Abstract
Background
Vascular endothelial growth factor A (VEGFA) is a major regulator of both physiological and pathological angiogenesis. Associations between polymorphisms in VEGFA and complex disease have been inconsistent. The parent from whom the allele was inherited may account for these inconsistencies. This study examined the parent of origin effect on the expression of murine Vegfa.
Methods
Two homozygous, inbred mouse strains A/J (AJ) and 129x1/SvJ (129) were crossed to produce reciprocal AJ129 and 129AJ offspring, respectively. RNA was extracted from cardiac tissue of 6 week old male (n = 8) and female (n = 8) parental, and male and female F1 offspring mice (AJ129 n = 8 and 129AJ n = 8). Vegfa and Hif1a expression levels were measured by qPCR and compared between the F1 offspring from the reciprocal crosses.
Results
We found significant differences in the expression of Vegfa in F1 offspring (AJ129 and 129AJ mice) of the reciprocal crosses between AJ and 129 mice. Offspring of male AJ mice had significantly higher expression of Vegfa than offspring of male 129 mice (p = 0.006). This difference in expression was not the result of preferential allele expression (allelic imbalance). Expression of Hif1a, a transcriptional regulator of Vegfa expression, was also higher in F1 offspring of an AJ father (p = 0.004).
Conclusion
Differences in Vegfa and Hif1a gene expression are likely the result of an upstream angiogenic regulator gene that is influenced by the parent of origin. These results highlight the importance of including inheritance information, such as parent of origin, when undertaking allelic association studies.
Keywords: Parent of origin, Epigenetics, Vascular endothelial growth factor
Background
Angiogenesis is the growth of capillary sized vessels. It plays a critical role in wound healing and embryological development, and in the pathogenesis of diseases such as cancer. Promoting angiogenesis can aid in the treatment of ischemia and myocardial infarction, while inhibition of angiogenesis has been effective in the treatment of cancer. Vascular endothelial growth factor A (VEGFA) is one of the most potent inducers of angiogenesis, and is a major regulator of both physiological and pathological angiogenesis. It has been shown to stimulate blood vessel formation in animal models of ischemic cardiovascular disease [1], has been trialled as a potential therapeutic for myocardial ischemia [2], and therapies that inhibit VEGF are used in the treatment of cancer [3].
A number of studies have investigated associations between polymorphisms within, or nearby, the VEGFA gene and complex disease, including coronary artery disease [4], atherosclerosis [5], myocardial infarction [6], and hypertension [7]. However, these sequence variants explain only a small fraction of the estimated heritability of the diseases studied, and the ability to replicate these findings can prove difficult. For example, inconsistent results have been shown when studying the association between the development of coronary artery lesions in individuals with Kawasaki disease with VEGFA polymorphisms [8,9]. Population differences, study power, and choice of polymorphism may explain inconsistent results. Additionally, the parent from whom the variant was inherited may also contribute to the inability to replicate results across studies.
Parent of origin effects influence gene expression in offspring and result in the preferential expression of the paternally or maternally inherited allele. It is caused by a variety of mechanisms including imprinting, sex, epigenetic effects, and environmental influences during development [10]. In genetic studies, parent of origin effects are largely ignored; however, recent studies have found evidence of parent of origin effects in breast cancer [11], colorectal cancer [12], and type 2 diabetes [11]. These findings highlight the importance of investigating the effects of parent of origin on phenotype and disease susceptibility.
Due to the inconsistency in results when studying the association between VEGFA and disease, this study aimed to determine whether parent of origin effects influence the expression of Vegfa. This study was carried out in cardiac tissue from F1 hybrids resulting from reciprocal crosses of two inbred mouse strains.
Methods
All experiments were carried out at the School of Medicine Animal Facility, University of Western Sydney and approved by the University of Western Sydney Animal Ethics Committee. Two homozygous, inbred mouse strains A/J (AJ) and 129x1/SvJ (129) were used. Crosses between AJ male and 129 female, and 129 male and AJ female produced reciprocal AJ129 and 129AJ offspring, respectively. Male (n = 8) and female (n = 8) mice from each of the parental strains were euthanised at six weeks of age by cervical dislocation. Male and female offspring were collected from each of the crosses (AJ129 n = 8 per sex and 129AJ n = 8 per sex). The hearts were excised, rinsed with PBS and flash frozen in LN2.
