Abstract
Background
Dilated cardiomyopathy (DCM) has been extensively investigated for many years, but its pathogenesis remains uncertain. The ACTC1 gene was the first sarcomeric gene whose mutation was shown to cause DCM; recent studies have indicated that the HSPB7 and ZBTB17 genes are also associated with DCM. To assess the potential role of these three genes in DCM, we examined 11 single nucleotide polymorphisms (SNPs) in the ZBTB17, HSPB7 and ACTC1 genes: namely, rs10927875 in ZBTB17; rs1739843, rs7523558, and rs6660685 in HSPB7; rs533021, rs589759, rs1370154, rs2070664, rs3759834, rs525720 and rs670957 in ACTC1.
Methods
A total of 97 DCM patients and 189 controls were included in the study. All SNPs were genotyped by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS).
Results
The genotype of SNP rs10927875 in ZBTB17 (OR=5.19, 95% CI =1.00 to 27.03, P=0.05) was associated with DCM in a Han Chinese population. There was no difference in genotype or allele frequencies in ACTC1 or HSPB7 between DCM patients and control subjects.
Conclusion
The ZBTB17 polymorphism rs10927875 appears to play a role in the susceptibility of the Han Chinese population to DCM.
Keywords: actci, Dilated cardiomyopathy, hspb7, Single nucleotide polymorphisms, zbtb17
Background
Dilated cardiomyopathy (DCM) is a primary myocardial disease and genetically heterogeneous disorder. It is characterized by progressive systolic dysfunction due to cardiac chamber dilatation and inefficient myocardial contractility. The phenotype in idiopathic DCM is characterized by cardiac muscle dysfunction in the absence of secondary causes. A majority of DCM cases are sporadic, but familial transmissions are observed in 25% to 35% of cases and are inherited as autosomal dominant, recessive, or X-linked traits with variable expressivity and penetrance [1]. Studies have led to the identification of mutations in more than 15 different genes coding for sarcomeric, cytoskeletal, or regulatory proteins as a primary cause of DCM [2,3].
HSPB7 encodes a cardiovascular small heat shock protein, HSP70, which belongs to the small HSP (sHSP) family [4]. HSPB7 is also called cardiovascular HSP, because of its selective expression in cardiovascular tissues [5]. In general, the expression and activation of heat shock proteins are influenced by elevated temperatures as well as ischemia, hypoxia and acute cellular stress [6,7]. In human beings, genetic variants in HSPB7 have been reported to be associated with advanced heart failure and systolic dysfunction of unspecific origin [8,9]. The single nucleotide polymorphism (SNP) is the most common type of genetic variation in the human genome, and recent DCM study has used large-scale screening on SNPs in European populations to show that HSPB7 gene SNPs (rs1739843) are associated with DCM [10].
The ACTC1 gene encodes skeletal muscle α-actin, which is the predominant actin isoform in the sarcomeric thin filaments of adult skeletal muscle; α-actin is also essential for cardiac muscle contraction. Each myosin head interacts with two adjacent actin monomers along the cardiac filament structure. The ACTC1 gene was the first sarcomeric gene whose mutation was shown to cause DCM [11]. To our knowledge, however, there have been no reports on the association between ACTC1 SNPs and DCM.
The ZBTB17 gene encodes protein 17, which contains both zincfinger and BTB domains. Protein 17 is also known as myc-interacting protein 1 (MIZ-1) and is a transcription factor of 87 kDa containing 13 zinc finger domains at its carboxy-terminal end and a BTB/POZ domain at its N-terminus [12]. MIZ-1 was originally identified as an interacting partner of the c-Myc proto-oncogene [12] and, depending on its interacting partner, MIZ-1, it can either activate or repress the transcription of its target genes [13-16]. Recently, a GWAS (genome-wide association study) study on DCM indicated that the ZBTB17 gene SNP rs10927875 was associated with DCM [17].
Based on these findings, we hypothesized that some DCM incidences in patients are associated with certain polymorphisms of ZBTB17, HSPB7 and ACTC1 genes. To our knowledge, there is no interaction among the three genes. To test this hypothesis, we used matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) to investigate 11 SNPs in ZBTB17, HSPB7 and ACTC1 in both DCM patients and normal subjects from a Han Chinese population.
Methods
Subjects
This case control study enrolled 97 unrelated DCM patients from the Fuwai Hospital in northern China from January 2006 to January 2007. Clinical diagnoses were made in accordance with the revised criteria [18], in which primary DCM was defined as systolic dysfunction (left ventricular (LV) ejection fraction <50%) with LV dilation in the absence of an apparent secondary cause of cardiomyopathy. In addition, 189 healthy unrelated individuals, without any sign or history of cardiovascular disease, were enrolled from a routine health survey as controls in this study. Patients with a history of hypertension, coronary heart disease, cardiac valve disease, diabetes, acute viral myocarditis, systemic diseases of putative autoimmune origin and a family history of DCM were intentionally excluded. This study was approved by the ethics committee of Fuwai Hospital; the subjects involved were from the Han nation in northern China. All subjects involved were aware of the study and gave written informed consent.
