Abstract
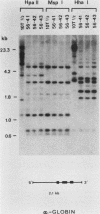
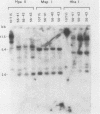
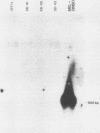
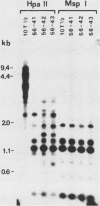
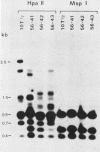
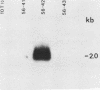
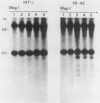
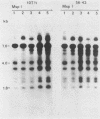
In previous work, three clonal cell lines with extremely low DNA methylation levels were derived by multiple consecutive treatments of C3H 10T1/2 C18 (10T1/2) cells with 5-aza-2'-deoxycytidine (5-aza-CdR). In this study we examined the methylation status of genes in these three methyl-deficient clones to assess the specificity of the induced hypomethylation. Complete demethylation of virtually all 5'-CCGG-3' sites was observed in four genes examined, but some sites common to all three clones were persistently methylated even after further exhaustive 5-aza-CdR treatment. Thus, there is a subset of methylation sites within these cells which can never be stably demethylated. The extensive demethylation was not always associated with changes in the level of RNA expression of the genes examined but was strongly correlated with an altered chromatin structure of the unexpressed alpha 1-globin gene and the muscle determination gene MyoD1. These results provide a direct correlation between hypomethylation and the induction of a transcriptionally competent chromatin state.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Becker P. B., Ruppert S., Schütz G. Genomic footprinting reveals cell type-specific DNA binding of ubiquitous factors. Cell. 1987 Nov 6;51(3):435–443. doi: 10.1016/0092-8674(87)90639-8. [DOI] [PubMed] [Google Scholar]
- Bird A. P. DNA methylation--how important in gene control? Nature. 1984 Feb 9;307(5951):503–504. doi: 10.1038/307503a0. [DOI] [PubMed] [Google Scholar]
- Bolden A. H., Nalin C. M., Ward C. A., Poonian M. S., McComas W. W., Weissbach A. DNA methylation: sequences flanking C-G pairs modulate the specificity of the human DNA methylase. Nucleic Acids Res. 1985 May 24;13(10):3479–3494. doi: 10.1093/nar/13.10.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolden A. H., Nalin C. M., Ward C. A., Poonian M. S., Weissbach A. Primary DNA sequence determines sites of maintenance and de novo methylation by mammalian DNA methyltransferases. Mol Cell Biol. 1986 Apr;6(4):1135–1140. doi: 10.1128/mcb.6.4.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschhausen G., Wittig B., Graessmann M., Graessmann A. Chromatin structure is required to block transcription of the methylated herpes simplex virus thymidine kinase gene. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1177–1181. doi: 10.1073/pnas.84.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouse G. F., Simonsen C. C., McEwan R. N., Schimke R. T. Structure of amplified normal and variant dihydrofolate reductase genes in mouse sarcoma S180 cells. J Biol Chem. 1982 Jul 10;257(13):7887–7897. [PubMed] [Google Scholar]
- Davis R. L., Weintraub H., Lassar A. B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987 Dec 24;51(6):987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Doerfler W. DNA methylation and gene activity. Annu Rev Biochem. 1983;52:93–124. doi: 10.1146/annurev.bi.52.070183.000521. [DOI] [PubMed] [Google Scholar]
- Ehrlich M., Gama-Sosa M. A., Huang L. H., Midgett R. M., Kuo K. C., McCune R. A., Gehrke C. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982 Apr 24;10(8):2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Flatau E., Gonzales F. A., Michalowsky L. A., Jones P. A. DNA methylation in 5-aza-2'-deoxycytidine-resistant variants of C3H 10T1/2 C18 cells. Mol Cell Biol. 1984 Oct;4(10):2098–2102. doi: 10.1128/mcb.4.10.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Sosa M. A., Midgett R. M., Slagel V. A., Githens S., Kuo K. C., Gehrke C. W., Ehrlich M. Tissue-specific differences in DNA methylation in various mammals. Biochim Biophys Acta. 1983 Jun 24;740(2):212–219. doi: 10.1016/0167-4781(83)90079-9. [DOI] [PubMed] [Google Scholar]
- Groudine M., Eisenman R., Weintraub H. Chromatin structure of endogenous retroviral genes and activation by an inhibitor of DNA methylation. Nature. 1981 Jul 23;292(5821):311–317. doi: 10.1038/292311a0. [DOI] [PubMed] [Google Scholar]