Results
Heterozygous F1 hybrids (AJ129 and 129AJ) resulting from reciprocal crosses of two inbred homozygous strains (AJ and 129) were used in this study. Direct sequencing of the full Vegfa mRNA transcript in homozygous parental strains and heterozygous F1 offspring confirmed a single nucleotide polymorphism (SNP) at the 3′UTR of the gene (AJ = GG, 129 = AA, AJ129 and 129AJ = GA).
Vegfa expression
This study compared the expression of Vegfa in AJ and 129 parental mice, and in male F1 offspring from reciprocal crosses (AJ129 and 129AJ), to determine whether parent of origin influences mRNA expression. Strain specific differences in Vegfa expression were seen between AJ and 129 mice, with Vegfa expression 1.5 fold higher in male AJ mice compared to male 129 mice (Figure 1a). No strain specific differences in Vegfa expression were observed between female AJ and female 129 mice (Figure 1b). In the male F1 offspring from the reciprocal crosses, Vegfa expression was 1.7 fold higher in AJ129 mice compared with 129AJ mice (Figure 1c). To ascertain whether this difference in Vegfa expression tracked with Y chromosome transmission, the expression of Vegfa was measured in female parentals and offspring. There was no difference in Vegfa expression between parental AJ males versus females (p = 0.15), and female offspring of AJ males had 1.3 fold increased expression of Vegfa (p = 0.03) compared with females of the reciprocal cross.
Figure 1.
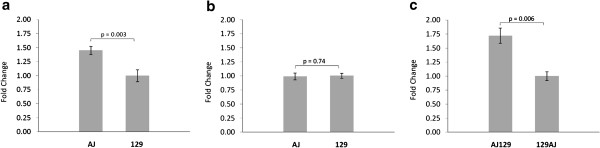
Relative expression of Vegfa. a. Parental: AJ male compared to 129 male. b. Parental: AJ female compared to 129 female. c. F1: AJ129 (offspring of AJ male) compared to 129AJ (offspring of 129 male).
Measurement of allelic expression
To determine whether differences in Vegfa mRNA expression between the F1 offspring from the reciprocal crosses were the result of unequal expression of the maternally or paternally inherited allele, the expression of each allele was measured using quantitative ARMS PCR. The rs16821436 SNP in the 3′UTR of Vegfa (G/A) was used to differentiate expression of alleles in the F1 heterozygous mice. In the F1 offspring, the expression of each allele relative to the homozygous parental strain was significantly lower, as expected. The expression of the G allele in the heterozygous AJ129 and 129AJ mice was significantly lower, 43% and 43%, than the homozygous AJ mice (Figure 2ai). Similarly, the expression of the A allele in the heterozygous offspring was significantly lower, 47% and 45%, than the homozygous 129 parental mice (Figure 2aii). No difference in the expression of the G allele relative to the A allele was seen within the F1 offspring (Figure 2bi and ii). Additionally, there was no difference in the expression ratio of the G allele to the A allele between the AJ129 and 129AJ offspring.
Figure 2.

Vegfa allelic expression. ai. Relative expression of the G allele in males. aii. Relative expression of the A allele in males. bi. Expression ratio of the G allele to the A allele in AJ129 mice. bii. Expression ratio of the G allele to the A allele in 129AJ mice.
Hif1a expression
To determine whether differences in Vegfa expression were the result of trans-acting regulatory elements, the expression of Hif1a, a transcription factor and known activator of Vegfa was measured. The expression of Hif1a was measured in the AJ and 129 parental strains and in the F1 offspring of reciprocal crosses. The expression of Hif1a was 1.4 fold higher in male AJ compared to male 129 mice (Figure 3a). No difference in Hif1a expression was observed between the female parental strains (Figure 3b). In the male F1 mice, the expression of Hif1a was 1.4 fold higher in AJ129 compared to 129AJ mice (Figure 3c).
Figure 3.
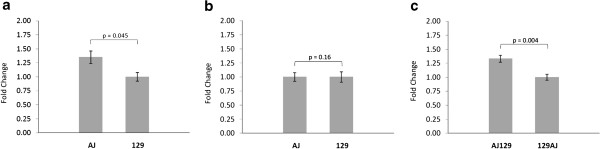
Relative expression of Hif1a. a. Parental: AJ male compared to 129 male. b. Parental: AJ female compared to 129 female. c. F1: AJ129 (offspring of AJ male) compared to 129AJ (offspring of 129 male).