Isolation of DNA and genotyping by MALDI-TOF-MS
Blood samples were collected from patients using tubes containing ethylenediaminetetraacetic acid (EDTA). Genomic DNA was isolated from whole blood, and genotyping was performed by MALDI-TOF-MS, as described previously [19]. SNP genotyping was performed using the MassARRAY system (Sequenom, San Diego, California) using the MALDI-TOF-MS method. Completed genotyping reactions were spotted onto a 384-well spectroCHIP (Sequenom) using a MassARRAY Nanodispenser (Sequenom), and the genotype was determined by MALDI-TOF-MS. Genotype calling was performed in real time with MassARRAY RT software version 3.1 (Sequenom) and analyzed using the MassARRAY Typer software version 4.0 (Sequenom) (Table 1).
Table 1.
Sequences of PCR primers, amplicon sizes, temperatures and GC content in SNPs of ZBTB17, HSPB7 , and ACTC1 in DCM patients and control subjects
| Markers | Forward primer (5′-3′) | Reverse primer (5′-3′) | Amplicon size (bp) | Temperature (°C) | GC (%) |
|---|---|---|---|---|---|
|
ZBTB17 |
|
|
|
|
|
| rs10927875 |
GCCAGAGTGGATGATCACTG |
TTCTGTTTCCACCACTGTAG |
91 |
54.2 |
55 |
|
HSPB7 |
|
|
|
|
|
| rs1739843 |
TGTCCTCACTCTGCCATCAC |
TGGGCAGAGGGAGCCTGAG |
99 |
50.8 |
50 |
| rs7523558 |
GCCCAGCACCTATTTATAGC |
CTCATAGGCCAGTGATGAAG |
94 |
46.6 |
35 |
| rs6660685 |
TACTGTCCCACAGCCAGCAC |
TCTTCTCAGCCTCCTGGTG |
98 |
50.8 |
58.8 |
|
ACTC1 |
|
|
|
|
|
| rs533021 |
GGCCTTCCATTTGAATATGC |
GTATGAGAGCACATTTTCTG |
96 |
45.1 |
33.3 |
| rs589759 |
CTTCTCACCTCCCTAATTCC |
TGTGGCCTGGAGGCTTTAAG |
96 |
49.4 |
45 |
| rs1370154 |
GCAGCAACTCATTCTAGAG |
TTTCATACCTGAAAAGCAG |
108 |
45.2 |
24 |
| rs2070664 |
CATAACAATGACTGCTGCAC |
ATAGCTTGTGGAGATAGGTC |
100 |
46.9 |
60 |
| rs3759834 |
TCTCCATCAAAGTATTTGCC |
GATGAGCATCTTTAAACTGG |
100 |
52 |
45 |
| rs525720 |
GTTCTTGACTGGAGCTTTG |
GCCAGTGCCCAGTTTTCTAT |
103 |
45.6 |
33.3 |
| rs670957 | ATGCCAAGAGTAGAACTGCC | TCTGACACCAGTCCCATTTC | 97 | 45.5 | 50 |
Statistical analyses
Differences in the distributions of demographic characteristics, selected variables, and genotypes of the ZBTB17, HSPB7, and ACTC1 gene variants between the cases and controls were evaluated using the chi-square (χ2) test; continuous variables were analyzed by an unpaired Student’s t test. Associations between ZBTB17, HSPB7, and ACTC1 genotypes and the risk of DCM were estimated by computing the odds ratios (ORs), and their 95% confidence intervals (CIs) using logistic regression analysis. Hardy-Weinberg equilibrium was tested by a goodness-of fit χ2 test to compare the observed genotype frequencies with those expected among control subjects. All statistical analyses were performed with SPSS 16.0 (IBM; Armonk, USA), and a P value ≤0.05 was considered statistically significant.
Results
The study included 286 subjects, with 97 patients with DCM and 189 healthy control subjects. The mean age of control subjects was 54.0±3.6 years; 79.4% were men. Patients with DCM were of similar age (51.6±12.0 years), and there was a similar percentage of men (77.3%) (t=1.700, P=0.092 and χ2 =0.160, P=0.689, respectively). The percentage of smokers among DCM patients was 43.3%, but no data on smoking status were collected from healthy subjects. No subjects in the present study were pregnant or peripartum. Dilated cardiomyopathy was diagnosed as systolic dysfunction (LV ejection fraction <50%) with LV dilation detected by echocardiogram in the absence of an apparent secondary cause of cardiomyopathy. In DCM patients, the mean LV ejection fraction was 32.0±8.4%, the LV diameter was 67.7±8.6 mm, and the left atrium diameter 42.7±7.6 mm. The SNP genotype calling was performed in real time with the Mass ARRAY system and MALDI-TOF-MS. A few patients and controls whose genotypes could not be called: their genotypic data were omitted from analysis,thus the total number of called genotypes for some SNPs totaled less than 286. The observed and expected genotype frequencies of each SNP were not significantly different between DCM patients and control subjects, indicating that the samples fit the assumption of Hardy-Weinberg equilibrium (Table 2).
