Google Scholar]
- 115.Berglund EC, et al. Run-off replication of host-adaptability genes is associated with gene transfer agents in the genome of mouse-infecting Bartonella grahamii. PLoS Genet. 2009;5:e1000546. doi: 10.1371/journal.pgen.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Umemori E, Sasaki Y, Amano K, Amano Y. A phage in Bartonella bacilliformis. Microbiol Immunol. 1992;36:731–736. doi: 10.1111/j.1348-0421.1992.tb02075.x. [DOI] [PubMed] [Google Scholar]
- 117.Anderson B, Goldsmith C, Johnson A, Padmalayam I, Baumstark B. Bacteriophage-like particle of Rochalimaea henselae. Mol Microbiol. 1994;13:67–73. doi: 10.1111/j.1365-2958.1994.tb00402.x. [DOI] [PubMed] [Google Scholar]
- 118.Barbian KD, Minnick MF. A bacteriophage-like particle from Bartonella bacilliformis. Microbiology. 2000;146:599–609. doi: 10.1099/00221287-146-3-599. [DOI] [PubMed] [Google Scholar]
- 119.Okamoto K, et al. Properties of the defective phage of Bacillus subtilis. J Mol Biol. 1968;34:413–428. doi: 10.1016/0022-2836(68)90169-1. [DOI] [PubMed] [Google Scholar]
- 120.Seaman E, Tarmy E, Marmur J. Inducible phages of Bacillus subtilis. Biochemistry. 1964;3:607–613. doi: 10.1021/bi00893a001. [DOI] [PubMed] [Google Scholar]
- 121.Wood HE, Dawson MT, Devine KM, McConnell DJ. Characterization of PBSX, a defective prophage of Bacillus subtilis. J Bacteriol. 1990;172:2667–2674. doi: 10.1128/jb.172.5.2667-2674.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]