Discussion
This study examined the parent of origin effect on the expression of Vegfa, an important regulator of angiogenesis. We found significant differences in the expression of Vegfa in F1 offspring (AJ129 and 129AJ mice) of the reciprocal crosses between AJ and 129 mice. Specifically, offspring of male AJ mice had significantly higher expression of Vegfa than offspring of male 129 mice. This difference in expression was not the result of preferential allele expression (allelic imbalance), as no difference in the expression of the paternally or maternally inherited allele was observed in the F1 heterozygotes. Expression of Hif1a, a transcriptional regulator of Vegfa expression, was also found to be significantly higher in F1 offspring of an AJ father. These results cannot be explained by Y chromosome transmission, as female offspring of AJ fathers had increased expression of Vegfa. In addition, the AJ female parentals had no difference in expression of Vegfa when compared with the male AJ parentals. The DNA sequences of F1 heterozygotes are identical on autosomal chromosomes and differ only on their sex chromosomes. As Vegfa and Hif1a are not located on sex chromosomes, this suggests that the observed differences in Vegfa and Hif1a gene expression are not a result of DNA sequence variation and may be explained by a parent of origin effect.
Angiogenesis is a strictly regulated process that results from a balance between pro and inhibitory angiogenic factors. VEGF is a potent pro-angiogenic factor and its expression is tightly controlled at a number of levels. Cis-acting SNPs have been shown to alter Vegfa promoter activity [13-15], while SNPs in the 3′UTR have been reported to affect expression via the binding of micro RNAs which affect mRNA stability [16,17]. The AJ and 129 parental strains differed at a single nucleotide in the 3′UTR of Vegf[18]. In this study, the paternally and maternally inherited alleles within the F1 offspring were expressed equally, indicating that cis-acting regulatory elements within, or nearby, the Vegfa gene were unlikely to account for the observed differences in Vegfa gene expression. However, the presence of cis- acting regulatory elements in genes known to regulate Vegfa expression, which are influenced by parent of origin, may explain the difference in Vegfa expression.
A number of trans-acting factors are known to regulate the expression of Vegfa. Transcription factors that bind to response elements within the Vegfa promoter include hypoxia-inducible factor (HIF1) [19], nuclear factor (NF)-κB [20], activator protein (AP1) [21], and specificity protein (SP1) [22]. HIF1 is a transcription factor that functions to restore oxygen homeostasis during hypoxia via the regulation of genes involved in angiogenesis (Vegfa), glycolysis and vasodilation [23]. In this study, Hif1a expression was significantly higher in the F1 offspring of AJ males which corresponded to increased Vegfa expression within these same animals. Imbalanced expression of alleles within the Hif1a gene could not be assessed as the F1 mice were homozygous [18]. No studies have reported a role for epigenetic mechanisms in the regulation of Hif1 expression however, epigenetic regulation of von Hippel-Lindau (VHL), an ubiquitin ligase that targets HIF1a for proteosomal destruction under normoxic conditions, has been reported. Hypermethylation of the VHL promoter in renal cell carcinoma has been correlated with increased HIF1a expression [24], and deletion of miR-23b has been reported to reduce HIF1a and VEGFA expression via its targeting of VHL[25]. Further work is needed to address whether epigenetic mechanisms are involved in the regulation of genes known to regulate Vegfa and Hif1a expression in this mouse model.
Transgenerational epigenetic inheritance is another mechanism that may explain the differences in expression patterns between the F1 offspring from the reciprocal crosses. It refers to genetic or environmental exposures in the parent, which affect patterns of gene expression in the offspring, without the offspring having inherited the allele or been exposed to the environmental agent [26]. Examples of transgenerational epigenetic inheritance include DNA methylation, histone modifications, changes in chromatin structure and the action of small non-coding RNA all of which can result in epialleles. That is, alleles that are variably expressed in the absence of genetic heterogeneity [27]. The epigenetic state is established during early embryogenesis and can be influenced by whether the allele is inherited from the mother or the father, as is the case with the agouti viable yellow allele (Avy) in mice [28]. Parent of origin effects may be observed for a number of genes, if these genes belong to pathways that are ultimately regulated by epialleles. This may be the case in the present study, where a regulator of genes involved in angiogenesis is influenced by the parent of origin, and downstream this alters the expression of both Hif1a and Vegfa. Further work is required to establish which genes are controlled by epigenetic mechanisms within the pathway controlling angiogenesis.