Table 2.
DNA variants identified inZBTB17,HSPB7, andACTC1
| Markers | Location of nucleotide change | Amino acid change | Note | ObsHET | PretHET | HWE(P) |
|---|---|---|---|---|---|---|
|
ZBTB17 |
|
|
|
|
|
|
| rs10927875 |
Intron 16299312 C>T |
Non-coding |
Nor-reported non-coding SNP |
0.025 |
0.025 |
0.9417 |
|
HSPB7 |
|
|
|
|
|
|
| rs1739843 |
Intron 16343254 C>T |
Non-coding |
Reported non-coding SNP |
0.413 |
0.381 |
0.7898 |
| rs7523558 |
5′ near gene 16346732 G>A |
Non-coding |
Nor-reported non-coding SNP |
0.156 |
0.15 |
0.7933 |
| rs6660685 |
5′ near gene 16346988 G>A |
Non-coding |
Nor-reported non-coding SNP |
0.029 |
0.028 |
0.9112 |
|
ACTC1 |
|
|
|
|
|
|
| rs533021 |
3′ UTR 35080931 T>C |
Non-coding |
Reported non-coding SNP |
0.446 |
0.453 |
1.0000 |
| rs589759 |
3′ UTR 35081574 C>T |
Non-coding |
Reported non-coding SNP |
0.524 |
0.497 |
0.2017 |
| rs1370154 |
3′ UTR 35082225 G>A |
Non-coding |
Reported non-coding SNP |
0.45 |
0.466 |
0.8150 |
| rs2070664 |
Intron 35085201 A>G |
Non-coding |
Nor-reported non-coding SNP |
0.47 |
0.491 |
0.9324 |
| rs3759834 |
5′ near gene 35088705 T>C |
Non-coding |
Nor-reported non-coding SNP |
0.06 |
0.058 |
0.6522 |
| rs525720 |
5′ near gene 35089134 G>A |
Non-coding |
Nor-reported non-coding SNP |
0.254 |
0.248 |
0.4169 |
| rs670957 | 5′ near gene 35089432 A>G | Non-coding | Nor-reported non-coding SNP | 0.482 | 0.5 | 0.5963 |
HWE(P), P value from Hardy-Weinberg equilibrium test in the control group; ObsHET, observed heterozygosity; PretHET, present heterozygosity.
Using the χ2 test, we compared genotypes and allele frequencies between cases and controls. Our results showed that the minor T allele frequency of SNP rs10927875 in ZBTB17 (OR=5.19, 95% CI =1.00 to 27.03, P=0.05) was associated with DCM in the Chinese population. In addition, as the number of homozygote ‘TT’ genotype carriers was zero among patients and controls and the minor T allele was considered a risky exposure factor, we compared the frequency of the heterozygous ‘CT’ genotype and the homozygous ‘CC’ genotype. The results showed that the ‘CT’ genotype might also be associated with DCM (OR=5.31, 95% CI =1.01 to 27.92, P=0.049). Multivariate analysis (logistic regression analysis) was also used to estimate the association of the T allele in rs10927875 and DCM. Logistic regression analysis indicated that the T allele was an independent risk factor for DCM after excluding some confounding factors, such as age and sex (OR= 6.42, 95% CI: 1.19 to 34.77; P= 0.031).
There was no difference in genotype or allele frequencies in ACTC1 or HSPB7 between DCM patients and control subjects. The allelic and genotypic frequencies of SNPs in cases and controls and the statistical analysis results are shown in Table 3.
Table 3.
