Abstract
Background
Cryptorchidism is the most frequent congenital disorder in male children; however the genetic causes of cryptorchidism remain poorly investigated. Comparative integratomics combined with systems biology approach was employed to elucidate genetic factors and molecular pathways underlying testis descent.
Methods
Literature mining was performed to collect genomic loci associated with cryptorchidism in seven mammalian species. Information regarding the collected candidate genes was stored in MySQL relational database. Genomic view of the loci was presented using Flash GViewer web tool (http://gmod.org/wiki/Flashgviewer/). DAVID Bioinformatics Resources 6.7 was used for pathway enrichment analysis. Cytoscape plug-in PiNGO 1.11 was employed for protein-network-based prediction of novel candidate genes. Relevant protein-protein interactions were confirmed and visualized using the STRING database (version 9.0).
Results
The developed cryptorchidism gene atlas includes 217 candidate loci (genes, regions involved in chromosomal mutations, and copy number variations) identified at the genomic, transcriptomic, and proteomic level. Human orthologs of the collected candidate loci were presented using a genomic map viewer. The cryptorchidism gene atlas is freely available online: http://www.integratomics-time.com/cryptorchidism/. Pathway analysis suggested the presence of twelve enriched pathways associated with the list of 179 literature-derived candidate genes. Additionally, a list of 43 network-predicted novel candidate genes was significantly associated with four enriched pathways. Joint pathway analysis of the collected and predicted candidate genes revealed the pivotal importance of the muscle-contraction pathway in cryptorchidism and evidence for genomic associations with cardiomyopathy pathways in RASopathies.
Conclusions
The developed gene atlas represents an important resource for the scientific community researching genetics of cryptorchidism. The collected data will further facilitate development of novel genetic markers and could be of interest for functional studies in animals and human. The proposed network-based systems biology approach elucidates molecular mechanisms underlying co-presence of cryptorchidism and cardiomyopathy in RASopathies. Such approach could also aid in molecular explanation of co-presence of diverse and apparently unrelated clinical manifestations in other syndromes.
Keywords: Cryptorchidism, Muscle-contraction pathway, Cardiomyopathy, Comparative integratomics, Protein-protein interactions, Systems biology, Undescended testes, RASopathy
Background
Cryptorchidism (CO) is the most frequent congenital disorder in male children (2-4% of full-term male births) and is defined as incomplete descent of one (unilateral) or both (bilateral) testes and associated structures. Cryptorchidism has a potential effect on health; defects in testes descent usually cause impaired spermatogenesis, resulting in reduced fertility and increased rates of testicular neoplasia, and testicular torsion (reviewed in [1]). Cryptorchidism is common in human, pigs, and companion animals (2–12%) but relatively rare in cattle, and sheep (≤ 1%) [2].
Testicular descent is a complex series of events which requires concerted action of hormones, constitutive mechanisms, and the nervous system. In most species, including human, the complete descent of testes usually occurs prenatally, while in some (e.g. dogs), postnatally. Beside environmental factors like endocrine disruptors, CO is at least in part determined by genetic causes (chromosome or gene mutations), and is often a common feature of different syndromes. For example, Klinefelter syndrome and mutations in INSL3 gene have already been recognized as a cause of CO in some cases [3].
The comparative knowledge attained through study of animal models has been of great importance in understanding complex disease etiology, suggesting several candidate genes involved also in the pathogenesis of human diseases [4]. Therefore, the use of comparative genomics approach, integrating and cross-filtering the available knowledge from different species seems highly justified. Different animal models for CO exist; for example natural mutants or transgenic mice, rat, rabbit, dog, pig and rhesus monkeys are used to elucidate the role of different factors involved in CO [5]. Based on mouse knock-out models from Mouse Genome Informatics (MGI) database, several genes appear as possible candidates (AR, HOX genes, INSL3, RXFP2, and WT1). Additionally, the technological progress in the last years enabled the use of high-throughput omics-information, at coding (DNA), expression (RNA), and proteomic level. This technological revolution creates a vast amount of data, which increases the need for application of bioinformatics tools that are able to connect omics data with phenotype and enable search for overlapping pathogenetic mechanisms in different genetic diseases [6]. However, this existing technology hasn’t been significantly employed in human CO research on a genome and transcriptome-wide scale; to date only one genome-wide expression study has been performed in rat [7].
Integratomics represents a novel trend in the omics-research and is based on the integration of diverse omics-data (genomic, transcriptomic, proteomic, etc.), regardless of the study approach or species [8-10]. High genetic homology between mammals and the availability of well annotated genomes from different species allows the assembled data to be presented in a form of a comparative genomic view, displaying candidate genes as a single species orthologs.
Information extracted from diverse and methodologically focused studies are often fragmented and controversial. To overcome this problem we integrated the collected data, using a holistic (map-driven) approach, and developed freely available interactive genomic visualization tool. Such map-based approach allows identification and prioritization of candidate genes [11] based on a number of literature sources (references), genomic position, and pathway analyses, employing all currently available knowledge in different species. However, extrapolating the gained knowledge from one species to another is often difficult due to different anatomical and physiological characteristics, which should be considered when comparing pathology of the disease in different species.
To identify genetic factors potentially involved in CO pathogenesis in human we 1) applied comparative integratomics approach and assembled the database of all CO-associated genomic loci reported in the literature, regardless of the study approach and species, 2) presented the loci on a genomic map as human orthologs, and 3) prioritized the collected data using systems biology approach. The collected candidate genes were classified in corresponding biological pathways and the most significant CO-enriched pathways were proposed. Such classification of candidate genes allowed us to prioritize biological pathways (characterized by genes involved in the pathogenesis of CO), which revealed importance of several pathways (for example muscle contraction mechanisms) that may also play a role in the pathogenesis of other clinical features distinctive for different syndromes often concurrent with CO. In order to additionally illuminate the CO-associated pathways we performed a network-based protein-protein interaction analysis, which resulted in prediction of 43 additional CO candidate genes.
Methods
In search for CO associated candidate loci seven different research approaches were considered: (i) chromosomal abnormalities associated with CO, (ii) copy number variations, (iii) clinical syndromes with known genetic mutations that feature CO, (iv) transgenes and knock-outs that result in CO associated phenotypes, (v) association studies/mutation screening that show association between sequence variation/mutation screening and CO, (vi) expression patterns associated with CO, and (vii) candidates associated with CO at proteomic level.
Data mining
We reviewed the literature published up to 9/2012 searching for the relevant publications through PubMed (http://www.ncbi.nlm.nih.gov/pubmed/) and Web of Science (http://isiknowledge.com) using key phrases: genetics, gene candidates, cryptorchidism, testicular descent, undescended testes, male infertility, QTL, microarray, association, microRNA, non-coding RNA, epigenetic, reproduction, and assisted reproduction. CO-associated candidate genes from different sources and species were retrieved from the literature search. Human clinical syndromes that may cause or feature CO were retrieved from Online Mendelian Inheritance in Man (OMIM) database (http://www.ncbi.nlm.nih.gov/sites/entrez?db=omim) and Disease database (http://www.diseasesdatabase.com/). The data for CO-related experiments on mouse models were retrieved from the Mouse Genome Informatics (MGI) database (http://www.informatics.jax.org/). Human orthologs for the CO associated genes were extracted from the MGI database, which contains information about mammalian ortholog genes for different species. Overlap analysis of the CO candidate genes with genomic regions involved in chromosome mutations was performed using data retrieved from Ensembl via BioMart data mining tool.