Conclusions
Our study demonstrates that parent of origin influences Vegfa expression in mouse cardiac tissue, and the genetic factors which determine gene expression are more complex than simple Mendelian inheritance. Additionally, given the common use of A/J and 129x1/SvJ strains to generate genetically engineered mice, studies using mixed genetic background mice, or studies using mice after backcrossing, need to consider parent of origin in addition to genetic variation. Finally, the identification of epigenetic regulation in genes involved in angiogenesis has important implications in how future genome-wide association studies need to incorporate technologies, which detect epigenetic and parent of origin information, to provide more informative associations.
RNA extraction, quantification and cDNA synthesis
Hearts were mechanically disrupted in the deep frozen state using the ‘Mikro Dismembrator’ automated frozen tissue disruptor (Sartorius, Germany), as previously described [29]. Total RNA was extracted from powderised tissue using TRIzol (Sigma Aldrich, USA) as per manufacturer’s protocol, followed by DNase I treatment using the ‘DNA-free’ kit (Applied Biosciences, USA) according to the manufacturer’s protocol. RNA were quantified using the NanoPhotometer (Implen, Germany) and cDNA was synthesised from 500 ng of RNA using Bioscript™ Reverse Transcriptase and random hexamers (Bioline Pty Ltd, Australia) according to the manufacturer’s protocol. Genomic DNA was extracted using the QIAamp DNA Mini Kit (Qiagen, Germany) as per the manufacturer’s protocol.
Quantitative PCR
Quantitative PCR (qPCR) was used to measure mRNA expression of Vegfa and hypoxic inducible factor 1 alpha (Hif1a). Mitochondrial ribosomal protein S28 (Mrps28S), 18S ribosomal RNA (Rn18S), and hypoxanthine guanine phosphoribosyl transferase (Hprt1) were used as normalising genes. Primer sequences are described in Table 1. Individual reactions (10 μl) contained 5× MyTaq PCR buffer (Bioline), 0.4 U of MyTaq (Bioline), 1x SYBR Green I dye, forward and reverse primers (10 μM), and 4 μl of cDNA (or 4 μl water for no template controls). A pooled sample of cDNA was serially diluted 1 in 5 to generate a standard curve for each primer set, and the efficiency of the reaction was calculated. The PCR reactions were carried out in a MxPro3005P Real Time PCR System (Stratagene, Agilent Technologies, USA) under the conditions listed in Table 1, followed by a dissociation curve. All samples were run in triplicate.
Table 1.
Primers and PCR conditions
| Primer | bp | Primer sequence 5′ to 3′ | PCR conditions |
|---|---|---|---|
|
Hprt1F |
94 |
TGACACTGGCAAAACAATGCA |
95°C 10′ (95°C 40″; 60°C 30″; 72°C 30″) × 40 |
|
Hprt1R |
GGTCCTTTTCACCAGCAAGCT |
Dissociation curve |
|
|
Rn18SF |
178 |
CTCTGGTTGCTCTGTGCAGT |
95°C 10′ (95°C 40″; 62°C 30″; 72°C 30″) × 40 |
|
Rn18SR |
GGCTCCTTGTAGGGGTTCTC |
Dissociation curve |
|
|
Mrps28SF |
223 |
GAAATGCAAGCACGGAGAGT |
95°C 10′ (95°C 40″; 62°C 30″; 72°C 30″) × 40 |
|
Mrps28SR |
CCGATGACCAGTTTGTCCTT |
Dissociation curve |
|
|
VegfaF |
200 |
GCTACTGCCGTCCGATTGAGAC |
95°C 10′ (95°C 40″; 65°C 30″; 72°C 30″) × 40 |
|
VegfaR |
GTGCTGGCTTTGGTGAGGTTTG |
Dissociation curve |
|
|
Hif1aF |
209 |
GCTTCTGTTATGAGGCTCACC |
95°C 10′ (95°C 40″; 62°C 30″; 72°C 30″) × 40 |
| Hif1aR | TCAAACTGAGTTAACCCCATGT | Dissociation curve |
Direct sequencing
Sanger sequencing was used to confirm the presence of a polymorphism in the 3′UTR of Vegfa (NCBI Reference Sequence: NM_001025250.3). Total RNA from the parental strains and F1 offspring was amplified and the purified PCR products were sequenced (Australian Genome Research Facility, AUST). Sequence data was aligned and analysed using Sequencher v5.0.1 (Gene Codes Corp, MI).