Genotypic and allelic frequencies of SNPs fromZBTB17, HSPB7, and ACTC1 genes in DCM patients and controls
| Marker | Genotype | Allele | P value | OR(95% CI) | |||
|---|---|---|---|---|---|---|---|
|
ZBTB17 |
|
|
|
|
|||
| rs10927875 |
C/C |
C/T |
TT |
C |
T |
0.05 |
5.19(1.00, 27.03) |
| Patients |
88(0.946) |
5(0.054) |
0 |
181(0.973) |
5(0.054) |
|
|
| Controls |
187(0.989) |
2(0.011) |
0 |
376(0.995) |
2(0.011) |
|
|
|
HSPB7 |
|
|
|
|
|
|
|
| rs1739843 |
C/C |
C/T |
T/T |
C |
T |
0.81 |
0.95(0.64, 1.42) |
| Patients |
49(0.521) |
43(0.457) |
2(0.021) |
141(0.75) |
47(0.250) |
|
|
| Controls |
103(0.545) |
74(0.392) |
12(0.063) |
280(0.741) |
98(0.259) |
|
|
| rs7523558 |
A/A |
A/G |
G/G |
A |
G |
0.96 |
0.98(0.52, 1.87 ) |
| Patients |
0(0) |
15(0.161) |
78(0.839) |
15(0.081) |
171(0.919) |
|
|
| Controls |
1(0.005) |
29(0.153) |
159(0.841) |
31(0.082) |
347(0.918) |
|
|
| rs6660685 |
A/A |
A/G |
G/G |
A |
G |
0.11 |
3.29(0.78, 13.91) |
| Patients |
0 |
5(0.053) |
90(0.947) |
5(0.026) |
185(0.974) |
|
|
| Controls |
0 |
3(0.016) |
181(0.984) |
3(0.008) |
365(0.992) |
|
|
|
ACTC1 |
|
|
|
|
|
|
|
| rs533021 |
C/C |
C/T |
T/T |
C |
T |
0.36 |
1.19(0.82, 1.73) |
| Patients |
13(0.149) |
39(0.448) |
35(0.402) |
65(0.374) |
109(0.626) |
|
|
| Controls |
21(0.111) |
84(0.444) |
84(0.444) |
126(0.333) |
252(0.667) |
|
|
| rs589759 |
C/C |
C/T |
T/T |
C |
T |
0.25 |
1.22(0.87, 1.73) |
| Patients |
25(0.258) |
48(0.495) |
24(0.247) |
98(0.505) |
96(0.495) |
|
|
| Controls |
54(0.286) |
102(0.540) |
33(0.175) |
210(0.556) |
168(0.444) |
|
|
| rs1370154 |
A/A |
A/G |
G/G |
A |
G |
0.77 |
0.93(0.58, 1.50) |
| Patients |
7(0.156) |
18(0.4) |
20(0.444) |
32(0.356) |
58(0.644) |
|
|
| Controls |
26(0.141) |
85(0.462) |
73(0.397) |
137(0.372) |
231(0.628) |
|
|
| rs2070664 |
A/A |
A/G |
G/G |
A |
G |
0.84 |
1.04(0.73, 1.47) |
| Patients |
33(0.344) |
42(0.438) |
21(0.219) |
108(0.562) |
84(0.438) |
|
|
| Controls |
62(0.328) |
92(0.487) |
35(0.185) |
216(0.571) |
162(0.429) |
|
|
| rs3759834 |
C/C |
C/T |
T/T |
C |
T |
0.72 |
0.82(0.29, 2.37) |
| Patients |
0 |
5(0.053) |
90(0.947) |
5(0.026) |
185(0.974) |
|
|
| Controls |
0 |
12(0.063) |
177(0.937) |
12(0.032) |
366(0.968) |
|
|
| rs525720 |
A/A |
A/G |
G/G |
A |
G |
0.42 |
0.81(0.48, 1.36) |
| Patients |
2(0.022) |
19(0.211) |
69(0.767) |
23(0.128) |
157(0.872) |
|
|
| Controls |
3(0.016) |
52(0.275) |
134(0.709) |
58(0.153) |
320(0.847) |
|
|
| rs670957 |
A/A |
A/G |
G/G |
A |
G |
0.09 |
1.36(0.95, 1.94) |
| Patients |
19(0.211) |
44(0.489) |
27(0.3) |
82(0.456) |
98(0.544) |
|
|
| Controls | 55(0.293) | 90(0.479) | 43(0.229) | 200(0.532) | 176(0.468) | ||
CI, confidence interval; OR, odds ratio in which the minor allele was viewed as an exposure factor in the case-controlled study.
Because some studies have documented that men account for a much larger percentage of DCM patients [18,20], we compared the frequencies of the genotype of SNP rs10927875 between cases and controls within sexes. For both men and women, the genotype of SNP rs10927875 showed no significant difference between patients and control subjects (χ2=3.374, P=0.066; χ2=1.889, P=0.169).
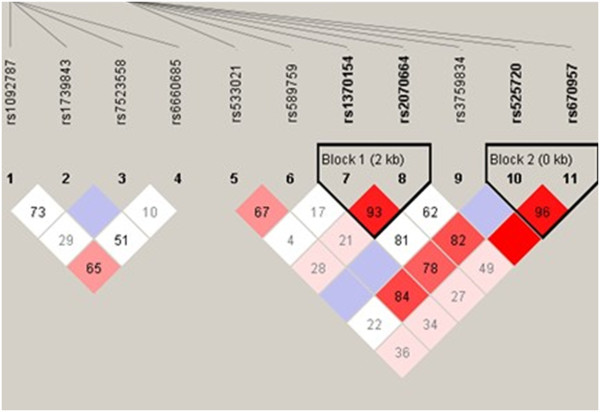
Non-random associations between polymorphic variants at different loci on the three genes were then measured by the degree of linkage disequilibrium (LD). Analysis of LD showed that none of the SNPs in the three genes was in high LD in the DCM patients (Figure 1, D′<0.80). Haplotype analysis showed that GG, AA, GA, and AG in block 1 and GG, GA, and AA in block 2 were not correlated significantly with DCM (Figure 1, Table 4).
Figure 1.
