Abstract
Background
This study is aimed at the analysis of genetic and physiological effects of myostatin on economically relevant meat quality traits in a genetic background of high muscularity. For this purpose, we generated G3 populations of reciprocal crosses between the two hypermuscular mouse lines BMMI866, which carries a myostatin mutation and is lean, and BMMI806, which has high intramuscular and body fat content. To assess the relationship between muscle mass, body composition and muscle quality traits, we also analysed intramuscular fat content (IMF), water holding capacity (WHC), and additional physiological parameters in M. quadriceps and M. longissimus in 308 G3-animals.
Results
We found that individuals with larger muscles have significantly lower total body fat (r = −0.28) and IMF (r = −0.64), and in females, a lower WHC (r = −0.35). In males, higher muscle mass was also significantly correlated with higher glycogen contents (r = 0.2) and lower carcass pH-values 24 hours after dissection (r = −0.19). Linkage analyses confirmed the influence of the myostatin mutation on higher lean mass (1.35 g), reduced body fat content (−1.15%), and lower IMF in M. longissimus (−0.13%) and M. quadriceps (−0.07%). No effect was found for WHC. A large proportion of variation of intramuscular fat content of the M. longissimus at the myostatin locus could be explained by sex (23%) and direction-of-cross effects (26%). The effects were higher in males (+0.41%). An additional locus with negative over-dominance effects on total fat mass (−0.55 g) was identified on chromosome 16 at 94 Mb (86–94 Mb) which concurs with fat related QTL in syntenic regions on SSC13 in pigs and BTA1 in cattle.
Conclusion
The data shows QTL effects on mouse muscle that are similar to those previously observed in livestock, supporting the mouse model. New information from the mouse model helps to describe variation in meat quantity and quality, and thus contribute to research in livestock.
Background
In livestock production, there is a high interest in controlling meat quantity and quality; knowledge about genes affecting muscle size and other meat properties can help breeders to select animals according to desired traits. Myostatin (Mstn, or growth and differentiation factor 8 - Gdf8) was first identified in knock-out mice as being a gene responsible for regulation of muscle growth [1]. However, the hypermuscular phenotype that originates from a hyperplasia and hypertrophy of muscle fibres was known long before its molecular-genetic background was elucidated [2]. In sheep, pigs and cattle, several mutations of myostatin or its promoter region have been identified as affecting muscle size [3-6]. Furthermore, myostatin also influences glucose metabolism and fat accumulation, as shown in knock-out mice that had smaller adipocytes [7] and did not develop obesity on a high-fat diet. These results suggested an altered metabolism for the utilization of lipids and glucose as energy fuel that prevented insulin resistance in mice [8,9]. A decreasing effect on intramuscular fat (IMF) content and carcass fat proportion was also described for the “double-muscled” phenotypes in cattle [10,11], which was later found to be caused by a myostatin mutation as well. However, different myostatin mutations can cause different degrees of hypermuscularity. The most extreme form of this phenotype in cattle is seen in the Belgian Blue, while different mutations with less extreme phenotypes were identified in other breeds [12].
Due to its large implication, not only on skeletal muscles, a lot of research has been undertaken on the effects of myostatin and its possible applications in health care and livestock production. The myostatin-gene, for instance, appears to be a promising candidate for the treatment of muscle dystrophy diseases [13,14], cardiac tissue regeneration or metabolic syndrome [15,16]. In livestock, efforts to manipulate the gene’s expression through immunization have been made for the purposes of higher lean mass production [17]. In accordance with this, modifiers of myostatin, such as follistatin (Fst), which inhibits myostatin, are currently under review as therapeutics [18,19].
In the present study, we generated G3-populations of reciprocal crosses between two hypermuscular Berlin Muscle Mouse Inbred (BMMI) lines to examine the genetic characteristics of myostatin and to find additional genes that could influence muscle growth and composition. The BMMI lines were hypermuscular as a result of long term selection that mirrors the selection process for high meat yield in livestock. The parental BMMI866 line originates from a population with the “compact” phenotype and therefore carries the known MstnCmpt-dl1Abc mutation [20,21]. Although the BMMI806 line originates from the same founder population, it does not carry this mutation; BMMI806 animals display very high intramuscular fat contents, fat mass and fat proportion, especially in males [22]. Previously, genetic modifier regions for the effect of the myostatin MstnCmpt-dl1Abc mutation on muscle mass have been identified on chromosomes 3, 5, 7, 11, 16, and X in a cross between Comp9 and CAST/Ei lines [23].
Given that the BMMI lines were originally developed as supporting genetic models for livestock research, we were particularly interested in myostatin effects on intramuscular fat content (IMF) and water holding capacity (WHC) on the genetic background of high muscularity. We also analysed the extent to which sex and the direction of the reciprocal cross impacted on the traits of interest. The latter could indicate parent-of-origin effects, where the impact on the phenotype can be different depending on the parent from which an allele was inherited. For example, the polar over-dominance caused by the ovine callipyge locus, where a hypermuscular phenotype only occurs if the mutated allele is inherited from the sire [24,25]. Parent-of-origin effects have also been described for body composition and fat-related traits in mice, pigs and cattle [26-29].
In addition to the relationship between muscle mass and meat quality traits, we were also interested in certain parameters of the muscle and whole body metabolism such as muscle glycogen and lactate contents, blood glucose levels, and the carcass pH-values. For this purpose, we present the correlations between these traits in the G3-population. The linkage study did not reveal genomic loci accounting for variation of those metabolic traits.
Results and discussion
Phenotypes
As shown in Table 1, significant differences were found between the two parental lines. Averaged over both sexes, the MstnCmpt-dl1Abc mutant BMMI866 animals showed 42%, 42%, 99% and 94% higher values for body weight, lean mass, M. longissimus and M. quadriceps masses than the BMMI806, respectively. BMMI866 mice had 30% lower total fat percentage than BMMI806. The IMF contents of the M. longissimus and the M. quadriceps were 52% and 40% lower, respectively, as compared to the BMMI806 line. These data confirm the hypertrophic effect of the MstnCmpt-dl1Abc mutation and its impact on fat accumulation [7]. Furthermore, fasting blood glucose levels of the BMMI866 line were 14% below the levels of BMMI806. The decreased glucose levels of BMMI866 mice support the model of a metabolic shift towards the utilization of glucose as energy fuel if myostatin is not fully functional, as shown by experiments in cell cultures [30]. Regarding differences between the sexes, male BMMI866 mice had lower carcass pH-values after 1 hour and female BMMI866 mice showed lower carcass pH-values after 24 hours post-mortem compared to BMMI806 (p < 0.05).
Table 1.