Measurement of allelic expression using amplification refractory mutation systems (ARMS) PCR
Heterozygous F1 samples were evaluated for Vegfa allelic expression imbalance using quantitative ARMS PCR. The amplification primers (forward: CTTTCATCCCATTGTCTACC(G/A); reverse: TGTTATTGGTGTCTTCACTGGA) used in the assay generated a 188 bp amplicon. The allele specific primers were run in parallel reactions. Reactions were run under the following conditions: 95°C 10 min; 95°C 30 sec, 62°C 30 sec, 72°C 30 sec for 30 cycles; followed by a dissociation curve. Individual reactions (10 μl) contained 5× MyTaq PCR buffer (Bioline), 0.4 U of MyTaq (Bioline), 1x SYBR Green I dye, forward and reverse primers (10 μM), and 4 μl of cDNA (or 4 μl water for no template controls). All reactions were performed in triplicate. The ability of the primers to distinguish between the two alleles with high specificity was assessed using cDNA from parental strains. Under these conditions neither primer exhibited cross-reactivity with the opposite allele. A standard curve was generated for each primer pair using a dilution series (200 ng to 0.20 ng, 1 in 4) of heterozygous genomic DNA. This served as a reference for the 50:50 allelic ratio as we would expect a perfect 50:50 ratio of the two alleles in genomic DNA from a heterozygote with a diploid genome. We avoided using cDNA as a control as this would bias our results towards the allele expression ratio of the reference sample.
Data analysis
For qPCR, cycle threshold (Ct) values for each sample were calculated using the MxPRo QPCR software (Stratagene, Agilent Technologies, USA). Triplicate Ct values were averaged and the quantity (Q) of each sample was calculated using the delta-delta Ct method [30]. Q values from the normaliser genes were input into geNorm [31] and the geometric means from these genes were used to generate a normalisation factor (NF). The Q values of the genes of interest were normalised by dividing by the NF value. Results are expressed as mean ± SE fold changes in expression.
Statistical significance between groups was determined using a general linear model. Statistical analysis was performed with PASW Statistics version 20. A P value of <0.01 was considered significant to account for multiple testing.
Competing interests
The authors declare that they have no completing interest.
Authors’ contributions
CLC carried out sample collection and preparation, gene expression studies, allele specific expression studies, data analysis and interpretation, and drafted the manuscript. CTM carried out sample collection and preparation, and gene expression studies. SJL carried out sample collection and preparation, and gene expression studies. JML conceived the study, participated in data analysis and interpretation, and helped to draft the manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Christine L Chiu, Email: c.chiu@uws.edu.au.
Chloe T Morgan, Email: 16356131@student.uws.edu.au.
Samantha J Lupton, Email: s.lupton@uws.edu.au.
Joanne M Lind, Email: j.lind@uws.edu.au.
Acknowledgements
The authors would like to thank the UWS Animal Facilities Manager, Mr Brad Harper, for his assistance in the collection of mouse tissue. The authors would like to acknowledge the contribution of Dr. Jost Preis in the selection of mouse strains. JML is supported by a National Health and Medical Research Council, Australian Biomedical Fellowship.
Financial disclosures
No external funding was received for this study.