Pairwise linkage disequilibrium (LD) values calculated between tagging SNPs in ZBTB17, HSPB7, and ACTC1. The value within each diamond represents the pairwise correlation between tagging SNPs (measured as D′), defined by the upper left and the upper right sides of the diamond. The diamond without a number corresponds to D′= 1. Shading represents the magnitude and significance of the pairwise LD, with darker red reflecting higher LD values and white indicating lower LD values.
Table 4.
Haplotype analysis of SNPs between DCM patients and control subjects
| Haplotype | Frequency (DCM patients) | Frequency (control subjects) | χ2 | Pvalue | |
|---|---|---|---|---|---|
|
Block 1 |
GG |
0.426 |
0.418 |
0.028 |
0.8681 |
| |
AA |
0.371 |
0.363 |
0.038 |
0.8456 |
| |
GA |
0.192 |
0.209 |
0.211 |
0.6460 |
| |
AG |
0.011 |
0.010 |
0.008 |
0.9293 |
|
Block 2 |
GG |
0.544 |
0.464 |
3.166 |
0.0752 |
| |
GA |
0.328 |
0.383 |
1.589 |
0.2075 |
| AA | 0.127 | 0.150 | 0.524 | 0.4691 |
Discussion
We have analyzed the relationship between DCM and SNPs in the genes HSPB7, ZBTB17, and ACTC1. It has been shown that the T allele of ZBTB17 (rs10927875) and the ‘CT’ genotype are associated with DCM in a subgroup of patients within the Chinese population. Moreover, the T allele in rs10927875 was determined to be an independent risk factor for DCM, suggesting that ZBTB17 polymorphisms (rs10927875) play an important role in the susceptibility to DCM in the Chinese population.
Dilated cardiomyopathy is a common form of heart muscle disease with a prevalence of 1/2,500 in the general population. It represents a major cause of cardiovascular morbidity and mortality and is characterized by systolic dysfunction, dilation, and impaired contraction of the ventricles, often leading to chronic heart failure and eventually requiring cardiac transplantation [18]. In approximately 35% of cases, DCM is a familial disease [20]. Mutations in both sarcomeric and cytoskeletal genes have been implicated in DCM, but the variable expression and penetrance of each gene that harbors a different mutation result in vast clinical heterogeneity among patients. Knowledge of the genetic risk factors for DCM is important to initiate treatment prior to symptomatic onset of the disease to delay its occurrence or possibly halt its progression. To date, only a few common susceptibility alleles for sporadic DCM have been identified from candidate-gene approaches [20,21].
The most common form of genetic variation is the SNP, defined according to the variation of a single nucleotide occurring in more than 1% of the population. The majority of SNPs are likely to be allelic variants that do not affect the expression or function of a protein. Such SNPs are commonly used as genetic markers to localize nearby disease-causing variations in linkage and association analyses. Single nucleotide polymorphisms that directly influence phenotype may be located within coding or regulatory regions of genes and can result in disease. In contrast, SNPs within regulatory regions tend to have more quantitative effects; for example, they may alter the expression level of a receptor or signaling protein, resulting in a more subtle variation in the associated phenotype [22].
Understanding the genetic heterogeneity of complex polygenic diseases like DCM is challenging. One of the many achievements in medical genetics over the past decades has been the ability to visualize sequence differences directly in DNA. These differences, or polymorphisms, serve as genetic markers of disease [23]. Recently, a GWAS was performed on a sporadic form of DCM in a European population. Another study screened approximately 2,000 candidate genes previously implicated in cardiovascular disease in more than 1,900 sporadic DCM cases in German and French populations. These studies showed that the SNPs rs10927875 and rs1739843 in ZBTB17 and HSPB7, respectively, were associated with DCM [10,17].
rs10927875 is located in an intron of ZBTB17 on chromosome 1p36.2-p36.1. The locus of interest covers approximately 210 kb in a genomic region; exhibits strong LD; and spans several other genes, including SPEN (spen homolog, transcriptional regulator), HSPB7, CLCNKA (chloride channel Ka), and CLCNKB (chloride channel Kb) [17]. Our data, together with the results from previous GWAS on DCM, substantiate the importance of rs10927875 and related polymorphisms in the ZBTB17 locus for DCM susceptibility [17]. However, the biological mechanism explaining the association between the polymorphism rs10927875 and DCM risk remains unclear. Preliminary data suggest that MIZ-1 (a transcription factor encoded by ZBTB17) may play a role in the transcriptional activation of numerous other genes [13-16]. MIZ-1 is composed of 13 zinc finger domains at its C terminus and a BTB/POZ (Broad-complex, Tramtrack, and Bric-a-brac/pox virus zinc finger) domain at its N-terminus [13]. Whether MIZ-1 activates or represses the transcription of its target genes depends on its interacting partner. The genes that encode the negative cell cycle regulators Cdkn2b [13,16] or Cdkn1a [24,25] have been validated as direct MIZ-1 targets, and c-Myc has been shown to be recruited to the Cdkn1a promoter by MIZ-1; this interaction blocks Cdkn1a induction by p53 and other activators in cancer cells [13,16,24,25]. As a result of Miz-1 actions, c-Myc switches the cell fate from cell cycle arrest to apoptosis in response to p53-dependent activation [24]. In vitro studies have shown that the MIZ-1 pathway is indispensable for the ability of c-Myc to act as an oncogene, inducing both cell cycle progression and transformation. The ability of c-Myc to bind and inactivate MIZ-1 is required for c-Myc’s induction of apoptosis in growth factor-deprived primary human fibroblasts [26]. In a recent study, MIZ-1 was shown to have an additional function in pre-T cell differentiation at the b-selection stage of DN3 cells, the first critical checkpoint in the maturation of pre-T cells [27]. As it is well known that both immunity and apoptosis play an important role in the pathology of DCM, the polymorphism in the ZBTB17 gene might be associated with DCM. Clearly, functional studies are required to support these hypotheses.