Body traits of parental, F1 and G3 animals (means and standard deviations)
| |
P |
F1 |
G3 |
|||||
|---|---|---|---|---|---|---|---|---|
| |
BMMI866 |
BMMI806 |
|
|
|
|||
| Trait | Males | Females | Males | Females | Males | Females | Males | Females |
| Body weight (g) |
49.31 (2.36)*** |
40.35 (2.47)*** |
36.43 (4.69) |
26.77 (2.16) |
42.17 (3.44)aaa,bb |
34.38 (2.12)aaa,bbb |
44.98 (4.21)aaa,bbb |
36.69 (4.63)aaa,bbb |
| Lean mass (g) |
41.89 (2.43)*** |
33.79 (2.12)*** |
29.31 (2.29) |
23.87 (1.99) |
32.30 (2.72)aaa,bbb |
29.04 (2.03)aaa,bbb |
36.12 (4.03)aaa,bbb |
30.61 (4.42)aaa,bbb |
| ML-Mass (g) |
1.33 (0.13)*** |
1.08 (0.1)*** |
0.67 (0.08) |
0.54 (0.05) |
0.89 (0.09)aaa,bbb |
0.75 (0.08)aaa,bbb |
0.95 (0.22)aaa,bbb |
0.85 (0.23)aaa,bbb |
| MQ-Mass (g) |
0.84 (0.05)*** |
0.67 (0.05)*** |
0.44 (0.04) |
0.34 (0.03) |
0.55 (0.05)aaa,bbb |
0.47 (0.03)aaa,bbb |
0.61 (0.15)aaa,bbb |
0.55 (0.15)aaa,bbb |
| Fat mass (g) |
3.53 (1.67) |
3.50 (1.35) |
4.41 (1.46) |
3.13 (1.19) |
4.41 (0.98)a |
3.76 (0.94) |
5.00 (1.96)aa |
2.88 (1.39)a |
| Fat mass (%) |
7.77 (3.65)*** |
9.36 (3.49) |
12.90 (3.52) |
11.41 (3.47) |
11.98 (2.35)aaa |
11.40 (2.31)a |
12.16 (4.50) aaa |
8.70 (4.31)b |
| IMF-ML (%) |
1.17 (0.23)*** |
1.28 (0.23)*** |
2.64 (0.34) |
2.45 (0.49) |
1.89 (0.32)aaa,bbb |
1.50 (0.2)a,bbb |
1.69 (0.39)aaa,bbb |
1.38 (0.33)bbb |
| IMF-MQ (%) |
1.07 (0.15)*** |
1.11 (0.12)*** |
1.81 (0.30) |
1.80 (0.29) |
1.46 (0.15)aaa,bbb |
1.38 (0.16)aaa,bbb |
1.34 (0.21)aaa,bbb |
1.22 (0.18)aa,bbb |
| WHC-ML (%) |
1.00 (0.58) |
1.03 (0.39) |
1.09 (0.37) |
1.01 (0.39) |
0.90 (0.25)b |
1.08 (0.35) |
1.24 (0.64)aa |
1.23 (0.41)a |
| WHC-MQ (%) |
0.75 (0.21) |
0.69 (0.17) |
0.85 (0.33) |
0.81 (0.26) |
0.86 (0.17)a |
0.95 (0.33)aa |
0.93 (0.29)aa |
0.93 (0.23)aaa,b |
| Glucose (mg/dL) |
105 (24)* |
88 (17)** |
120 (26) |
104 (17) |
108 (14) |
87 (12)bbb |
106 (13)bb |
90 (12)bbb |
| Glycogen (mg/g) |
1.94 (0.75) |
2.24 (1.51) |
2.47 (0.76) |
2.45 (1.38) |
1.52 (0.52)b |
1.73 (0.54) |
1.37 (0.90)bb |
1.18 (0.76)b |
| Lactate (mg/g) |
0.068 (0.008) |
0.091 (0.014) |
0.064 (0.003) |
0.070 (0.025) |
0.035 (0.010)aa,bb |
0.035 (0.012)a,b |
0.048 (0.017)aa,bb |
0.045 (0.013)aa,bb |
| pH 1 hour |
6.67 (0.11)* |
6.63 (0.08) |
6.74 (0.09) |
6.71 (0.12) |
- |
- |
6.77 (0.15)aaa |
6.73 (0.15)aa |
| pH 24 hours | 6.16 (0.10) | 6.15 (0.12)* | 6.12 (0.12) | 6.25 (0.12) | - | - | 6.12 (0.11) | 6.13 (0.09)bbb |
Trait abbreviations: IMF = intramuscular fat content, WHC = water holding capacity (measured as ‘drip loss’ in percent of muscle weight), ML = Musculus longissimus, MQ = Musculus quadriceps, Glucose = blood glucose level. Phenotypes of the parental lines are compared with each other and the F1 and G3, separated for sex. Asterisks indicate significant differences between the parental lines; letters indicate significant differences between the F1 or G3 and BMMI866 (a) or BMMI806 (b) lines. Number of asterisks and letters show levels of significance (***/aaa/bbbp < 0.001, **/aa/bbp < 0.01, */a/bp < 0.05).
Comparing the F1 and G3-population with the parental lines, we found intermediate values for body weight, lean mass and muscle masses. Dominance of the BMMI806 characteristics was indicated for total fat percentage in the F1 population. While the IMF values of both sexes of F1 animals and the males of the G3 population were intermediate, in female G3 animals, the measurements of total fat percentage and IMF of the M. longissimus were similar to the lower values of the BMMI866 line. Fasting blood glucose levels indicated a dominance of the BMMI866 line alleles, whereas the BMMI806 phenotype was found prevalent for the higher carcass pH-values for 1 hour post mortem.
Correlations
In the G3-population, high positive correlations in males and females were observed between muscle mass and total lean mass (r > 0.79) and negative correlations with body fat percentage (r < −0.46) and IMF (r < −0.53; Table 2). In females, muscle mass was also negatively correlated with WHC in M. quadriceps (r = −35), whilst in males, no correlation was found. Furthermore, in males, higher muscle mass was associated with lower pH-values 24 hours post mortem (r < −0.16). Nevertheless, in both sexes negative correlations were evident between muscle glycogen contents and the pH-values of 1 and 24 hours post mortem (r < −0.15), and between WHC and the pH-value after 24 hours (r < −0.14). The muscle glycogen content was associated with the body fat content (r < −0.17), but was not correlated with the blood glucose levels; higher muscle glycogen contents were found in leaner animals.
Table 2.