References
- Gowdak LH, Poliakova L, Wang X, Kovesdi I, Fishbein KW, Zacheo A, Palumbo R, Straino S, Emanueli C, Marrocco-Trischitta M. Adenovirus-mediated VEGF(121) gene transfer stimulates angiogenesis in normoperfused skeletal muscle and preserves tissue perfusion after induction of ischemia. Circulation. 2000;102:565–571. doi: 10.1161/01.CIR.102.5.565. [DOI] [PubMed] [Google Scholar]
- Henry TD, Annex BH, McKendall GR, Azrin MA, Lopez JJ, Giordano FJ, Shah PK, Willerson JT, Benza RL, Berman DS. The VIVA trial: vascular endothelial growth factor in Ischemia for vascular angiogenesis. Circulation. 2003;107:1359–1365. doi: 10.1161/01.CIR.0000061911.47710.8A. [DOI] [PubMed] [Google Scholar]
- Van Meter ME, Kim ES. Bevacizumab: current updates in treatment. Curr Opin Oncol. 2010;22:586–591. doi: 10.1097/CCO.0b013e32833edc0c. [DOI] [PubMed] [Google Scholar]
- Lin TH, Su HM, Wang CL, Voon WC, Shin SJ, Lai WT, Sheu SH. Vascular endothelial growth factor polymorphisms and extent of coronary atherosclerosis in Chinese population with advanced coronary artery disease. Am J Hypertens. 2010;23:960–966. doi: 10.1038/ajh.2010.104. [DOI] [PubMed] [Google Scholar]
- Howell WM, Ali S, Rose-Zerilli MJ, Ye S. VEGF polymorphisms and severity of atherosclerosis. J Med Genet. 2005;42:485–490. doi: 10.1136/jmg.2004.025734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic D, Verhovec R, Globocnik Petrovic M, Osredkar J, Peterlin B. Association of vascular endothelial growth factor gene polymorphism with myocardial infarction in patients with type 2 diabetes. Cardiology. 2007;107:291–295. doi: 10.1159/000099064. [DOI] [PubMed] [Google Scholar]
- Hamedian AA, Esteghamati A, Noshad S, Mozafari M, Moin-Tavakkoli H, Nakhjavani M, Mahmoudi T, Nikzamir M, Safary R, Nikzamir A. Vascular endothelial growth factor (VEGF) +405 C/G polymorphism is associated with essential hypertension in a population from Tehran of Iran. Mol Biol Rep. 2012;39:6213–6218. doi: 10.1007/s11033-011-1440-8. [DOI] [PubMed] [Google Scholar]
- Huang FY, Chang TY, Chen MR, Lee HC, Chi H, Chiu NC, Hsu CH, Lin SP, Kao HA, Chen WF. Lack of association of the vascular endothelial growth factor gene polymorphisms with Kawasaki disease in Taiwanese children. J Clin Immunol. 2008;28:322–328. doi: 10.1007/s10875-008-9185-4. [DOI] [PubMed] [Google Scholar]
- Kariyazono H, Ohno T, Khajoee V, Ihara K, Kusuhara K, Kinukawa N, Mizuno Y, Hara T. Association of vascular endothelial growth factor (VEGF) and VEGF receptor gene polymorphisms with coronary artery lesions of Kawasaki disease. Pediatr Res. 2004;56:953–959. doi: 10.1203/01.PDR.0000145280.26284.B9. [DOI] [PubMed] [Google Scholar]
- Guilmatre A, Sharp AJ. Parent of origin effects. Clin Genet. 2012;81:201–209. doi: 10.1111/j.1399-0004.2011.01790.x. [DOI] [PubMed] [Google Scholar]
- Kong A, Steinthorsdottir V, Masson G, Thorleifsson G, Sulem P, Besenbacher S, Jonasdottir A, Sigurdsson A, Kristinsson KT, Frigge ML. Parental origin of sequence variants associated with complex diseases. Nature. 2009;462:868–874. doi: 10.1038/nature08625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet CM, Dowty JG, van Vliet JL, Smith L, Mead LJ, Macrae FA, St John DJ, Giles GG, Southey MC, Jenkins MA. Dependence of colorectal cancer risk on the parent-of-origin of mutations in DNA mismatch repair genes. Hum Mutat. 2011;32:207–212. doi: 10.1002/humu.21408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TF, Spindler KL, Lorentzen KA, Olsen DA, Andersen RF, Lindebjerg J, Brandslund I, Jakobsen A. The importance of −460 C/T and +405 G/C single nucleotide polymorphisms to the function of vascular endothelial growth factor A in colorectal cancer. J Cancer Res Clin Oncol. 2010;136:751–758. doi: 10.1007/s00432-009-0714-1. [DOI] [PubMed] [Google Scholar]