Database implementation
CO-associated candidate genes database is a web resource, which provides integrated and curated information on molecular components involved in the pathogenesis of CO. Information regarding collected CO-associated candidate genes has been stored in relational MySQL database, which is publicly available for search, data entry and update at http://www.integratomics-time.com/cryptorchidism/. Search interface enables users to find specific CO-associated candidate genes based on the number of criteria. Online data entry interface enables users to update or submit new CO-associated candidate genes.
Genomic view of the CO associated loci
Overview of the chromosomal locations of CO associated loci is graphically represented in genomic view, as previously described [12]. It is possible to visualize the literature-collected and network-predicted CO genes on the same genomic view or separately. Genomic view is visible through the web-based interactive visualization tool Flash GViewer (http://gmod.org/wiki/Flashgviewer/), which was developed by the GMOD project.
Pathway and network analysis
In the first pathway analysis we considered human orthologs of the literature-collected candidate genes (179 genes). DAVID Bioinformatics Resources 6.7 [13] was employed for the enrichment (overrepresentation) analysis. The background for the analysis was defined using the 179 candidate genes plus their first neighbours (5018 proteins) selected in the human protein-protein interaction network (PPIN). The result of the enrichment analysis was obtained using Bonferroni multiple test correction and a p-value significant threshold of 0.01. The human PPIN was obtained by fusion of the following human networks: IRefIndex [14], Chuang et al. article [15], Ravasi et al. article [16], Consensus-PathDB [17].
A new cohort of 43 candidate genes was predicted using PiNGO 1.11 [18]. PiNGO is a tool designed to find candidate genes in biological networks and it is freely provided as a plug-in for Cytoscape 2.8 [19], which is an open source software platform for visualizing and integrating molecular interaction networks. PiNGO predicts the categorization of a candidate gene based on the annotations of its neighbors, using enrichment statistics. In our analysis we quested which first-neighbour-genes significantly interact with the original cohort of 179 literature-collected genes in the human PPIN. We adopted: hypergeometric statistical test, Bonferroni multiple testing correction and p-value significant threshold of 0.01. The cohort of 43 network-predicted genes resulted strongly significant (Bonferroni p-value < 0.0095) for being new candidate genes.
In order to evaluate the importance of this new cohort of 43 candidate genes we performed the pathway analysis according to the procedure already described for the 179 literature-collected candidate genes.
Finally, in order to investigate the biological relations between the 179 literature-collected and 43 network-predicted genes, we repeated the pathway analysis in DAVID (using the same procedure previously described) considering the 222 (179 + 43) candidate genes. The background for the analysis was defined using the 222 candidate genes plus their first neighbours in the human PPIN. In addition, we visualized the protein-protein interactions occurring between the genes present in at least two pathways using the STRING database (version 9.0) [20] and selecting only interactions with high confidence score.
Genetic variability of candidate genes
Genetic variability for the most promising CO candidate genes was extracted from the Ensembl database (http://www.ensembl.org/). Probably damaging genetic variations were predicted by PolyPhen-2, version 2.1.0, provided by Ensembl database. Putative polymorphic miRNA target sites in candidate genes were obtained from Patrocles database (http://www.patrocles.org/) [21].
Results and discussion
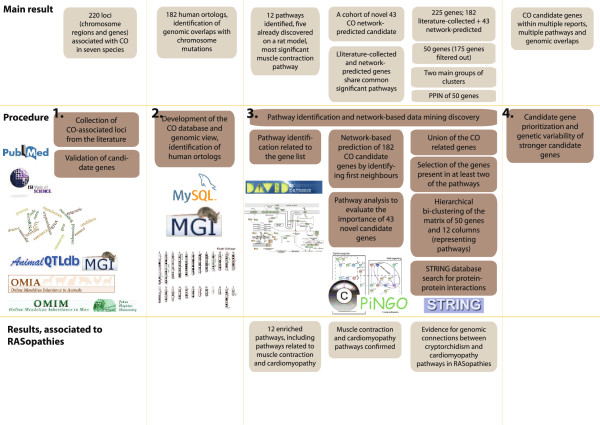
Extensive literature mining was performed resulting in 217 collected candidate loci (chromosome regions and genes) reported to be involved in CO in human or/and animals. The generated database served as the foundation for the development of freely available interactive genomics viewer designed to integrate multi-species data from various research approaches. Enriched biological pathways and 43 additional CO candidate genes were suggested, based on protein-protein interaction network (PPIN) analysis. The workflow of the study is presented in the Figure 1.
Figure 1.
Workflow of the study.
Collection of the cryptorchidism associated loci from the literature
The collected data incorporates genomic loci associated with cryptorchidism by seven different types of research approaches (chromosomal mutations, copy number variations, clinical syndromes, transgenes and knock-outs, association studies/mutation screening, transcriptomic/expression studies, and proteomic studies). The collected data originates from seven different species (human, cattle, horse, sheep, dog, rat, and mouse) (Table 1). The collected CO data is available in Additional file 1: Table S1, Additional file 2: Table S2, Additional file 3: Table S3, Additional file 4: Table S4 and Additional file 5: Table S5 and include physical locations of the candidate loci in human and species of origin.
Table 1.
The summary of CO associated candidate loci
| Locus type / study approach | Number of loci |
|---|---|
|
DNA level | |
| Chromosomal aberrations |
32 |
| Copy number variants (CNVs) |
2 |
| Clinical syndromes |
42 |
| Knock-out and transgenic experiments |
40 |
| Association studies |
12 |
|
RNA level | |
| Expression study |
112 |
|
Protein level |
|
| Injection of exogenous protein |
1 |
| Total | 217* |
* Unique loci (individual locus/gene reported by multiple studies was counted only once).
Chromosomal aberrations and copy number variations
We reviewed studies reporting 32 different chromosomal mutations including numerical and structural aberrations associated with cryptorchidism [22-33]. Additionally, two de novo copy number variations (CNVs) - microduplications were found to be associated with CO using array-based comparative genomic hybridization (aCGH) [34]. The collected data is available in Additional file 1: Table S1.
Clinical syndromes
Studies of complex disease traits can be facilitated by analysis of the molecular pathways represented by genes responsible for monogenic syndromes that also exhibit these traits [7,35]. There are over 200 different human syndromes with known molecular basis in OMIM database that feature “cryptorchidism” or “undescended testis” as a possible feature in their clinical synopsis. Since cryptorchidism phenotype prevalence is low in some syndromes, and could only occur coincidentally, it is difficult to justify association of syndrome causative genes with a particular phenotype.
To collect CO candidate genes (Additional file 2: Table S2) we obtained list of syndromes from the literature [4,36,37], OMIM and Diseases database (“may be caused or feature”) and then further examined phenotype-gene relationships and clinical features for each of the syndromes. Only syndromes where cryptorchidism is present as a regular feature, described in multiple clinical cases, and where gene(s) causing the syndrome is/are known were included.
Transgenes and knock-outs
From the Mouse Genome Informatics (MGI) database and the literature [38-42] we retrieved 39 mouse and one rat KO and transgenic experiments that result in phenotypes associated with CO (Additional file 3: Table S3).