- Harrington M. A., Jones P. A., Imagawa M., Karin M. Cytosine methylation does not affect binding of transcription factor Sp1. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2066–2070. doi: 10.1073/pnas.85.7.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeveler A., Doerfler W. Specific factors binding to the late E2A promoter region of adenovirus type 2 DNA: no apparent effects of 5'-CCGG-3' methylation. DNA. 1987 Oct;6(5):449–460. doi: 10.1089/dna.1987.6.449. [DOI] [PubMed] [Google Scholar]
- Hsiao W. L., Gattoni-Celli S., Kirschmeier P., Weinstein I. B. Effects of 5-azacytidine on methylation and expression of specific DNA sequences in C3H 10T1/2 cells. Mol Cell Biol. 1984 Apr;4(4):634–641. doi: 10.1128/mcb.4.4.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao W. L., Gattoni-Celli S., Weinstein I. B. Effects of 5-azacytidine on expression of endogenous retrovirus-related sequences in C3H 10T1/2 cells. J Virol. 1986 Mar;57(3):1119–1126. doi: 10.1128/jvi.57.3.1119-1126.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. A., Taylor S. M. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980 May;20(1):85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- Keshet I., Lieman-Hurwitz J., Cedar H. DNA methylation affects the formation of active chromatin. Cell. 1986 Feb 28;44(4):535–543. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- Keshet I., Yisraeli J., Cedar H. Effect of regional DNA methylation on gene expression. Proc Natl Acad Sci U S A. 1985 May;82(9):2560–2564. doi: 10.1073/pnas.82.9.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leder A., Swan D., Ruddle F., D'Eustachio P., Leder P. Dispersion of alpha-like globin genes of the mouse to three different chromosomes. Nature. 1981 Sep 17;293(5829):196–200. doi: 10.1038/293196a0. [DOI] [PubMed] [Google Scholar]
- Lehrach H., Diamond D., Wozney J. M., Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977 Oct 18;16(21):4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- Lewis S. A., Balcarek J. M., Krek V., Shelanski M., Cowan N. J. Sequence of a cDNA clone encoding mouse glial fibrillary acidic protein: structural conservation of intermediate filaments. Proc Natl Acad Sci U S A. 1984 May;81(9):2743–2746. doi: 10.1073/pnas.81.9.2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock L. F., Takagi N., Martin G. R. Methylation of the Hprt gene on the inactive X occurs after chromosome inactivation. Cell. 1987 Jan 16;48(1):39–46. doi: 10.1016/0092-8674(87)90353-9. [DOI] [PubMed] [Google Scholar]
- McClelland M. The effect of sequence specific DNA methylation on restriction endonuclease cleavage. Nucleic Acids Res. 1981 Nov 25;9(22):5859–5866. doi: 10.1093/nar/9.22.5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalowsky L. A., Jones P. A. Differential nuclear protein binding to 5-azacytosine-containing DNA as a potential mechanism for 5-aza-2'-deoxycytidine resistance. Mol Cell Biol. 1987 Sep;7(9):3076–3083. doi: 10.1128/mcb.7.9.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka Y., Leder P. The complete sequence of a chromosomal mouse alpha--globin gene reveals elements conserved throughout vertebrate evolution. Cell. 1979 Nov;18(3):875–882. doi: 10.1016/0092-8674(79)90139-9. [DOI] [PubMed] [Google Scholar]
- Razin A., Szyf M. DNA methylation patterns. Formation and function. Biochim Biophys Acta. 1984 Sep 10;782(4):331–342. doi: 10.1016/0167-4781(84)90043-5. [DOI] [PubMed] [Google Scholar]
- Reeves R. Transcriptionally active chromatin. Biochim Biophys Acta. 1984 Sep 10;782(4):343–393. doi: 10.1016/0167-4781(84)90044-7. [DOI] [PubMed] [Google Scholar]
- Reznikoff C. A., Brankow D. W., Heidelberger C. Establishment and characterization of a cloned line of C3H mouse embryo cells sensitive to postconfluence inhibition of division. Cancer Res. 1973 Dec;33(12):3231–3238. [PubMed] [Google Scholar]
- Russell D. W., Zinder N. D. Hemimethylation prevents DNA replication in E. coli. Cell. 1987 Sep 25;50(7):1071–1079. doi: 10.1016/0092-8674(87)90173-5. [DOI] [PubMed] [Google Scholar]
- Selig S., Ariel M., Goitein R., Marcus M., Cedar H. Regulation of mouse satellite DNA replication time. EMBO J. 1988 Feb;7(2):419–426. doi: 10.1002/j.1460-2075.1988.tb02829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker E. U., Jensen B. C., Richardson G. A. A portable signal causing faithful DNA methylation de novo in Neurospora crassa. Science. 1987 Oct 2;238(4823):48–53. doi: 10.1126/science.2958937. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]