Spearman’s correlation coefficients between different traits in the G3 population
|
Males | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Body Weight (g) | Lean Mass (g) | ML-Mass (g) | MQ-Mass (g) | Fat Mass (g) | Fat Mass (%) | IMF-ML (%) | IMF-MQ (g) | Glucose (mg/dL) | WHC-ML (%) | WHC-MQ (%) | Glycogen (mg/g) | Lactate (mg/g) | pH, 1 hour | pH, 24 hours | |
| Body weight (g) |
|
0.88*** |
0.66*** |
0.66*** |
0.12 . |
−0.12 . |
−0.31*** |
−0.23** |
0.01 |
−0.01 |
−0.02 |
0.12 |
−0.02 |
−0.15* |
−0.19** |
| Lean mass (g) |
0.95*** |
|
0.80*** |
0.79*** |
−0.27*** |
−0.50*** |
−0.50*** |
−0.49*** |
−0.06 |
0.01 |
−0.04 |
0.15* |
−0.06 |
−0.07 |
−0.17* |
| ML-Mass (g) |
0.82*** |
0.87*** |
|
0.91*** |
−0.32*** |
−0.51*** |
−0.65*** |
−0.69*** |
−0.01 |
0.04 |
−0.04 |
0.20** |
−0.02 |
−0.12 |
−0.19** |
| MQ-Mass (g) |
0.82*** |
0.87*** |
0.96*** |
|
−0.28*** |
−0.46*** |
−0.66*** |
−0.64*** |
−0.03 |
0.05 |
−0.1 |
0.11 |
−0.04 |
−0.12. |
−0.16* |
| Fat mass (g) |
−0.10 |
−0.32*** |
−0.28** |
−0.29** |
|
0.96*** |
0.52*** |
0.62*** |
0.03 |
−0.01 |
−0.03 |
−0.13 . |
0.09 |
−0.14* |
−0.11 |
| Fat mass (%) |
−0.34*** |
−0.54*** |
−0.48*** |
−0.49*** |
0.96*** |
|
0.60*** |
0.70*** |
0.04 |
−0.02 |
−0.02 |
−0.17* |
0.08 |
−0.1 |
−0.05 |
| IMF-ML (%) |
−0.51*** |
−0.60*** |
−0.64*** |
−0.66*** |
0.45*** |
0.56*** |
|
0.67*** |
0.05 |
0.05 |
0.03 |
−0.15* |
−0.01 |
0.03 |
0.08 |
| IMF-MQ (g) |
−0.35*** |
−0.45*** |
−0.57*** |
−0.53*** |
0.37*** |
0.46*** |
0.52*** |
|
0.03 |
0.03 |
0.09 |
−0.12 |
−0.09 |
0 |
0.08 |
| Glucose (mg/dL) |
0.05 |
0.05 |
−0.08 |
−0.01 |
0.01 |
0.01 |
0.13 |
0.11 |
|
−0.01 |
0.01 |
0 |
−0.03 |
−0.05 |
−0.05 |
| WHC-ML (%) |
−0.16 . |
−0.13 |
−0.15 |
−0.09 |
−0.16 . |
−0.09 |
−0.05 |
0.03 |
0.15 |
|
0.35*** |
0.15* |
−0.02 |
0.08 |
−0.21** |
| WHC-MQ (%) |
−0.38*** |
−0.39*** |
−0.35*** |
−0.35*** |
0.04 |
0.13 |
0.09 |
0.1 |
0.04 |
0.42*** |
|
0.10 |
0.10 |
0.11 |
−0.14 . |
| Glycogen (mg/g) |
0.09 |
0.10 |
0.13 |
0.12 |
−0.24* |
−0.23* |
−0.04 |
−0.02 |
0.14 |
0.05 |
0.03 |
|
−0.05 |
−0.16* |
−0.27*** |
| Lactate (mg/g) |
0.04 |
0.06 |
0.10 |
0.08 |
−0.05 |
−0.07 |
−0.14 |
−0.20* |
0.06 |
0.14 |
0.15 |
0.07 |
|
−0.17* |
−0.15* |
| pH, 1 hours |
0.05 |
0.07 |
−0.11 |
−0.12 |
0.01 |
−0.01 |
0.12 |
0.08 |
0.01 |
−0.02 |
−0.06 |
−0.20* |
−0.17 . |
|
0.18** |
| pH, 24 hours |
0.03 |
0 |
−0.08 |
−0.07 |
−0.01 |
−0.01 |
0.04 |
0.01 |
0.01 |
−0.23* |
−0.16 . |
−0.31*** |
−0.23* |
0.23* |
|
| Females | |||||||||||||||
Trait abbreviations: IMF = intramuscular fat content, WHC = water holding capacity, measured as ‘drip loss’ in percent of muscle weight, ML = M. longissimus, MQ = M. quadriceps. Glucose = blood glucose level; Asterisks indicate the significance of correlations: *** p < 0.001, ** p < 0.01, * p < 0.05, .p < 0.10.
The results regarding the carcass pH-value and WHC confirm studies in pigs, where a low pH-value negatively affects WHC and leads to high drip loss. The low WHC is likely the result of a developing acidic environment that affects the proteolysis of cell scaffold proteins [31,32]. In turn, the acidic environment is caused by high intramuscular glycogen contents that promote the development of high lactate levels via glycolysis [31]. The negative correlation between muscle glycogen and body fat content as well as IMF of the M. longissimus in males supports the model of a metabolic shift towards glycolysis. The non-significant correlation with fasting blood glucose levels is likely due to the short fasting period of two hours before measurements leading to a high variability.
QTL analysis
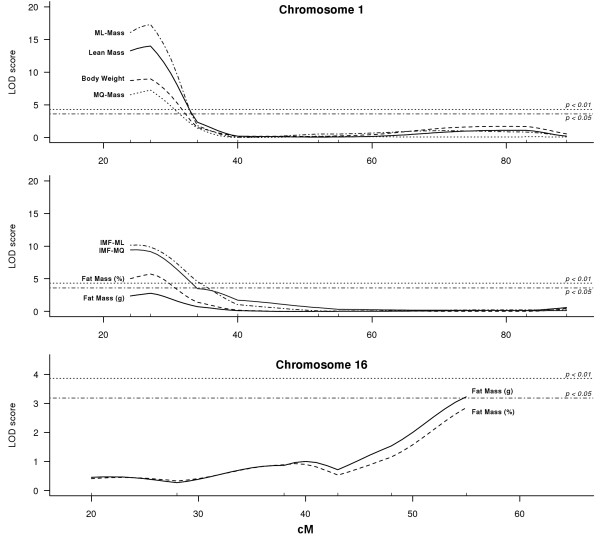
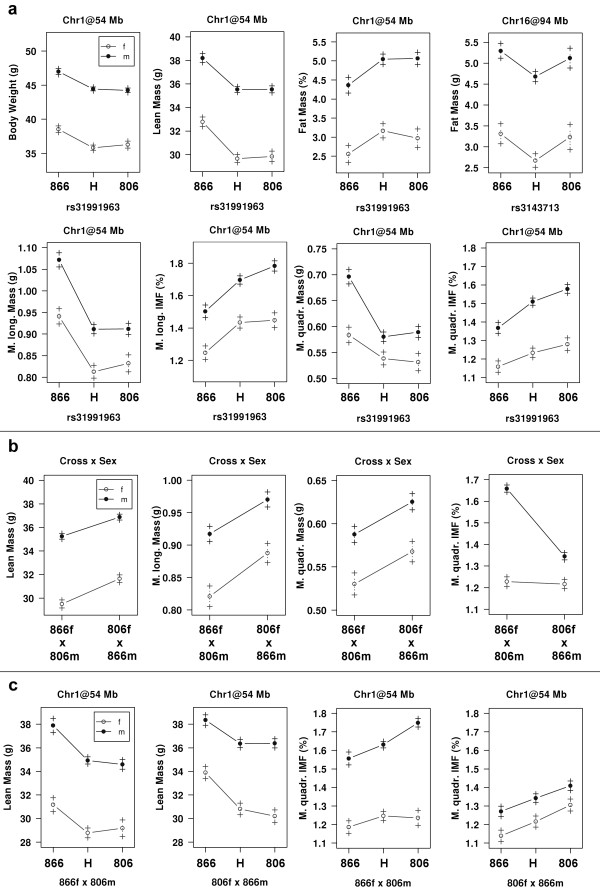
Due to the relative small number of animals in our G3-population, we could only detect regions with larger effects. Two genome-wide significant QTL were identified (Figure 1). The first one was located on chromosome (Chr) 1 at 54 Mb (47–59 Mb) in a region that contains the previously described myostatin MstnCmpt-dl1Abc mutation, located at 53 Mb (Figure 2). In the G3-population, the BMMI866 allele at this locus caused additive effects of 1.25 g for body weight and 1.35 g for lean mass. It also increased the masses of the M. longissimus and M. quadriceps muscles by 0.07 g and 0.04 g, respectively (Table 3). The body fat percentage was decreased by the BMMI866 allele by 1.15 points. A similar effect was observed for IMF where the BMMI866 allele had a decreasing effect of 0.13 and 0.07 percentage points for IMF values of the M. longissimus and M. quadriceps, respectively (Figure 3a, Table 3).
Figure 1.
MapChart plot of 138 reference single nucleotide polymorphisms used in this study. Positions are given in Mb (NCBI Build 37). Bars indicate LOD support intervals of identified significant QTL.
Figure 2.
QTL scans for different traits on chromosome 1 and 16. Sex and direction-of-cross were used as additive covariates and genome-wide significance thresholds of p < 0.05 and p < 0.01.
Table 3.