Association studies/mutation screening
Nine genes (AR, BMP7, ESR1, HOXA10, INSL3, KISS1R, NR5A1, RXFP2, and TGFBR3) in human [43-59], INSL3 in sheep [60] and dog [61], and COL2A1 in dog [62] showed positive association between sequence polymorphisms/mutations and CO susceptibility (Additional file 4: Table S4). In the case of androgen receptor (AR) gene Ferlin et al. [45] found no difference between the numbers of CAG and GGC repeats, resulting in variable lengths of PolyGln/PolyGly in the AR gene and cryptorchidism; however, it has been proposed that a particular combination of the PolyGln/PolyGly polymorphisms may be linked to CO. In some cases opposing results have been found; for example, no association between the sequence polymorphisms and CO have been found for the genes ESR1[63-65], INSL3[66-68], HOXA10[69], and RXFP2[70]. The LHCGR has been excluded as a CO candidate gene in an association study in men [71], although KO of this gene in mice showed cryptorchid phenotype (MGI) and is causative gene of Leydig cell hypoplasia-a syndrome that features CO as one of the clinical signs (OMIM). In addition, Y chromosome microdeletions have been found to be present in patients with CO, but are not likely to be a common etiological cause of CO [72-74].
Expression patterns
There are several studies comparing expression profiles in testes between cryptorchid and normal males investigating the resulting effects of but not causes for development of CO (e.g.[75,76]). To our knowledge, there is only one microarray study that analyzed transcript profiles in gubernaculum during normal and abnormal testicular descent and reported 3589 differentially expressed genes between inherited cryptorchydism orl rats and a control group [7]. We included a subset of 112 promising candidate genes to our candidate gene list that were selected by the authors of the study based on expression levels, inclusion in specific pathways of interest and/or previous reports showing association with cryptorchidism (Additional file 5: Table S5).
Protein level
Hutson et al. (1998) [77] investigated the effect of exogenous calcitonin gene-related peptide (CGRP) in neonatal pigs. They found that exogenous CGRP, in pigs also known as calcitonin gene-related peptide B (CALCB), stimulated migration of inguinal testes that had been arrested in the line of descent, while ectopic testes did not respond. The results support the role for this protein in testicular descent. However, mutation screening performed by Zuccarello et al. (2004) [78] failed to confirm CGRP (in human also known as CALCA) pathway genes as a major players in human sporadic CO.
Development of the CO database and genomic viewer
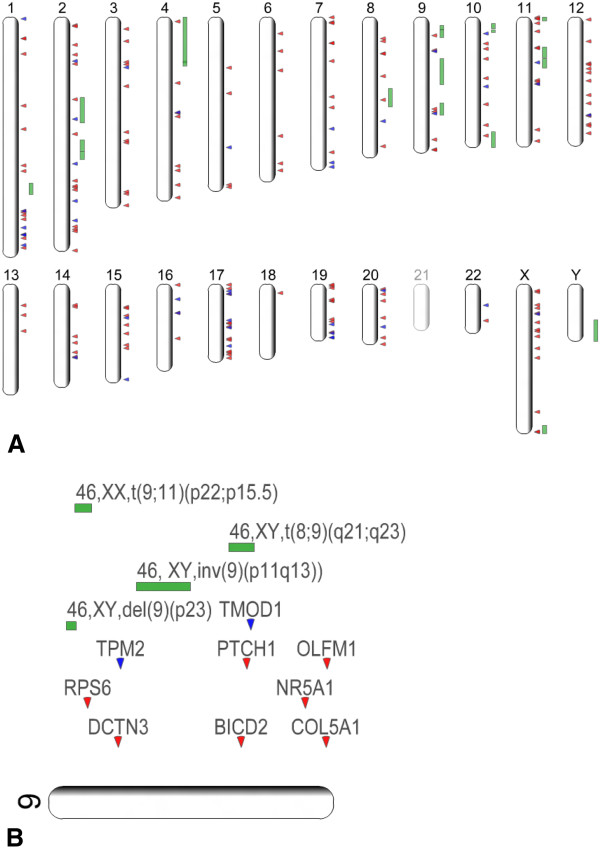
The CO-associated loci, obtained by comparative integratomics approach, were assembled into a freely accessible database available at http://www.integratomics-time.com/cryptorchidism/. The curated database is open for public data entry. Researchers are invited to submit new cryptorchidism candidate genes from their research or other publications by filling data entry form on our web site. The collected loci from human and animal species were presented as genomic view for human orthologs (in a form of a human genomic view) (Figure 2).
Figure 2.
Genomic view of the cryptorchidism candidate genes. A. Genomic view of the literature-collected (red) and network-predicted (blue) CO associated candidate loci presented as human orthologs. The view includes syndromes with known genetic mutations that feature CO, mouse transgenic and knock-out experiments, chromosomal abnormalities, genes tested for association with CO, genes with expression patterns associated with CO, and genes associated with CO on proteomic level. Loci are placed at approximate positions on chromosome map. B. Enlargement of the chromosome 9.
Some candidate genes have been associated with CO by multiple independent literature reports in multiple species. For example, twenty genes (AMH, AMHR2, AR, ARID5B, BMP7, EPHA4, ESR1, FGFR2, HOXA10, HRAS, INSL3, LHCGR, MAP2K1, MSX1, NR5A1, RXFP2, SOS1, TNNI2, TNNT3, and WT1) have been associated with CO in at least two independent studies using different study approaches (Table 2). These genes are denoted in bold in the online database (http://www.integratomics-time.com/cryptorchidism/candidate_genes/).
Table 2.
Literature-collected candidate genes associated with CO in at least two independent literature reports
| Gene | Clinical syndromes | KOs and transgenes | Overlapping chromosome mutations | Association studies (number of studies) | Expression experiments |
|---|---|---|---|---|---|
|
AMH |
human |
mouse |
|
|
|
|
AMHR2 |
human |
mouse |
|
|
|
|
AR |
human |
mouse |
|
human (2) |
|
|
ARID5B |
|
mouse |
|
|
rat |
|
BMP7 |
|
|
|
human |
rat |
|
EPHA4 |
|
mouse |
|
|
rat |
|
ESR1 |
|
mouse |
|
human (2) |
|
|
FGFR2 |
human |
|
human |
|
rat |
|
HOXA10 |
|
mouse |
|
human |
rat |
|
HRAS |
human |
|
human |
|
rat |
|
INSL3 |
|
mouse |
|
human (5), sheep, dog |
|
|
LHCGR |
human |
mouse |
|
|
|
|
MAP2K1 |
human |
|
|
|
rat |
|
MSX1 |
|
|
human |
|
rat |
|
NR5A1 |
|
mouse |
|
human |
|
|
RXFP2 |
|
mouse |
|
human (4) |
|
|
SOS1 |
human |
|
|
|
rat |
|
TNNI2 |
human |
|
human |
|
|
|
TNNT3 |
human |
|
human |
|
|
| WT1 | human | mouse |
The CO associated loci mapped to all human chromosomes, except HSA21. Genomic distribution of the selected loci revealed several overlapping areas between the candidate loci. Overlaps between structural chromosomal mutations and candidate genes can be observed in Figure 2 or by using interactive genomic view available on the website (http://www.integratomics-time.com/cryptorchidism/genomic_view/). Genomic regions involved in chromosome mutations on chromosomes 2, 4, 8, 9, 11, and X [22-33] overlapped with 13 literature-collected candidate genes: CAPG, MSX1, E2F5, PTCH1, BICD2, RPS6, FGFR2, HRAS, PAX6, WT1, TNNI2, TNNT3, FLNA, and MECP2. For instance, a breakpoint on 11p15.5 overlapped with three CO candidate genes: HRAS, TNNI2, and TNNT3. Additionally, in some cases two regions involved in chromosome mutations overlapped; duplication on position 4p overlapped with MSX1 gene and the region involved in chromosomal translocation on position 4p12. Interestingly, three network-predicted candidate genes, FHL2, TMOD1 and MYBPC3, also overlapped with genomic regions involved in the chromosome mutations.