It has become clear that certain heat shock proteins functioning as molecular chaperones form a potent natural defense against proteotoxic stress induced by protein misfolding diseases. HSPB7 is known to be expressed in cardiovascular and insulin-sensitive tissues [5]. In general, the expression and activation of heat shock proteins are influenced by elevated temperatures as well as ischemia, hypoxia and acute cellular stress [6,7]. In aging skeletal muscle, an increase in HSP70 protein content has been observed [28]. Recently, Cappola reported an association between rs1739843 and heart failure [9]. The findings on HSPB7 are also in line with a previously reported large-scale resequencing approach of four biologically relevant cardiac signaling genes, in which HSPB7 sequence diversity was detected in sporadic cardiomyopathy [8,10]. However, in the present case control study, we identified a polymorphism (rs1739843) in intron 2 of the HSPB7 gene that had no association with susceptibility to DCM in a Han Chinese population. Many factors account for the difference between the results of our study with the abovementioned studies: the subjects differed in race, the enrolment criteria for DCM patients differed, the patient statuses differed, and so on. In addition, we did not find any association between SNPs in ACTC1 and DCM.
To our knowledge, this study is the first to describe the association of SNPs in ZBTB17, HSPB7, and ACTC1 with DCM in a Han Chinese population. This study has provided the first evidence that a ZBTB17 gene SNP (rs10927875) is a risk factor for susceptibility to DCM in Chinese populations. The present study found no association between SNPs in HSPB7 and ACTC1 genes with DCM.
Conclusion
In conclusion, we have provided the first evidence that a ZBTB17 gene SNP (rs10927875) is a risk factor for susceptibility to DCM in Chinese populations. However, genetic polymorphisms vary greatly among different ethnic populations; therefore, further studies in other populations are needed to exclude a population-oriented association.
Limitations
Some limitations may have affected the accuracy of the results. One limitation was the relatively small sample size. In addition, as there were a few patients and controls whose genotypes could not be called, their genotypic data were omitted from analysis, and thus, the total number of called genotypes for some SNPs was less than 286. Secondly, no data on body mass index, smoking status, or pregnancy history were obtained from healthy subjects. Thirdly, as the control subjects were recruited using a routine health survey and lacked any signs or history of cardiovascular disease, we did not obtain echocardiograph parameters to fully exclude cardiovascular diseases in the healthy subjects. Last but not least, a possible limitation of our study is the lack of a more comprehensive genetic analysis of other SNPs in ZBTB17, which may also play a role in DCM susceptibility. Taken together, all of these limitations may have affected the results of this study.
Abbreviations
CI: Confidence interval; DCM: Dilated cardiomyopathy; EDTA: Ethylenediaminetetraacetic acid; GWAS: Genome-wide association study; HSP: Heat shock protein; LD: Linkage disequilibrium; LV: Left ventricular; MALDI-TOF-MS: Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry; MIZ-1: Myc-interacting protein 1; OR: odds ratio; PCR: Polymerase chain reaction; sHSP: small HSP; SNP: Single nucleotide polymorphism
Competing interests
The authors declare no conflict of interest.
Authors’ contributions
XL and RL performed all in vitro experimental studies and data acquisition, and contributed to the study conception, design, analysis, and data interpretation. XM, RJ, and HK collected serum samples, performed data analysis, and drafted the manuscript. WH and XW designed, set up, and monitored the study from which the serum samples were obtained. All authors read and approved the final manuscript.
Contributor Information
Xiaoping Li, Email: lixiaoping0119@163.com.
Rong Luo, Email: luorong77@126.com.
Xiaoyang Mo, Email: moxiaoyang@yahoo.com.cn.
Rongjian Jiang, Email: RongjianJiang@yahoo.cn.
Hong Kong, Email: khong1212@yahoo.cn.
Wei Hua, Email: drhua@yahoo.cn.
Xiushan Wu, Email: xiushanwu@yahoo.com.