QTL, sex and direction-of-cross effects (p < 0.001) in the G3population at age of 10 weeks
| Trait | QTL/Effect1 | Chr | Mb2 | CI3 | Marker4 | LOD5 | a (s.e.)6 | d (s.e.)6 | % G3 var7 |
|---|---|---|---|---|---|---|---|---|---|
| Body weight (g) |
sex |
|
|
|
|
83.01 |
8.43 (0.31) |
|
68.35 |
| |
pgm |
|
|
|
|
6.44 |
1.67 (0.3) |
|
2.81 |
| |
1@27.0 |
1 |
54 |
47-59 |
rs31991963 |
8.93 |
−1.25 (0.21) |
−1.03 (0.31) |
3.97 |
| Lean mass (g) |
sex |
|
|
|
|
62.43 |
5.74 (0.27) |
|
55.22 |
| |
pgm |
|
|
|
|
7.78 |
1.59 (0.26) |
|
4.42 |
| |
1@27.0 |
1 |
54 |
47-55 |
rs31991963 |
13.91 |
−1.35 (0.18) |
−1.2 (0.26) |
8.27 |
| ML-Mass (g) |
sex |
|
|
|
|
14.30 |
0.1 (0.01) |
|
15.31 |
| |
pgm |
|
|
|
|
3.43 |
0.04 (0.01) |
|
3.38 |
| |
1@27.0 |
1 |
54 |
47-59 |
rs31991963 |
17.26 |
−0.07 (0.01) |
−0.07 (0.01) |
18.90 |
| MQ-Mass (g) |
sex |
|
|
|
|
8.68 |
0.07 (0.01) |
|
10.86 |
| |
pgm |
|
|
|
|
2.50 |
0.03 (0.01) |
|
2.98 |
| |
1@27.0 |
1 |
54 |
47-55 |
rs31991963 |
7.17 |
−0.04 (0.01) |
−0.04 (0.01) |
8.88 |
| Fat mass (g) |
sex |
|
|
|
|
35.31 |
1.98 (0.15) |
|
40.23 |
| |
16@55.0 |
16 |
94 |
86-94 |
rs3143713 |
3.57 |
−0.07 (0.12) |
−0.55 (0.15) |
3.16 |
| Fat mass (%) |
sex |
|
|
|
|
15.49 |
2.97 (0.36) |
|
18.85 |
| |
1@27.0 |
|
54 |
47-59 |
rs31991963 |
6.32 |
1.03 (0.24) |
1.15 (0.35) |
7.17 |
| IMF-ML (%) |
sex |
|
|
|
|
20.04 |
0.28 (0.03) |
|
22.55 |
| |
1@27.0 |
1 |
54 |
47-55 |
rs31991963 |
10.15 |
0.13 (0.02) |
0.06 (0.03) |
10.56 |
| IMF-MQ (%) |
sex |
|
|
|
|
50.46 |
0.41 (0.02) |
|
37.56 |
| |
pgm |
|
|
|
|
39.19 |
−0.01 (0.03) |
|
26.50 |
| |
1@26.0 |
1 |
51 |
47-59 |
rs31991963 |
10.05 |
0.07 (0.01) |
0.00 (0.02) |
5.36 |
| sex:pgm | 16.08 | −0.29 (0.03) | 9.00 |
1 QTL positions are given as chromosome number, @, and the genetic distance from the centromer in cM.
2 Most likely chromosomal location given as Mb-position.
3 1-LOD support interval in Mb.
4 Marker closest to the chromosomal position with the highest LOD score.
5 LOD scores from the full model estimations.
6 additive (a) and dominance (d) effect and their standard errors (s.e.) determined with the non-transformed raw trait values, therefore given in the respective unit; the direction of a and d, respectively given as the effect of the BMMI866-allele.
7 G3 phenotypic variance (%) explained by the QTL; QTL effect given as the reduction of the residual sum of squares fitting 1 vs. 0 QTL.
Figure 3.
Effect plots showing means and standard errors of the three genotype-classes at significant QTL positions. a) QTL significant under 95% and b) sex and direction-of-cross effects; c) directions-of-cross effects for the three genotypes for lean mass and IMF of the M. quadriceps; 866 = homozygous BMMI866-allele, 806 = homozygous BMMI806-allele, H = heterozygous animals; 806f x 866m = G3-animals originating from mating a female BMMI806 and a male BMMI866, 866f x 806m G3-animals originating from mating a female BMMI866 and a male BMMI806 in the parental generations.
An additional QTL was identified on Chr16 at 94 Mb (86–94 Mb; Figure 2) that decreased the total fat mass by 0.55 g in heterozygous animals (Figure 3a, Table 3). This is a negative heterosis effect where heterozygous animals have significantly lower t values than either homozygous class. This significant negative dominance for fat mass at the Chr. 16 QTL is interesting because it is in contrast to the phenotypic observation that male F1 and G3 individuals tended to have a higher fat mass than the mean of to the parental lines (Table 1). The general positive heterosis of the offspring lines points to other loci with effects on fat mass that could not be detected in this study. The QTL on chromosome 16 showed no linkage with IMF, which indicates a specific role in adipose tissue for this region in mice. An interaction between the Chr1 and the Chr16 locus was not observed, and thus our data provides evidence that the region on Chr16 is an independent regulator of total fat mass. Up to now only one marker in that region, the D16Mit51 at 93 Mb, was proposed provisionally by Srivastava et al. (2006) as causative for variation in total fat mass in mice.
The QTL for fat mass on Chr16 is located within a region from 75–98 Mb that is syntenic with a region in pigs on Sus scrofa chromosome (SSC) 13 (129–145 Mb). According to the animal genome database (animalgenome.org) this region contains QTL for marbling, back fat and for IMF [32-35]. Dominance effects for fat mass, as they were found in our study, were not significant for the syntenic region in pigs, however a positive dominance was found for ham weight [36-38]. Thus, this syntenic locus could possibly have a negative dominance effect on fat mass because muscle weight and fat weight are often negatively correlated. Furthermore, similar effects were observed for the syntenic region in cattle. In cattle, this region is divided into two parts which reside on chromosome 1 (BTA1) between 0–23 Mb and 141–155 Mb. Within those two regions on BTA1, fat related QTL were mapped for milk fat yield, marbling and fat thickness at the 12th rib [39-42]. A positive but non-significant dominance effect on adjusted subcutaneous fat thickness was reported in cattle [40]. Finally, the syntenic region in the human genome resides on chromosome 21 at 15–43 Mb. The Human Obesity Gene Map counts four obesity-associated loci in this syntenic region [43,44]. Assuming that the same gene is underlying the variation across species, further refinement of the murine Chr16 regions seems promising and our study adds to the confirmation of mice as a model animal to study meat quality traits in livestock or muscular disorders in humans.
As we did not find any other loci affecting muscle mass apart from the myostatin locus, we assume that several or many small effect alleles that contribute to these traits and that have been shown in other crosses [45] are hidden in our study behind the strong effect of myostatin, in particular in the hypermuscular line BMMI806. Another reason could be that the statistical power was not sufficient in the examined population.
Effects of sex and direction-of-cross
As shown in Table 3, sex affected all traits for all identified QTL. Males significantly increased body weight, lean mass, and the masses of the M. longissimus and M. quadriceps by 8.43 g, 5.74 g, 0.10 g and 0.07 g, respectively. In males, the total fat mass and the total fat percentage were also raised by 1.98 g and 2.97 points, respectively. Sex also affected IMF values of the two examined muscles, which were increased by 0.28 points and 0.41 points for the M. longissimus and the M. quadriceps in males, respectively. The increase in the lean mass related traits was expected as a sex difference; however, the increase in fat mass percentage in male mice was a new observation. An explanation could be that male BMMI806 mice display very high fat values (Table 1) and the high-fat phenotype in G3-males is thought to be inherited by the BMMI806 alleles of both QTLs.