Pathway identification and network-based data mining discovery
Pathway analysis of the cryptorchidism associated candidate genes
We performed pathway analysis of the 179 literature-collected CO-candidate genes (refer to Methods). This pathway enrichment analysis, conducted by applying very stringent criteria (Bonferroni multiple test correction and p-value significant threshold of 0.01), yielded the presence of twelve significant pathways associated with the list of our CO candidate genes in human (Table 3). The literature-collected candidate genes involved in multiple (at least four) pathways are presented in Additional file 6: Table S6 and marked with an asterisk in the online database.
Table 3.
Pathway analysis of the literature-collected and network-predicted candidate genes, respectively
| Database | Pathway | Bonferonni p-value | Candidate genes involved in the pathway |
|---|---|---|---|
|
Literature-collected candidate genes | |||
| KEGG |
Regulation of actin cytoskeleton |
4.70E-06 |
ACTB, BRAF, CDC42, CFL1, CHRM3, EZR, FGD1, FGF9, FGFR1, FGFR2, HRAS, ITGB1, KRAS, MAP2K1, MAP2K2, MYL2, MYL9, PDGFA, PFN1, PPP1CA, PPP1CB, PXN, RAC1, RAF1, RHOA, RRAS, SOS1 |
| REACTOME |
Muscle contraction |
5.39E-06 |
DES, MYH3, MYL2, MYL3, TNNI2, TNNT2, TNNT3, TPM1, TPM3, TPM4, TTN |
| KEGG |
Focal adhesion |
1.05E-05 |
ACTB, BRAF, CCND1, CDC42, COL1A2, COL2A1, COL5A1, FLNA, GRB2, GSK3B, HRAS, IGF1, ILK, ITGB1, MAP2K1, MYL2, MYL9, PDGFA, PPP1CA, PPP1CB, PXN, RAC1, RAF1, RHOA, SOS1, THBS4 |
| REACTOME |
Signaling by PDGF |
3.61E-5 |
COL1A2, COL2A1, COL5A1, GRB2, HRAS, KRAS, MAP2K1, MAP2K2, PDGFA PTPN11, RAF1, SOS1, STAT3, THBS4 |
| REACTOME |
Signaling by insulin receptor |
7.02E-05 |
EIF4E, EIF4EBP1, GRB2, HRAS, KRAS, MAP2K1, MAP2K2, RAF1, RPS6, RPS6KB1, SOS1 |
| REACTOME |
Signaling by EGFR |
0.0022 |
CDC42, GRB2, HRAS, KRAS, MAP2K1, MAP2K2, PTPN11, PXN, RAF1, SOS1 |
| PANTHER |
RAS pathway |
0.0025 |
BRAF, CDC42, GRB2, GSK3B, HRAS, KRAS, MAP2K1, MAP2K2, RAC1, RAF1, RHOA, RRAS, SOS1, STAT3 |
| KEGG |
Hypertrophic cardiomyopathy (HCM) |
0.0035 |
ACTB, DES, IGF1, ITGB1, MYH7, MYL2, MYL3, TNNT2, TPM1, TPM3, TPM4, TTN |
| BIOCARTA |
Role of MAL in Rho-mediated activation of SRF |
0.044 |
ACTA1, CDC42, MAP2K1, MAP2K2, RAC1, RAF1, RHOA |
| BIOCARTA |
IGF-1 signaling |
0.0067 |
FOS, GRB2, HRAS, IGF1, MAP2K1, PTPN11, RAF1, SOS1 |
| PANTHER |
Integrin signaling |
0.0070 |
BRAF, CDC42, COL1A2, COL2A1, COL5A1, FLNA, GRB2, HRAS, ILK, ITGB1, KRAS, MAP2K1, MAP2K2, PXN, RAC1, RAF1, RHOA, RND2, RRAS, SOS1 |
| KEGG |
Dilated cardiomyopathy |
0.0099 |
ACTB, DES, IGF1, ITGB1, MYH7, MYL2, MYL3, TNNT2, TPM1, TPM3, TPM4, TTN |
|
Network-predicted candidate genes | |||
| REACTOME |
Muscle contraction |
2.71E-24 |
ACTN2, DMD, MYBPC1, MYBPC2, MYBPC3, MYH8, MYL1, MYL4, NEB, TCAP, TMOD1, TNNC1, TNNC2, TNNI1, TNNI3, TNNT1, TPM2, VIM |
| KEGG |
Hypertrophic cardiomyopathy (HCM) |
9.68E-6 |
ACTC1, DMD, MYBPC3, TGFB1, TGFB2, TGFB3, TNNC1, TNNI3, TPM2, |
| PANTHER |
TGF-beta signaling pathway |
1.07E-5 |
BMP2, LEFTY1, LEFTY2, LOC100271831, MAPK1, MAPK3, MSTN, NODAL, TGFB1, TGFB2, TGFB3 |
| KEGG | Dilated cardiomyopathy | 2.35E-5 | ACTC1, DMD, MYBPC3, TGFB1, TGFB2, TGFB3, TNNC1, TNNI3, TPM2 |
The presence of pathways related to “cytoskeleton”, “muscle development”, “muscle contraction”, “focal adhesion”, and “insulin signaling” was previously reported in rat [7]. In addition to these pathways, our analysis showed new pathways: “cardiomyopathy” (hypertrophic and dilated),“RAS signaling”, “signaling by PDGF”, “signaling by EGFR”, “role of MAL in Rho-mediated activation of SRF”, “IGF-1 signaling pathway”, and “integrin signaling”. The results represent a valid example of pathway-based data mining discovery.
As an additional validation analysis, we excluded the 112 candidate genes proposed by Barthold et al. (2008) [7] from the overall candidate genes list (consisting of 179 unique human genes) and repeated the pathway analysis. Nine genes from Barthold et al. (2008) [7] were reported as CO candidate genes also in other studies, therefore we retained them in the analysis, so that the new list of candidate genes consisted of 79 genes. The pathway analysis of these remaining 79 candidate genes returned similar results as were obtained when using the overall 179 candidate gene list. In fact, 10 of the 12 enriched pathways were the same after excluding the discussed data from the candidate gene list. In particular, the five pathways reported by Barthold et al. (2008) [7] in rat (“cytoskeleton”, “muscle development”, “muscle contraction”, “focal adhesion”, and “insulin signaling”) were all confirmed in this independent validation analysis. The main effect of the gene removal were higher, but still significant, p-values in the pathway analysis. According to these results we can infer that inclusion of the candidate genes from Barthold et al. (2008) [7] is not the reason for the substantial overlap of the five pathways identified in both studies. On the contrary, the findings proposed here are a further confirmation of the validity of the conclusions made by Barthold et al. (2008) [7].