Acknowledgements
This study was supported in part by grants from the National Natural Science Foundation of China (no. 81000104, 31171402, 81160141), the China Postdoctoral Science Foundation (2012M510356) and the Postdoctoral Fellows Foundation of the Chinese Academy of Medical Sciences (2011-XH-9).
References
- Kamisago M, Sharma SD, DePalma SR, Solomon S, Sharma P, McDonough B, Smoot L, Mullen MP, Woolf PK, Wigle ED, Seidman JG, Seidman CE. Mutations in sarcomere protein genes as a cause of dilated cardiomyopathy. N Engl J Med. 2000;343:1688–1696. doi: 10.1056/NEJM200012073432304. [DOI] [PubMed] [Google Scholar]
- Burkett EL, Hershberger RE. Clinical and genetic issues in familial dilated cardiomyopathy. J Am Coll Cardiol. 2005;45:969–981. doi: 10.1016/j.jacc.2004.11.066. [DOI] [PubMed] [Google Scholar]
- Murphy RT, Mogensen J, Shaw A, Kubo T, Hughes S, McKenna WJ. Novel mutation in cardiac troponin I in recessive idiopathic dilated cardiomyopathy. Lancet. 2004;363:371–372. doi: 10.1016/S0140-6736(04)15468-8. [DOI] [PubMed] [Google Scholar]
- Krief S, Faivre JF, Robert P, Le Douarin B, Brument-Larignon N, Lefre’re I, Bouzyk MM, Anderson KM, Greller LD, Tobin FL, Souchet M, Bril A. Identification and characterization of cvHsp. A novel human small stress protein selectively expressed in cardiovascular and insulin-sensitive tissues. J BiolChem. 1999;274:36592–36600. doi: 10.1074/jbc.274.51.36592. [DOI] [PubMed] [Google Scholar]
- Kappé G, Franck E, Verschuure P, Boelens WC, Leunissen JA, de Jong WW. The human genome encodes 10 α-crystallin-related small heat shock proteins: HspB1-10. Cell Stress Chaperones. 2003;8:53–61. doi: 10.1379/1466-1268(2003)8<53:THGECS>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S, Craig EA. The heat-shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- Anckar J, Sistonen L. Heat shock factor 1 as a coordinator of stress and developmental pathways. Adv Exp Med Biol. 2007;594:78–88. doi: 10.1007/978-0-387-39975-1_8. [DOI] [PubMed] [Google Scholar]
- Matkovich SJ, Van Booven DJ, Hindes A, Kang MY, Druley TE, Vallania FLM, Mitra RD, Reilly MP, Cappola TP, Dorn GW. Cardiac signaling genes exhibit unexpected sequence diversity in sporadic cardiomyopathy, revealing HSPB7 polymorphisms associated with disease. J Clin Invest. 2010;120:280–289. doi: 10.1172/JCI39085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappola TP, Li M, He J, Ky B, Gilmore J, Qu L, Keating B, Reilly M, Kim CE, Glessner J, Frackelton E, Hakonarson H, Syed F, Hindes A, Matkovich SJ, Cresci S, Dorn GW. Common variants in HSPB7 and FRMD4B associated with advanced heart failure. Circ Cardiovasc Genet. 2010;3:147–154. doi: 10.1161/CIRCGENETICS.109.898395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark K, Esslinger UB, Reinhard W, Petrov G, Winkler T, Komajda M, Isnard R, Charron P, Villard E, Cambien F, Tiret L, Aumont MC, Dubourg O, Trochu JN, Fauchier L, Degroote P, Richter A, Maisch B, Wichter T, Zollbrecht C, Grassl M, Schunkert H, Linsel-Nitschke P, Erdmann J, Baumert J, Illig T, Klopp N, Wichmann HE, Meisinger C, Koenig W. Genetic association study identifies HSPB7 as a risk gene for idiopathic dilated cardiomyopathy. PLoS Genet. 2010;6:e1001167. doi: 10.1371/journal.pgen.1001167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson TM, Michels VV, Thibodeau SN, Tai YS, Keating MT. Actin mutations in dilated cardiomyopathy, a heritable form of heart failure. Science. 1998;280:750–752. doi: 10.1126/science.280.5364.750. [DOI] [PubMed] [Google Scholar]
- Peukert K, Staller P, Schneider A, Carmichael G, Hänel F, Eilers M. An alternative pathway for gene regulation by Myc. EMBO J. 1997;16:5672–5686. doi: 10.1093/emboj/16.18.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staller P, Peukert K, Kiermaier A, Seoane J, Lukas J, Karsunky H, Möröy T, Bartek J, Massagué J, Hänel F, Eilers M. Repression of p15INK4b expression by Myc through association with Miz-1. Nat Cell Biol. 2001;3:392–399. doi: 10.1038/35070076. [DOI] [PubMed] [Google Scholar]