Interestingly, for the myostatin locus, we also found significant effects of the direction-of-cross and an interaction between sex and the direction-of-cross on IMF of the M. quadriceps (LOD = 13.62). Animals derived from mating a female BMMI806 with a male BMMI866 showed higher body weight, lean mass and muscle mass at the Chr1 locus in comparison to animals of the reciprocal cross. While total fat mass, fat percentage and IMF of the M. longissimus were not significantly affected, we observed a large increase of IMF in the M. quadriceps by 0.29 points in male G3-animals that were derived from a mating between a female BMMI866 and a male BMMI806 (Figure 3b,c). This means that in male G3-animals descending from a BMMI806 grand-grandfather, myostatin expressed less of its IMF decreasing effect. Furthermore, the effects of myostatin were generally larger in G3-animals descending from a male BMMI866 parent of the initial cross.
As a likely result of the recent selection and inbreeding process, SNP data of the mitochondrial DNA and the Y-chromosome did not differ between the two parental lines. Therefore, we assumed that mitochondria and Y-chromosomes did not account for trait differences between reciprocal crosses. Nevertheless, we cannot completely exclude hidden mutations derived during the breeding history. Such hidden mutations do not play a role for mitochondria, but might affect Y-chromosomal effects. However, so far, no Y-chromosomal effects on traits that we analysed in this study have been identified. Therefore, the phenotypic differences between the reciprocal crosses are likely parent-of-origin effects.
The high IMF values for the QTL on Chr1, for G3-males of the cross between a BMMI866 female and a BMMI806 male, were very similar to males of the parental line BMMI806. This was not observed in the reciprocal cross and therefore, might result from parent-of-origin effects. Since evidence is growing that parent-of-origin effects could exist over several generations [46] our findings suggest epigenetic patterns, which alter the gene expression of the chromosomes according to the ancestor they were inherited from. Whether the myostatin locus alone underlies genetic imprinting, or additional loci that did not directly affect the analysed traits, still has to be tested in a genome-wide screen.
Recently, we have identified bidirectional parent-of-origin effects for muscle glycogen content and glycolytic potential as well as body weight in a population between two other hypermuscular mouse lines BFMI806 × BMMI816 [47]. The imprinted loci were not discovered by a genome-wide QTL search directly for additive and dominance effects.
In sheep with the callipyge mutation, the callipyge phenotype was not expressed in homozygous mutated animals [24,25]. It was suggested that the maternal inactivation of the locus also negatively affected the paternal expression, but a paternally inherited mutated locus escapes the inactivation in the presence of a maternally inherited wild-type allele and leads to the callipygous phenotype. Phenomenon like this have their molecular origin in a DNA methylation ‘reset’ that occurs during gametogenesis, the fusion of oocyte and spermatocyte in the zygote and the following early embryogenesis [48,49]. As seen in the callipyge sheep example, small genetic differences can affect the reprogramming processes and lead to different gene expression patterns and phenotypes.
Conclusions
The strong effects of the myostatin mutation MstnCmpt-dl1Abc on high muscle and low fat mass were confirmed, even in a background of high muscularity in a cross between two hypermuscular mouse lines. The data provides evidence for sex and very likely parent-of-origin effects modifying the direct effects of the myostatin locus on muscle mass and IMF in M. quadriceps. However, further tests are necessary to confirm imprinting effects. In addition to the myostatin locus, we have shown that a region on Chr 16 affecting fat mass is conserved between mice, pig and cattle showing effects on body fat, marbling and milk fat. Furthermore, many reported observations and correlations of the post mortem physiology of livestock were reproduced in our mice.
The results obtained in this study help to justify the mouse model to study economically relevant livestock traits. New information from the mouse model helps to describe variation in meat quantity and quality and thus supports research in livestock.
Methods
Animals
The Berlin Muscle Mouse population had been long term selected for high body weight and high muscle mass to reflect the selective mechanisms in livestock breeding. Founder animals of the Berlin Muscle Mouse (BMM) population were originally purchased in several pet shops in Berlin, Germany. The selection process comprised several distinct phases. The beginning of the selection process constituted a phase of 23 generations of selection for high protein content of the carcass at the age of 60 days. Protein content was determined by chemical analyses. In a second phase, mice were selected for high body weight and low fat content at 42 days for 10 generations. Afterwards, mice were monitored for high muscularity by palpating and mice with highest muscularity on a scale between 1 and 5 were selected for the next generation. As a result of selection and likely the occurrence of natural mutation(s) over 25 generations, a mouse population with a high muscular phenotype had been generated. A high compact sub-line was perpetuated through random mating of selected animals [21]. Sequencing of the myostatin gene in this line revealed a 12 bp deletion [20] leading to a loss of function. Scale and weight based selection continued for another 28 generations. After 86 generations of selection, full-sibs with distinct phenotypes were mated. These founder animals became the basis of seven Berlin Muscle Mouse inbred lines (BMMI). Four lines display the MstnCmpt-dl1Abc mutation and three are wild type. In this study we used the lines BMMI866, which carries the described MstnCmpt-dl1Abc mutation, and BMMI806, which is hypermuscular but wild type for myostatin and shows high IMF. At the time of setting up the crossbred experiment, the lines were in generation 20 (BMMI866) and 21 (BMMI806) of inbreeding.
Pedigree structure
Two pairs of full sibs of the Berlin Muscle Mouse inbred lines BMMI866 and BMMI806 were crossed reciprocally to generate F1, F2 and G3-intercross populations. F2-animals within every direction-of-cross were randomly mated under avoidance of sibling-mating [50], to generate 308 G3-animals. Altogether, there were 195 males and 113 females. From these 308 animals, 97 males and 53 females came from the cross between a female BMMI866 and a male BMMI806 mouse, while the reciprocal cross between a male BMMI866 and a female BMMI806 mouse consisted of 98 males and 60 females. Using this design, we would expect different mitochondria and Y-chromosomes segregation in the reciprocal crosses. Since high-density SNP data of the inbred strains BMMI866 and BMMI806 provided evidence for no variation in both mitochondria and Y-chromosomes, we assume that the two parental mouse strains have the same mitochondria and Y-chromosomes as a result of their breeding history (See also under Genotyping).
Husbandry and feeding conditions
The mice were treated in accordance to, and all experimental protocols were approved by, the German Animal Welfare Authorities (approval no. G0405/08). The animals were maintained under conventional conditions at 22 ± 2°C and controlled lightning with a 12:12 hour light:dark cycle. They were kept in groups of two to four animals of the same sex per Macrolon cage and given ad libitum access to food and water. Until the age of 70 days, the animals were fed a standard breeding diet (‘Altromin standard breeding diet no. 1314 TPF’, Lage, Germany). This diet contained 27.0% crude protein, 5.0% crude fat, 4.5% crude fibre, 6.5% crude ash, 50.5% nitrogen free extract (starch and sugar), vitamins, trace elements and minerals (2988 kcal/kg metabolizable energy; thereof 27.0% energy from proteins, 13.0% from fat and 60.0% from carbohydrates).