- Summers AM, Coupes BM, Brennan MF, Ralph SA, Short CD, Brenchley PE. VEGF −460 genotype plays an important role in progression to chronic kidney disease stage 5. Nephrol Dial Transplant. 2005;20:2427–2432. doi: 10.1093/ndt/gfi029. [DOI] [PubMed] [Google Scholar]
- Awata T, Kurihara S, Takata N, Neda T, Iizuka H, Ohkubo T, Osaki M, Watanabe M, Nakashima Y, Inukai K. Functional VEGF C-634G polymorphism is associated with development of diabetic macular edema and correlated with macular retinal thickness in type 2 diabetes. Biochem Biophys Res Commun. 2005;333:679–685. doi: 10.1016/j.bbrc.2005.05.167. [DOI] [PubMed] [Google Scholar]
- Yin KJ, Olsen K, Hamblin M, Zhang J, Schwendeman SP, Chen YE. Vascular endothelial cell-specific microRNA-15a inhibits angiogenesis in hindlimb ischemia. J Biol Chem. 2012;287:27055–27064. doi: 10.1074/jbc.M112.364414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z, Lv Q, Ye W, Wong CK, Cai G, Gu D, Ji Y, Zhao C, Wang J, Yang BB, Zhang Y. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS One. 2006;1:e116. doi: 10.1371/journal.pone.0000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE. The Mouse Genome Database (MGD): comprehensive resource for genetics and genomics of the laboratory mouse. Nucleic Acids Res. 2012;40:D881–D886. doi: 10.1093/nar/gkr974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsythe JA, Jiang BH, Iyer NV, Agani F, Leung SW, Koos RD, Semenza GL. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol Cell Biol. 1996;16:4604–4613. doi: 10.1128/mcb.16.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leychenko A, Konorev E, Jijiwa M, Matter ML. Stretch-induced hypertrophy activates NFkB-mediated VEGF secretion in adult cardiomyocytes. PLoS One. 2011;6:e29055. doi: 10.1371/journal.pone.0029055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz BV, Lenoir MC, Ladoux A, Frelin C, Demarchez M, Michel S. Regulation of vascular endothelial growth factor expression in human keratinocytes by retinoids. J Biol Chem. 2000;275:642–650. doi: 10.1074/jbc.275.1.642. [DOI] [PubMed] [Google Scholar]
- Schafer G, Cramer T, Suske G, Kemmner W, Wiedenmann B, Hocker M. Oxidative stress regulates vascular endothelial growth factor-A gene transcription through Sp1- and Sp3-dependent activation of two proximal GC-rich promoter elements. J Biol Chem. 2003;278:8190–8198. doi: 10.1074/jbc.M211999200. [DOI] [PubMed] [Google Scholar]
- Loor G, Schumacker PT. Role of hypoxia-inducible factor in cell survival during myocardial ischemia-reperfusion. Cell Death Differ. 2008;15:686–690. doi: 10.1038/cdd.2008.13. [DOI] [PubMed] [Google Scholar]
- Schmitt AM, Schmid S, Rudolph T, Anlauf M, Prinz C, Kloppel G, Moch H, Heitz PU, Komminoth P, Perren A. VHL inactivation is an important pathway for the development of malignant sporadic pancreatic endocrine tumors. Endocr Relat Cancer. 2009;16:1219–1227. doi: 10.1677/ERC-08-0297. [DOI] [PubMed] [Google Scholar]
- Chen L, Han L, Zhang K, Shi Z, Zhang J, Zhang A, Wang Y, Song Y, Li Y, Jiang T. VHL regulates the effects of miR-23b on glioma survival and invasion via suppression of HIF-1alpha/VEGF and beta-catenin/Tcf-4 signaling. Neuro Oncol. 2012;14:1026–1036. doi: 10.1093/neuonc/nos122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daxinger L, Whitelaw E. Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nat Rev Genet. 2012;13:153–162. doi: 10.1038/nrm3288. [DOI] [PubMed] [Google Scholar]
- Rakyan VK, Blewitt ME, Druker R, Preis JI, Whitelaw E. Metastable epialleles in mammals. Trends Genet. 2002;18:348–351. doi: 10.1016/S0168-9525(02)02709-9. [DOI] [PubMed] [Google Scholar]
- Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- Butt RH, Pfeifer TA, Delaney A, Grigliatti TA, Tetzlaff WG, Coorssen JR. Enabling coupled quantitative genomics and proteomics analyses from rat spinal cord samples. Mol Cell Proteomics. 2007;6:1574–1588. doi: 10.1074/mcp.M700083-MCP200. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034-RESEARCH0034.11. [DOI] [PMC free article] [PubMed] [Google Scholar]