Surprisingly, when we searched the medical literature for articles that describe pathologies where CO, cardiomyopathy, and RAS signaling are common features, we found a perfect matching with Noonan, Cardiofaciocutaneous, LEOPARD, and Costello syndrome that all belong to the class of RASopathies [79,80]. Features of all four syndromes are different physical anomalies including concomitant presence of cardiomyopathy due to heart defects and, in males, cryptorchidism [79]. Noonan syndrome (NS) is the most common single gene cause of congenital heart disease, and NS subjects also present other features as leukemia predisposition [81]. In particular, five different mutations in RAF1 were identified in individuals with NS; four mutations causing changes in the CR2 domain of RAF1 were associated with hypertrophic cardiomyopathy (HCM), whereas mutations in the CR3 domain were not [82]. Additionally, PTPN11, RAF1, and SOS1 mutants were identified as a major cause of Noonan syndrome, BRAF of Cardiofaciocutaneous, PTPN11 of LEOPARD, and HRAS of Costello syndrome, providing new insights into RAS regulation [80,81]. These genes have also been found to be mutated in patients with RASopathies having cryptorchidism in a clinical picture. In NS patients having CO in their clinical picture 11/14 had mutated PTPN11, 4/5 had mutated SOS1, and 1/2 had mutated RAF1. BRAF has been found to be mutated in 2/3 patients with Cardiofaciocutaneous syndrome having CO, PTPN11 in 1/4 patients with LEOPARD having CO, and HRAS in 2/4 patients with Costello syndrome and CO [80,81]. However, the genes responsible for the remainder are unknown, and the gene pathway relations responsible for potential connections between unrelated features such as cryptorchidism and HCM in RASopathies are not clear. Therefore, we performed a network-based prediction (see next paragraph) of CO candidate genes by identifying the most significant first neighbors (in the human protein-protein interaction network; PPIN) of the 179 literature-collected candidates.
Pathway analysis of the network-predicted candidate genes
A new cohort of 43 candidate genes (Additional file 7: Table S7) was predicted by PiNGO 1.11 [18], which is a Cytoscape plug-in (see Methods) [19]. The question we tried to address was which first-neighbor genes significantly interact with the original cohort of 179 literature-collected genes in the human PPIN. We adopted hypergeometric statistical test and Bonferroni multiple testing correction. The cohort of 43 network-predicted genes was strongly significant (Bonferroni p-value < 0.0095); therefore, we consider them as additional CO candidate genes.
In order to evaluate the importance of these new candidate genes we performed the pathway analysis (Table 3), according to the same procedure already used in the previous paragraph (and described in the methods). The most intriguing evidence is the presence of significant pathways related to cardiomyopathy and muscle contraction in both sets of candidate genes (i.e. literature-collected and network-predicted). Pathways common to both sets of candidate genes represent a confirmation of the validity and robustness of the results obtained in the first pathway analysis and regarding the hypothesis of connection between CO and cardiomyopathy, in NS. Yet, it is also a quality proof of the procedure adopted for network prediction of new candidate genes.
Pathway analysis of the overall CO candidate gene list (179 literature-collected and 43 network-predicted genes)
The first cohort of 179 literature-collected genes and the second one containing 43 network-predicted genes were condensed in a list of 222 unique genes - the overall candidate gene list. We repeated the pathway analysis on this list applying the same very stringent criteria used above (Bonferroni multiple test correction and p-value significant threshold of 0.01). The analysis suggested the presence of 12 significant pathways associated with the overall list of candidate genes in human (Table 4).
Table 4.
Pathway analysis of the overall candidate gene list (literature-collected and network-predicted)
| Database | Pathway | Bonnferoni p-value | Candidate genes involved in the pathway |
|---|---|---|---|
| REACTOME |
Muscle contraction |
4.55E-33 |
ACTN2, DES, DMD, MYBPC1, MYBPC2, MYBPC3, MYH3, MYH8, MYL1, MYL2, MYL3, MYL4, NEB, TCAP, TMOD1, TNNC1, TNNC2, TNNI1, TNNI2, TNNI3, TNNT1, TNNT2, TNNT3, TPM1, TPM2, TPM3, TPM4, TTN, VIM |
| KEGG |
Hypertrophic cardiomyopathy (HCM) |
1.21E-9 |
ACTB, ACTC1, DES, DMD,IGF1, ITGB1, MYBPC3, MYH7, MYL2, MYL3, TGFB1, TGFB2, TGFB3, TNNC1, TNNI3, TNNT2, TPM1, TPM2, TPM3, TPM4, TTN |
| KEGG |
Dilated cardiomyopathy |
1.21E-8 |
ACTB, ACTC1, DES, DMD, IGF1, ITGB1, MYBPC3, MYH7, MYL2, MYL3, TGFB1, TGFB2, TGFB3, TNNC1, TNNI3, TNNT2, TPM1, TPM2, TPM3, TPM4, TTN |
| KEGG |
Focal adhesion |
4.68E-7 |
ACTB, ACTN2, BRAF, CAV1, CCND1, CDC42, COL1A2, COL2A1, COL5A1, FLNA, GRB2, GSK3B, HRAS, IGF1, IGF1R, ILK, ITGB1, MAP2K1, MAPK1, MAPK3, MYL2, MYL9, PDGFA, PPP1CA, PPP1CB, PRKCA, PXN, RAC1, RAF1, RHOA, SOS1, THBS4 |