- Adhikary S, Marinoni F, Hock A, Hulleman E, Popov N, Beier R, Bernard S, Quarto M, Capra M, Goettig S, Kogel U, Scheffner M, Helin K, Eilers M. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell. 2005;123:409–421. doi: 10.1016/j.cell.2005.08.016. [DOI] [PubMed] [Google Scholar]
- Herold S, Wanzel M, Beuger V, Frohme C, Beul D, Hillukkala T, Syvaoja J, Saluz HP, Haenel F, Eilers M. Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol Cell. 2002;10:509–521. doi: 10.1016/S1097-2765(02)00633-0. [DOI] [PubMed] [Google Scholar]
- Seoane J, Pouponnot C, Staller P, Schader M, Eilers M, Massague J. TGFβ influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat Cell Biol. 2001;3:400–408. doi: 10.1038/35070086. [DOI] [PubMed] [Google Scholar]
- Villard E, Perret C, Gary F, Proust C, Dilanian G, Hengstenberg C, Ruppert V, Arbustini E, Wichter T, Germain M, Dubourg O, Tavazzi L, Aumont MC, DeGroote P, Fauchier L, Trochu JN, Gibelin P, Aupetit JF, Stark K, Erdmann J, Hetzer R, Roberts AM, Barton PJ, Regitz-Zagrosek V, Cardiogenics C, Aslam U, Duboscq-Bidot L, Meyborg M, Maisch B, Madeira H. A genome-wide association study identifies two loci associated with heart failure due to dilated cardiomyopathy. Eur Heart J. 2011;32:1065–1076. doi: 10.1093/eurheartj/ehr105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maron BJ, Towbin JA, Thiene G, Antzelevitch C, Corrado D, Arnett D, Moss AJ, Seidman CE, Young JB. Contemporary definitions and classification of the cardiomyopathies: an American Heart Association Scientific Statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee; quality of care and outcomes research and functional genomics and translational biology interdisciplinary working groups; and council on epidemiology and prevention. Circulation. 2006;113:1807–1816. doi: 10.1161/CIRCULATIONAHA.106.174287. [DOI] [PubMed] [Google Scholar]
- Schaeffeler E, Zanger UM, Eichelbaum M, Asante-Poku S, Shin JG, Schwab M. Highly multiplexed genotyping of thiopurine s-methyltransferase variants using MALD-TOF mass spectrometry: reliable genotyping in different ethnic groups. Clin Chem. 2008;54:1637–1647. doi: 10.1373/clinchem.2008.103457. [DOI] [PubMed] [Google Scholar]
- Richard P, Villard E, Charron P, Isnard R. The genetic bases of cardiomyopathies. J Am Coll Cardiol. 2006;48:A79–A89. doi: 10.1016/j.jacc.2006.09.014. [DOI] [Google Scholar]
- Rampersaud E, Kinnamon DD, Hamilton K, Khuri S, Hershberger RE, Martin ER. Common susceptibility variants examined for association with dilated cardiomyopathy. Ann Hum Genet. 2010;74:110–116. doi: 10.1111/j.1469-1809.2010.00566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho E, Bhindi R, Ashley EA, Figtree GA. Genetic analysis in cardiovascular disease: a clinical perspective. Cardiol Rev. 2011;19:81–89. doi: 10.1097/CRD.0b013e318207ffac. [DOI] [PubMed] [Google Scholar]
- Alameddine FM, Zafari AM. Genetic polymorphisms and oxidative stress in heart failure. Congest Heart Fail. 2002;8:157–164. doi: 10.1111/j.1527-5299.2002.00719.x. [DOI] [PubMed] [Google Scholar]
- Seoane J, Le HV, Massagué J. Myc suppression of the p21Cip1Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature. 2002;419:729–734. doi: 10.1038/nature01119. [DOI] [PubMed] [Google Scholar]
- Wu S, Cetinkaya C, Munoz-Alonso MJ, von der Lehr N, Bahram F, Beuger V, Eilers M, Leon J, Larsson LG. Myc represses differentiation-induced p21CIP1 expression via Miz-1-dependent interaction with the p21 core promoter. Oncogene. 2003;22:351–360. doi: 10.1038/sj.onc.1206145. [DOI] [PubMed] [Google Scholar]
- Patel JH, McMahon SB. Targeting of Miz-1 is essential for Myc-mediated apoptosis. J Biol Chem. 2006;281:3283–3289. doi: 10.1074/jbc.M513038200. [DOI] [PubMed] [Google Scholar]
- Saba I, Kosan C, Vassen L, Klein-Hitpass L, Möröy T. Miz-1 is required to coordinate the expression of TCRβ and p53 effector genes at the pre-TCR “β-selection” checkpoint. J Immunol. 2011;187:2982–2992. doi: 10.4049/jimmunol.1101451. [DOI] [PubMed] [Google Scholar]
- Doran P, Gannon J, O’Connell K, Ohlendieck K. Aging skeletal muscle shows a drastic increase in the small heat shock proteins αB-crystallin/HspB5 and cvHsp/HspB7. Eur J Cell Biol. 2007;86:629–640. doi: 10.1016/j.ejcb.2007.07.003. [DOI] [PubMed] [Google Scholar]