Phenotypic measures
After a fasting period of two hours, 71 day-old mice were anaesthetised under isoflurane and decapitated. The Musculus longissimus (ML) and Musculus quadriceps (MQ) were dissected and weighed. The summed muscle weights of the left and right M. longissimus and the left and right M. quadriceps were recorded as muscle mass (MM). The right muscles were immediately frozen in liquid nitrogen and subsequently stored at −80°C. The left muscles were cooled down to 6°C for one hour, subsequently frozen at −20°C and stored at the same temperature until WHC and IMF were measured. Carcasses were stored at 6°C and pH-values were taken within the M. biceps femoris at one and 24 hours post mortem (ebro PHT 810, Ingolstadt, Germany). For the determination of WHC, frozen muscles were thawed and stored at 6°C for 24 hours. Muscles were then centrifuged for 60 sec at 604 × g in invitek® 1.5 ml receiver tubes with filter inlays to collect the tissue fluid that was not held from the muscle (Eppendorf Minispin, Hamburg, Germany; Invitek, Berlin, Germany). The ratio of lost tissue fluid to tissue mass was designated as ‘drip loss’. IMF was measured as percentage of muscle weight using nuclear magnetic resonance technology (SMART Trac System, CEM, Kamp-Lintfort, Germany) [51]. Body weights were recorded weekly. At ten weeks, total fat and lean masses were measured by quantitative magnetic resonance (QMR) analysis, using the EchoMRI whole body composition analyser (Echo Medical Systems, Houston, Texas, USA) [22,52]. Body fat percentage is the percentage of total fat mass calculated from the sum of total fat and lean mass. Blood glucose levels were measured after two hours of fasting, prior to dissection at ten weeks (Bayer ‘Contour’, Leverkusen, Germany). Muscle glycogen content was determined colorimetrically in the right M. longissimus (GOD/PAP method ‘Glucose liquicolor’ by Human, Wiesbaden, Germany) as suggested by Barham and Trinder (1972). Lactate contents were determined colorimetrically in the right M. longissimus using the Lactate Assay Kit by Techung Lee [53].
Genotyping
Parental BMMI lines were genotyped with the Mouse-Diversity-Array [54] comprising 623,124 single-nucleotide polymorphisms (SNPs). The SNP information provided evidence for allele fixation of 98.1% and 98.3% in lines BMMI866 and BMMI806 after 20 and 21 generations of inbreeding, respectively. Both lines differed from each other by 4.8% at the SNP level. SNPs of the mitochondria and the Y-chromosome were not different between the lines. Using the information on diverse genomic regions between BMMI806 and BMMI866, 138 informative SNP markers covering all chromosomes (except Y) in an average distance of 24.8 Mb were selected for genotyping the parents, F2 and the G3-animals (Figure 1). Regions larger than 10 Mb that did not differ in SNP-alleles between parental lines could not be included into the linkage analysis. We assume that these regions have the same ancestral origin as a result of the breeding history. Genotyping was done at KBiosciences (Hoddesdon, U.K.). The physical map was converted into the genetic map using the ‘Mouse Map Converter’ software from The Jackson Laboratory [55].
QTL and statistical analyses
For the QTL analysis, 308 G3-animals were used. Analyses for single QTL detection and detection of interacting QTL were performed using R/qtl [56]. Unlike F2, the G3-animals may be unequally related to each other. Ignoring the unequal relatedness may result in a serious inflation of false positive rates. Therefore, we calculated environmental residuals for both sexes and each of the two crosses and corrected for litter size and genomic kinship. This part of the analysis was carried out with a Genome-wide Rapid Analysis using Mixed Models And Mixed Models and Regression (GRAMMAR) as implemented in GenABLE [57-60]. The environmental residuals were used to generate corrected phenotypes, which were applied in the actual QTL analysis. Direction-of-cross and sex were included as additive covariates in the model and used as interactive covariates to test their effects on QTL. All phenotypes were log-transformed to obtain normal distribution. Trait-specific significance thresholds were estimated with 1000 permutations [61]. On average, the thresholds for genome-wide significance and suggestive linkage results were LOD = 3.59 (p < 0.05) and LOD = 3.19 (p < 0.10), respectively. Genome-wide significant QTL (p < 0.05) were included into a mixed model for each trait to calculate the respective trait variance in the G3-population. Factors being significant under p < 0.001 were kept in the model. The genotype of the myostatin locus was accounted for by including it as a covariate in scans for additional QTL. Basic statistics were performed using the SAS software package (SAS System for Windows, Release 9.2). QTL support intervals were determined by bootstrapping as implemented in R/qtl [60].
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SK conceived of the study, carried out the data collection, molecular genetic studies, statistical analysis of the data and drafted the manuscript. EMS and AOS participated in the statistical analysis, particularly in correcting the data for population stratification and estimating heterozygosity, respectively. AW carried out lactate and glucose measurements, FPMV and HY provided the data of the mouse diversity array (MDA). GAB participated in conceiving of the study, its design and coordination and helped drafting the manuscript. All authors read and approved the final manuscript.
Contributor Information
Stefan Kärst, Email: stefan.kaerst@agrar.hu-berlin.de.
Eva M Strucken, Email: eva.mraria.strucken@agrar.hu-berlin.de.
Armin O Schmitt, Email: armin.schmitt@unibz.it.
Alexandra Weyrich, Email: weyrich@izw-berlin.de.
Fernando PM de Villena, Email: fernando@med.unc.edu.
Hyuna Yang, Email: Hyuna.Yang@jax.org.
Gudrun A Brockmann, Email: gudrun.brockmann@agrar.hu-berlin.de.
Acknowledgement
The project was supported by the Deutsche Forschungsgemeinschaft (Project BR1285/8-1). We thank Gary Churchill for the support of the Center for Genome Dynamics for high density SNP genotyping at The Jackson Laboratory. Furthermore, we thank Carsten Berndt, Melanie Riedel and Annett Kannegiesser for technical assistance and Yan C.S.M. Laurenson for orthographic review.