| REACTOME |
Signaling by insulin receptor |
3.83E-6 |
EIF4E, EIF4EBP1, GRB2, HRAS, KRAS, MAP2K1, MAP2K2, MAPK1, MAPK3, RAF1, RHEB, RPS6, RPS6KB1, SOS1 |
| KEGG |
Regulation of actin cytoskeleton |
4.29E-6 |
ACTB, ACTN2, BRAF, CDC42, CFL1, CHRM3, EZR, FGD1, FGF3, FGF9, FGFR1, FGFR2, HRAS, ITGB1, KRAS, MAP2K1, MAP2K2, MAPK1, MAPK3, MYL2, MYL9, PDGFA, PFN1, PPP1CA, PPP1CB, PXN, RAC1, RAF1, RHOA, RRAS, SOS1 |
| PANTHER |
TGF-beta signaling pathway |
7.01E-6 |
AMH, AMHR2, BMP2, BMP4, BMP5, BMP7, CDC42, FOS, FOXO1, FOXP3, HRAS, KRAS, LEFTY1, LEFTY2, MAPK1, MAPK3, MSTN, NODAL, RHEB, RRAS, TGFB1, TGFB2, TGFB3 |
| BIOCARTA |
Integrin signaling pathway |
1.35E-5 |
ACTA1, ACTN2, CAV1, GRB2, HRAS, ITGB1, MAP2K1, MAP2K2, MAPK1, MAPK3, PXN, RAF1, RHOA, SOS1 |
| REACTOME |
Signaling by PDGF |
7.27E-5 |
COL2A1, COL1A2, COL5A1, GRB2, HRAS, KRAS, MAP2K1, MAP2K2, MAPK1, MAPK3, PDGFA, PTPN11, RAF1, SOS1, STAT3, THBS4 |
| KEGG |
Cardiac muscle contraction |
0.0024 |
ACTC1, MYH7, MYL2, MYL3, TNNC1, TNNI3, TNNT2, TPM1, TPM2, TPM3, TPM4 |
| PANTHER |
RAS pathway |
0.0030 |
BRAF, CDC42, GRB2, GSK3B, HRAS, KRAS, MAP2K1, MAP2K2, MAPK1, MAPK3, RAC1, RAF1, RHOA, RRAS, SOS1, STAT3 |
| KEGG | Vascular smooth muscle contraction | 0.0058 | ACTA2, BRAF, MAP2K1, MAP2K2, MAPK1, MAPK3, MYH11, MYL9, PPP1CA, PPP1CB, PRKCA, PRKCE, RAF1, RHOA |
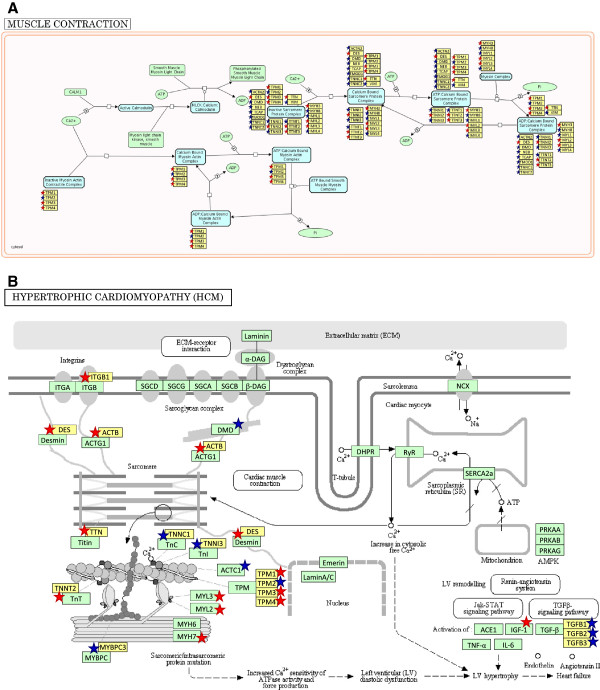
The “muscle contraction” pathway was the most significant (in absolute) with Bonferroni corrected p-value of 4.55E-33 (Figure 3A); while the “hypertrophic cardiomyopathy” was the second most significant pathway with Bonferroni corrected p-value of 1.21E-09 (Figure 3B). These results are crucial for our study because they suggest the presence of a strong genomic connection among diverse pathways associated with clinical features that seemed unrelated. To address relationship among these mechanisms we created a matrix merging the information related to the gene participation in several identified pathways. Of the 222 (179 + 43) candidate genes, 172 were filtered out because they were not present in at least two of the 12 significant pathways. The resulting matrix consists of 50 candidate genes in the rows and 12 enriched pathways in the columns (Additional file 8: Table S8). The matrix values are binary: 0 indicates that the gene is not present in a pathway, whereas 1 indicates that the gene is present.
Figure 3.
Pathways, containing literature-collected and network-predicted CO candidate genes. CO genes are marked with stars: red stars stand for literature-collected genes, blue stars stand for network-predicted genes. Gene names are added in yellow boxes to the original pathway images. A. Reactome pathway: Muscle contraction – human. B. KEGG pathway: Hypertrophic cardiomyopathy (HCM) – human.
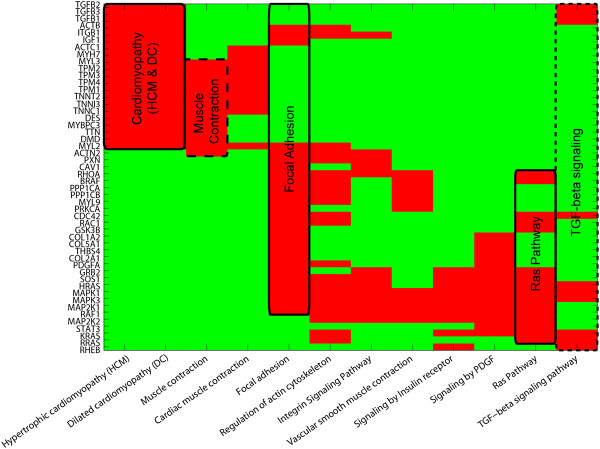
Hierarchical bi-clustering of the matrix [83], both in the rows and in the columns, was performed to detect clusters of genes which participated in common pathways, and clusters of pathways which share the same genes, respectively. The result of this analysis is provided in the Figure 4. The presence of two main groups of clusters is evident. The first group is constituted of “cardiomyopathy” (hypertrophic and dilated), “muscle contraction” and “cardiac muscle contraction” pathways. The second group is constituted of “focal adhesion”, “regulation of actin cytoskeleton, “integrin signaling”, “vascular smooth muscle contraction”, “signaling by insulin receptor”, “signaling by PDGF”, “RAS pathway”, and “TGF-beta signaling”.
Figure 4.
Hierarchical bi-clustering of the CO candidate genes. Hierarchical bi-clustering of the matrix of the CO candidate genes present in at least two pathways. The matrix consists of 50 candidate genes (rows) and 12 enriched pathways (columns). The black lines (full and dashed) are used to indicate the modules corresponding to clusters of interacting proteins in the respective pathways.
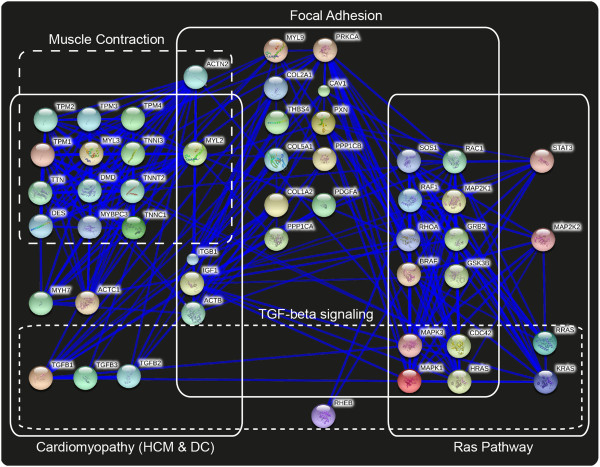
In order to further investigate the relation between the genes involved in the “cardiomyopathy” (hypertrophic and dilated), the “muscle contraction” and the “RAS pathway” and to interpret their role in creating connections between the diverse pathway modules, we searched the STRING database [20] for protein-protein interactions, selecting only the interactions with high confidence score. The outcome of this analysis is represented in Figure 5. All the 50 genes presented at least one interaction in the PPI network produced by the STRING database. This network is provided as a supplementary material (Additional file 9: Table S9).
Figure 5.
String protein-protein interaction network (PPIN) of the CO candidate genes. PPIN is obtained from the 50 CO candidate genes interacting in the STRING database. The lines (full and dashed) delimit the presence of the diverse overlapped protein-pathway-modules. The same line is used in the Figure 4 to indicate the modules corresponding to clusters of interacting proteins in the respective pathways.