References
- McPherron AC, Lawler AM, Lee S-J. Regulation of skeletal muscle mass in mice by a new TGF-p superfamily member. Nature. 1997;387(6628):83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- Bellinge RHS, Liberles DA, Iaschi SPA, O’Brien PA, Tay GK. Myostatin and its implications on animal breeding: a review. Anim Genet. 2005;36(1):1–6. doi: 10.1111/j.1365-2052.2004.01229.x. [DOI] [PubMed] [Google Scholar]
- Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibe B, Bouix J, Caiment F, Elsen J-M, Eychenne F. et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet. 2006;38(7):813–818. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- Kambadur R, Sharma M, Smith TP, Bass JJ. Mutations in myostatin (GDF8) in double-muscled Belgian Blue and Piedmontese cattle. Genome Res. 1997;7(9):910–916. doi: 10.1101/gr.7.9.910. [DOI] [PubMed] [Google Scholar]
- Guimaraes SEF, Stahl CH, Lonergan SM, Geiger B, Rothschild MF. Myostatin promoter analysis and expression pattern in pigs. Livest Sci. 2007;112(1–2):143–150. [Google Scholar]
- Hadjipavlou G, Matika O, Clop A, Bishop SC. Two single nucleotide polymorphisms in the myostatin (GDF8) gene have significant association with muscle depth of commercial Charollais sheep. Anim Genet. 2008;39(4):346–353. doi: 10.1111/j.1365-2052.2008.01734.x. [DOI] [PubMed] [Google Scholar]
- McPherron AC, Lee SJ. Suppression of body fat accumulation in myostatin-deficient mice. J Clin Invest. 2002;109(5):595–601. doi: 10.1172/JCI13562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wall RJ, Yang J. Transgenic expression of myostatin propeptide prevents diet-induced obesity and insulin resistance. Biochem Biophys Res Commun. 2005;337(1):248–255. doi: 10.1016/j.bbrc.2005.09.044. [DOI] [PubMed] [Google Scholar]
- Dilger AC, Spurlock ME, Grant AL, Gerrard DE. Myostatin null mice respond differently to dietary-induced and genetic obesity. Anim Sci J. 2010;81(5):586–593. doi: 10.1111/j.1740-0929.2010.00776.x. [DOI] [PubMed] [Google Scholar]
- Hocquette J-F, Bas P, Bauchart D, Vermorel M, Geay Y. Fat partitioning and biochemical characteristics of fatty tissues in relation to plasma metabolites and hormones in normal and double-muscled young growing bulls. Comp Biochem Physiol A Mol Integr Physiol. 1999;122(1):127–138. doi: 10.1016/S1095-6433(98)10172-1. [DOI] [PubMed] [Google Scholar]
- Shahin KA, Berg RT. Growth-Patterns of Muscle, Fat and Bone, and Carcass Composition of Double Muscled and Normal Cattle. Can J Anim Sci. 1985;65(2):279–294. doi: 10.4141/cjas85-035. [DOI] [Google Scholar]
- Smith JA, Lewis AM, Wiener P, Williams JL. Genetic variation in the bovine myostatin gene in UK beef cattle: allele frequencies and haplotype analysis in the South Devon. Anim Genet. 2000;31(5):306–309. doi: 10.1046/j.1365-2052.2000.00521.x. [DOI] [PubMed] [Google Scholar]
- Haidet AM, Rizo L, Handy C, Umapathi P, Eagle A, Shilling C, Boue D, Martin PT, Sahenk Z, Mendell JR. et al. Long-term enhancement of skeletal muscle mass and strength by single gene administration of myostatin inhibitors. Proc Natl Acad Sci. 2008;105(11):4318–4322. doi: 10.1073/pnas.0709144105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida K. Targeting myostatin for therapies against muscle-wasting disorders. Curr Opin Drug Discov Devel. 2008;11(4):487–494. [PubMed] [Google Scholar]
- Guo T, Jou W, Chanturiya T, Portas J, Gavrilova O, McPherron AC. Myostatin inhibition in muscle, but not adipose tissue, decreases fat mass and improves insulin sensitivity. PLoS One. 2009;4(3):e4937. doi: 10.1371/journal.pone.0004937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhfakh R, Michaud A, Tremblay JP. Blocking the Myostatin Signal With a Dominant Negative Receptor Improves the Success of Human Myoblast Transplantation in Dystrophic Mice. Mol Ther. 2011;19(1):204–210. doi: 10.1038/mt.2010.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D-b L, K-y Z, D-w C, X-m D, Yu B. Effects of active immunization against myostatin on carcass quality and expression of the myostatin gene in pigs. Anim Sci J. 2009;80(5):585–590. doi: 10.1111/j.1740-0929.2009.00666.x. [DOI] [PubMed] [Google Scholar]
- Benabdallah BF, Bouchentouf M, Rousseau J, Bigey P, Michaud A, Chapdelaine P, Scherman D, Tremblay JP. Inhibiting myostatin with follistatin improves the success of myoblast transplantation in dystrophic mice. Cell Transplant. 2008;17(3):337–350. doi: 10.3727/096368908784153913. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Lee YS, Zimmers TA, Soleimani A, Matzuk MM, Tsuchida K, Cohn RD, Barton ER. Regulation of muscle mass by follistatin and activins. Mol Endocrinol. 2010;24(10):1998–2008. doi: 10.1210/me.2010-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo G, Dallmann G, Muller G, Patthy L, Soller M, Varga L. A deletion in the myostatin gene causes the compact (Cmpt) hypermuscular mutation in mice. Mamm Genome. 1998;9(8):671–672. doi: 10.1007/s003359900843. [DOI] [PubMed] [Google Scholar]
- Varga L, Szabo G, Darvasi A, Muller G, Sass M, Soller M. Inheritance and mapping of Compact (Cmpt), a new mutation causing hypermuscularity in mice. Genetics. 1997;147(2):755–764. doi: 10.1093/genetics/147.2.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuschl C, Hantschel C, Wagener A, Schmitt AO, Illig T, Brockmann GA. A unique genetic defect on chromosome 3 is responsible for juvenile obesity in the Berlin Fat Mouse. Int J Obes. 2010;34:1706–1714. doi: 10.1038/ijo.2010.97. [DOI] [PubMed] [Google Scholar]
- Varga L, Müller G, Szabó G, Pinke O, Korom E, Kovács B, Patthy L, Soller M. Mapping Modifiers Affecting Muscularity of the Myostatin Mutant (MstnCmpt-dl1Abc) Compact Mouse. Genetics. 2003;165(1):257–267. doi: 10.1093/genetics/165.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockett NE, Jackson SP, Shay TL, Farnir F, Berghmans S, Snowder GD, Nielsen DM, Georges M. Polar overdominance at the ovine callipyge locus. Science. 1996;273(5272):236–238. doi: 10.1126/science.273.5272.236. [DOI] [PubMed] [Google Scholar]
- Freking BA, Murphy SK, Wylie AA, Rhodes SJ, Keele JW, Leymaster KA, Jirtle RL, Smith TP. Identification of the single base change causing the callipyge muscle hypertrophy phenotype, the only known example of polar overdominance in mammals. Genome Res. 2002;12(10):1496–1506. doi: 10.1101/gr.571002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud JM, Hager R, Roseman C, Fawcett G, Wang B, Wolf JB. Genomic imprinting effects on adult body composition in mice. Proc Natl Acad Sci U S A. 2008;105(11):4253–4258. doi: 10.1073/pnas.0706562105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Jennen DGJ, Tholen E, Juengst H, Kleinwächter T, Hölker M, Tesfaye D, Ün G, Schreinemachers HJ, Murani E. et al. A genome scan reveals QTL for growth, fatness, leanness and meat quality in a Duroc-Pietrain resource population. Anim Genet. 2007;38(3):241–252. doi: 10.1111/j.1365-2052.2007.01592.x. [DOI] [PubMed] [Google Scholar]
- Goodall JJ, Schmutz SM. IGF2 gene characterization and association with rib eye area in beef cattle. Anim Genet. 2007;38(2):154–161. doi: 10.1111/j.1365-2052.2007.01576.x. [DOI] [PubMed] [Google Scholar]
- Mantey C, Brockmann GA, Kalm E, Reinsch N. Mapping and Exclusion Mapping of Genomic Imprinting Effects in Mouse F2 Families. J Hered. 2005;96(4):329–338. doi: 10.1093/jhered/esi044. [DOI] [PubMed] [Google Scholar]
- Antony N, Bass JJ, McMahon CD, Mitchell MD. Myostatin regulates glucose uptake in BeWo cells. Am J Physiol-Endoc M. 2007;293(5):E1296–E1302. doi: 10.1152/ajpendo.00331.2007. [DOI] [PubMed] [Google Scholar]