The principal pathways involved in both, CO and RASopathies, are displayed on the PPIN (Figure 5), and also marked in the Figure 4 to facilitate the comparison. This figure addresses the question of the relation between the common genetic mechanisms underlying CO and RASopathies. Figure 5 provides a clear visualization of the overlapping pathways and of the integrated network of relations existing on proteomic level. At the best of our knowledge, this is the first time that such relation is presented, and it might help in understanding the relation between co-presence of CO and cardiomyopathy as clinical and apparently unrelated features in RASopathies. This fact is clarified by the layout offered in Figure 5 that reveals how the “cardiomyopathy” and the “RAS signaling” pathways are connected by a plethora of interactions with high confidence score in the STRING database. To investigate the precise type of intra- and inter- pathway interactions we suggest to mine the network that we provide in the supplementary material (Additional file 9: Table S9). Figure 5 further emphasizes how the “focal adhesion” and the “TGF-beta signaling” pathways overlap the “cardiomyopathy”, the “muscle contraction” and the “RAS signaling” pathways by connecting proteins at different metabolic levels. The relevance of the “focal adhesion” pathway, as well as the importance of “cytoskeleton”, “muscle development”, “muscle contraction”, and “insulin signaling” pathways in cryptorchidism were widely discussed [7]. However, the referred study was conducted on a rat model and all of the pathways were considered and treated separately. Here, for the first time, we proceed to an integratomic investigation of the genetic factors linked to CO in human. Meanwhile, we offer the holistic perspective that points out how clinical features apparently unrelated with CO might be generated by genetic mutation(s) which propagate at different pathway levels of the network. This propagation on different pathway-modules can justify the onset of multiple unrelated clinical features in complex diseases, such as RASopathies. The selection of 43 network-based predicted genes considered together with these last disease-related evidences are another proof that confirms the power of PPIN for association of genes with diseases [21,84].
Our results are in concordance with previous observations that alignment of human interactome with human phenome enables identification of causative genes (and networks) underlying disease families. Phenotypic overlap implies genetic overlap and human phenome can be viewed as a landscape of interrelated diseases, which reflects overlapping molecular causation [6,85-88]. In addition, it has been already shown that causative genes from syndromes that are phenotypically similar to a genetically uncharacterized syndrome can be used to query the gene network for functionally related candidate genes [89].
Candidate gene prioritization
Prioritization of candidate genes underlying complex traits remains one of the main challenges in molecular biology [11]. In this study we used three criteria for selecting the most promising candidate genes: 1) number of independent literature reports connecting the candidate gene with CO (Table 2), 2) involvement of candidate genes in enriched pathways (Table 3), and 3) position of candidate genes on the genomic map (genes positioned in regions where multiple CO associated data overlap were considered positional candidates) (Figure 2).
Twenty genes have been suggested as a genetic cause for CO in at least two independent studies (criterion 1) using different study approaches (AMH, AMHR2, AR, ARID5B, BMP7, EPHA4, ESR1, FGFR2, HOXA10, HRAS, INSL3, LHCGR, MAP2K1, MSX1, NR5A1, RXFP2, SOS1, TNNI2, TNNT3, and WT1). Among them, INSL3 has been associated with CO in eight, RXFP2 in five, and AR in four independent studies. However, this approach should be treated with some caution because of the possible bias towards research interest into more “popular” genes. The approach will be more reliable after significant amount of unbiased genome-wide studies is available.
Considering involvement in enriched pathways (criterion 2), the most promising candidates would be HRAS, MAP2K1, MAP2K2, GRB2, RAF1 and SOS1, which are all involved in seven or more enriched pathways. For the literature-collected candidate genes involved in multiple (four or more) CO-enriched pathways we assembled genetic information relevant for further functional analyses: assignment to corresponding biological pathways, genetic variability, and putative presence of polymorphic microRNA (miRNA) target sites (Additional file 6: Table S6). The importance of small non-coding RNAs (ncRNAs) in gene regulation and pathogenesis of the diseases, including reduced fertility, is today evident [90]. However, to our knowledge, there are no literature reports associating ncRNAs or epigenetic factors with CO.
The most promising candidates meeting both suggested criteria (1 and 2) are FGFR2 (reported in two CO-associated studies/ involved in one CO-associated pathway), HRAS (3/8), MAP2K1 (2/9), and SOS1 (2/5). Additionally, TNNI2 and TNNT3 are reported in the literature (once each), involved in one CO enriched pathway (i.e. “muscle contraction”), and positioned in a region overlapping chromosomal mutation.
Genomic regions involved in the chromosome mutations on chromosomes 2, 4, 8, 9, 11, and X overlapped with 14 candidate genes suggested as positional candidates (criterion 3): CAPG, MSX1, E2F5, PTCH1, BICD2, RPS6, FGFR2, HRAS, PAX6, WT1, TNNI2, TNN3, FLNA, and MECP2. Additionally, three network-predicted candidate genes, FHL2, TMOD1 and MYBPC3 overlapped with chromosomal mutations. Considering suggested prioritization criteria, HRAS gene meets all of them.
Reliability of such methodologically different approaches is not always comparable (for example, data from genome-wide expression experiments is much less validated than syndromic or transgenic data); therefore, ranking candidate genes based only on a number of different reports/approaches is not always feasible. However, less validated data may also be of high biological relevance and should not be discarded for hypothesis-driven approaches. To increase reliability of the collected heterogeneous data we tested in silico how candidate genes interact at the proteomic level. Although integratomic approaches are only partially established yet and have several drawbacks, including already mentioned heterogeneity of input data, we believe that such approaches are a reasonable and at the moment among the most promising ways for hypothesis generation, which should be further experimentally validated in animal and/or human populations. Similar integratomics approach was already used for identification of candidate loci for mammary gland associated phenotypes [8], male infertility [9], and obesity [10,91], and could be adapted to any other complex trait.
Conclusions
In this study we present an overview of CO associated candidate regions/genes and suggest pathways potentially involved in the pathogenesis of the disease. The integrative, comparative-genomics approach, and in silico analyses of the collected data aim to help solving the problem of fragmented and often contradictory data extracted from different methodologically focused studies. The protein-protein interactions analysis revealed the most relevant pathways associated with CO candidate gene list and enabled us to suggest additional candidate genes based on network prediction. Described systems biology approach will contribute to a better understanding of genetic causes for cryptorchidism and provides possible example how integration and linking of complex traits related data can be used for hypothesis generation. Publicly available online CO gene atlas and data entry option will allow researcher to enter, browse, and visualize CO associated data. The proposed network-based approach elucidates co-presence of similar pathogenetic mechanisms underlying diverse clinical syndromes/defects and could be of a great importance in research in the field of molecular syndromology. This approach has also a potential to be used for future development of diagnostic, prognostic, and therapeutic markers. The developed integratomics approach can be extrapolated to study genetic background of any other complex traits/diseases and to generate hypothesis for downstream experimental validation.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
CVC performed pathway and PPIN analyses. JO, TK, and MZ performed the data mining and established the database. TK, JO, and CVC interpreted the results and drafted the manuscript. MZ developed web-based interactive visualization tool. PD and TR provided feedback on the initial draft and contributed to the final editing of the manuscript. All authors have read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Chromosomal abnormalities and CNVs associated with cryptorchidism.