- Choe JH, Choi YM, Lee SH, Shin HG, Ryu YC, Hong KC, Kim BC. The relation between glycogen, lactate content and muscle fiber type composition, and their influence on postmortem glycolytic rate and pork quality. Meat Sci. 2008;80(2):355–362. doi: 10.1016/j.meatsci.2007.12.019. [DOI] [PubMed] [Google Scholar]
- Bee G, Anderson AL, Lonergan SM, Huff-Lonergan E. Rate and extent of pH decline affect proteolysis of cytoskeletal proteins and water-holding capacity in pork. Meat Sci. 2007;76(2):359–365. doi: 10.1016/j.meatsci.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Stearns TM, Beever JE, Southey BR, Ellis M, McKeith FK, Rodriguez-Zas SL. Evaluation of approaches to detect quantitative trait loci for growth, carcass, and meat quality on swine chromosomes 2, 6, 13, and 18. II. Multivariate and principal component analyses. J Anim Sci. 2005;83(11):2471–2481. doi: 10.2527/2005.83112471x. [DOI] [PubMed] [Google Scholar]
- Ma J, Ren J, Guo Y, Duan Y, Ding N, Zhou L, Li L, Yan X, Yang K, Huang L. et al. Genome-wide identification of quantitative trait loci for carcass composition and meat quality in a large-scale White Duroc x Chinese Erhualian resource population. Anim Genet. 2009;40(5):637–647. doi: 10.1111/j.1365-2052.2009.01892.x. [DOI] [PubMed] [Google Scholar]
- de Koning DJ, Pong-Wong R, Varona L, Evans GJ, Giuffra E, Sanchez A, Plastow G, Noguera JL, Andersson L, Haley CS. Full pedigree quantitative trait locus analysis in commercial pigs using variance components. J Anim Sci. 2003;81(9):2155–2163. doi: 10.2527/2003.8192155x. [DOI] [PubMed] [Google Scholar]
- Duthie C, Simm G, Doeschl-Wilson A, Kalm E, Knap PW, Roehe R. Epistatic analysis of carcass characteristics in pigs reveals genomic interactions between quantitative trait loci attributable to additive and dominance genetic effects. J Anim Sci. 2010;88(7):2219–2234. doi: 10.2527/jas.2009-2266. [DOI] [PubMed] [Google Scholar]
- Nezer C, Moreau L, Wagenaar D, Georges M. Results of a whole genome scan targeting QTL for growth and carcass traits in a Pietrain x Large White intercross. Genet Sel Evol. 2002;34(3):371–387. doi: 10.1186/1297-9686-34-3-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varona L, Ovilo C, Clop A, Noguera JL, Perez-Enciso M, Coll A, Folch JM, Barragan C, Toro MA, Babot D. et al. QTL mapping for growth and carcass traits in an Iberian by Landrace pig intercross: additive, dominant and epistatic effects. Genet Res. 2002;80(2):145–154. doi: 10.1017/s0016672302005803. [DOI] [PubMed] [Google Scholar]
- Höglund JK, Buitenhuis AJ, Guldbrandtsen B, Su G, Thomsen B, Lund MS. Overlapping chromosomal regions for fertility traits and production traits in the Danish Holstein population. J Dairy Sci. 2009;92(11):5712–5719. doi: 10.3168/jds.2008-1964. [DOI] [PubMed] [Google Scholar]
- Kim J-J, Farnir F, Savell J, Taylor JF. Detection of quantitative trait loci for growth and beef carcass fatness traits in a cross between Bos taurus (Angus) and Bos indicus (Brahman) cattle. J Anim Sci. 2003;81(8):1933–1942. doi: 10.2527/2003.8181933x. [DOI] [PubMed] [Google Scholar]
- McClure MC, Morsci NS, Schnabel RD, Kim JW, Yao P, Rolf MM, McKay SD, Gregg SJ, Chapple RH, Northcutt SL. et al. A genome scan for quantitative trait loci influencing carcass, post-natal growth and reproductive traits in commercial Angus cattle. Anim Genet. 2010;41(6):597–607. doi: 10.1111/j.1365-2052.2010.02063.x. [DOI] [PubMed] [Google Scholar]
- Nadesalingam J, Plante Y, Gibson JP. Detection of QTL for milk production on Chromosomes 1 and 6 of Holstein cattle. Mamm Genome. 2001;12(1):27–31. doi: 10.1007/s003350010232. [DOI] [PubMed] [Google Scholar]
- Li W-D, Dong C, Li D, Zhao H, Price RA. An Obesity-Related Locus in Chromosome Region 12q23-24. Diabetes. 2004;53(3):812–820. doi: 10.2337/diabetes.53.3.812. [DOI] [PubMed] [Google Scholar]
- Rankinen T, Zuberi A, Chagnon YC, Weisnagel SJ, Argyropoulos G, Walts B, Perusse L, Bouchard C. The Human Obesity Gene Map: The 2005 Update. Obesity. 2006;14(4):529–644. doi: 10.1038/oby.2006.71. [DOI] [PubMed] [Google Scholar]
- Carroll AM, Palmer AA, Lionikas A. QTL Analysis of Type I and Type IIA Fibers in Soleus Muscle in a Cross between LG/J and SM/J Mouse Strains. Frontiers Genetics. 2011;2:99. doi: 10.3389/fgene.2011.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud JM, Hager R, Roseman C, Fawcett G, Wang B, Wolf JB. Genomic imprinting effects on adult body composition in mice. Proc Natl Acad Sci. 2008;105(11):4253–4258. doi: 10.1073/pnas.0706562105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaerst S, Vahdati AR, Brockmann GA, Hager R. Genomic imprinting and genetic effects on muscle traits in mice. BMC Genomics. 2012;13(1):408. doi: 10.1186/1471-2164-13-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaf T. Methylation dynamics in the early mammalian embryo: implications of genome reprogramming defects for development. Curr Top Microbiol Immunol. 2006;310:13–22. doi: 10.1007/3-540-31181-5_2. [DOI] [PubMed] [Google Scholar]
- May A, Reifenberg K, Zechner U, Haaf T. Asynchronous replication dynamics of imprinted and non-imprinted chromosome regions in early mouse embryos. Exp Cell Res. 2008;314(15):2788–2795. doi: 10.1016/j.yexcr.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Schmitt A, Bortfeldt R, Neuschl C, Brockmann G. RandoMate: a program for the generation of random mating schemes for small laboratory animals. Mamm Genome. 2009;20(5):321–325. doi: 10.1007/s00335-009-9185-6. [DOI] [PubMed] [Google Scholar]
- Kaerst S, Schmitt A, Brockmann G. A novel method for measuring of fat content in low-weight tissue: A NMR study. WebmedCentral OBESITY 2010. 2010;1(12):WMC001368. [Google Scholar]
- Tinsley FC, Taicher GZ, Heiman ML. Evaluation of a quantitative magnetic resonance method for mouse whole body composition analysis. Obesity. 2004;12(1):150–160. doi: 10.1038/oby.2004.20. [DOI] [PubMed] [Google Scholar]
- Gordon GRJ, Choi HB, Rungta RL, Ellis-Davies GCR, MacVicar BA. Brain metabolism dictates the polarity of astrocyte control over arterioles. Nature. 2008;456(7223):745–749. doi: 10.1038/nature07525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Ding Y, Hutchins LN, Szatkiewicz J, Bell TA, Paigen BJ, Graber JH, de Villena FP, Churchill GA. A customized and versatile high-density genotyping array for the mouse. Nat Methods. 2009;6(9):663–666. doi: 10.1038/nmeth.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox A, Ackert-Bicknell CL, Dumont BL, Ding Y, Bell JT, Brockmann GA, Wergedal JE, Bult C, Paigen B, Flint J. et al. A new standard genetic map for the laboratory mouse. Genetics. 2009;182(4):1335–1344. doi: 10.1534/genetics.109.105486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19(7):889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Aulchenko YS, de Koning DJ, Haley C. Genomewide rapid association using mixed model and regression: a fast and simple method for genomewide pedigree-based quantitative trait loci association analysis. Genetics. 2007;177(1):577–585. doi: 10.1534/genetics.107.075614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin N, van Duijn CM, Aulchenko YS. A Genomic Background Based Method for Association Analysis in Related Individuals. PLoS One. 2007;2(12):e1274. doi: 10.1371/journal.pone.0001274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23(10):1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- Thompson EA, Shaw RG. Pedigree analysis for quantitative traits: variance components without matrix inversion. Biometrics. 1990;46(2):399–413. doi: 10.2307/2531445. [DOI] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138(3):963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]