Selected clinical syndromes that feature CO in their clinical picture.
Transgenic and knock-out murine models that display cryptorchid phenotype.
Genes tested for association with CO.
Genes with expression patterns associated with CO. Genes with expression patterns associated with CO in rat (adapted from [7]) and genomic location of their human orthologs.
The literature collected candidate genes involved in multiple (four or more) CO-associated pathways.
Forty-three network-predicted CO-associated candidate genes.
Protein network information matrix for candidate genes. Protein network information matrix for candidate genes involved in at least two pathways significant for literature-collected and network-predicted candidate genes. The matrix consists of 50 rows (each row corresponds to a different gene involved in at least two pathways) and 12 columns (each column corresponds to a different pathway), where 0 indicates that the gene is not present in a pathway and 1 indicates that the gene is present in it.
STRING network data: list of protein network interactions present in STRING with high confidence score.
Contributor Information
Carlo V Cannistraci, Email: kalokagathos.agon@gmail.com.
Jernej Ogorevc, Email: jernej.ogorevc@bf.uni-lj.si.
Minja Zorc, Email: minja.zorc@gmail.com.
Timothy Ravasi, Email: timothy.ravasi@kaust.edu.sa.
Peter Dovc, Email: peter.dovc@bf.uni-lj.si.
Tanja Kunej, Email: tanja.kunej@bf.uni-lj.si.
Acknowledgements
This work was supported by the Slovenian Research Agency (ARRS) through the Research programme Comparative genomics and genome biodiversity (P4-0220). C.V.C. received financial support from the Italian Inter-polytechnic School of Doctorate (SIPD) and from the King Abdullah University of Science and Technology.
References
- Dovc P, Kunej T, Williams GA. In: Reproductive Genomics of Domestic Animals. 1. Jiang Z, Ott TL, editor. Oxford, UK: Wiley-Blackwell; 2010. Genetics and genomics of reproductive disorders; pp. 67–97. [Google Scholar]
- Amann RP, Veeramachaneni DNR. Cryptorchidism in common eutherian mammals. Reproduction. 2007;133(3):541–561. doi: 10.1530/REP-06-0272. [DOI] [PubMed] [Google Scholar]
- Foresta C, Zuccarello D, Garolla A, Ferlin A. Role of hormones, genes, and environment in human cryptorchidism. Endocr Rev. 2008;29(5):560–580. doi: 10.1210/er.2007-0042. [DOI] [PubMed] [Google Scholar]
- Barthold JS. Undescended testis: current theories of etiology. Curr Opin Urol. 2008;18(4):395–400. doi: 10.1097/MOU.0b013e3283005869. [DOI] [PubMed] [Google Scholar]
- Mortell A, Montedonico S, Puri P. Animal models in pediatric surgery. Pediatr Surg Int. 2006;22(2):111–128. doi: 10.1007/s00383-005-1593-4. [DOI] [PubMed] [Google Scholar]
- Oti M, Huynen MA, Brunner HG. Phenome connections. Trends Genet. 2008;24(3):103–106. doi: 10.1016/j.tig.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Barthold JS, McCahan SM, Singh AV, Knudsen TB, Si X, Campion L, Akins RE. Altered expression of muscle- and cytoskeleton-related genes in a rat strain with inherited cryptorchidism. J Androl. 2008;29(3):352–366. doi: 10.2164/jandrol.107.003970. [DOI] [PubMed] [Google Scholar]
- Ogorevc J, Kunej T, Razpet A, Dovc P. Database of cattle candidate genes and genetic markers for milk production and mastitis. Anim Genet. 2009;40(6):832–851. doi: 10.1111/j.1365-2052.2009.01921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogorevc J, Dovc P, Kunej T. Comparative genomics approach to identify candidate genetic loci for male fertility. Reprod Domest Anim. 2011;46(2):229–239. doi: 10.1111/j.1439-0531.2010.01648.x. [DOI] [PubMed] [Google Scholar]
- Kunej T, Jevsinek Skok D, Zorc M, Ogrinc A, Michal JJ, Kovac M, Jiang Z. Obesity gene atlas in mammals. J Genomics. 2012;1:45–55. doi: 10.7150/jgen.3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau Y, Tranchevent LC. Computational tools for prioritizing candidate genes: boosting disease gene discovery. Nat Rev Genet. 2012;13(8):523–536. doi: 10.1038/nrg3253. [DOI] [PubMed] [Google Scholar]
- Zorc M, Jevsinek Skok D, Godnic I, Calin GA, Horvat S, Jiang Z, Dovc P, Kunej T. Catalog of MicroRNA Seed Polymorphisms in Vertebrates. PLoS One. 2012;7(1):e30737. doi: 10.1371/journal.pone.0030737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Huang W, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35(Web Server issue):W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razick S, Magklaras G, Donaldson IM. iRefIndex: a consolidated protein interaction database with provenance. BMC Bioinforma. 2008;9:405. doi: 10.1186/1471-2105-9-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol Syst Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasi T, Cannistraci CV, Suzuki H, Katayama S, Bajic VB, Tan K, Akalin A, Schmeier S, Kanamori-Katayama M, Bertin N. An Atlas of Combinatorial Transcriptional Regulation in Mouse and Man. Cell. 2010;140(5):744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamburov A, Wierling C, Lehrach H, Herwig R. ConsensusPathDB--a database for integrating human functional interaction networks. Nucleic Acids Res. 2009;37(Database issue):D623–D628. doi: 10.1093/nar/gkn698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot M, Ono K, Ideker T, Maere S. PiNGO: a Cytoscape plugin to find candidate genes in biological networks. Bioinformatics. 2011;27(7):1030–1031. doi: 10.1093/bioinformatics/btr045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27(3):431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39(Database issue):D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiard S, Charlier C, Coppieters W, Georges M, Baurain D. Patrocles: a database of polymorphic miRNA-mediated gene regulation in vertebrates. Nucleic Acids Res. 2010;38(Database issue):D640–D651. doi: 10.1093/nar/gkp926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake Y, Kaneda Y. A new type of Robertsonian translocation (1/26) in a bull with unilateral cryptorchidism, probably occurring de novo. Nihon juigaku zasshi J Vet Sci. 1987;49(6):1015–1019. doi: 10.1292/jvms1939.49.1015. [DOI] [PubMed] [Google Scholar]
- Sasagawa I, Nakada T, Ishigooka M, Sawamura T, Adachi Y, Hashimoto T. Chromosomal anomalies in cryptorchidism. Int Urol Nephrol. 1996;28(1):99–102. doi: 10.1007/BF02550145. [DOI] [PubMed] [Google Scholar]
- Ogata T, Muroya K, Matsuo N, Hata J, Fukushima Y, Suzuki Y. Impaired male sex development in an infant with molecularly defined partial 9p monosomy: implication for a testis forming gene(s) on 9p. J Med Genet. 1997;34(4):331–334. doi: 10.1136/jmg.34.4.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Sasagawa I, Nakada T, Onmura Y. Bilateral cryptorchidism associated with terminal deletion of 10q. Urol Int. 1998;61(3):186–187. doi: 10.1159/000030321. [DOI] [PubMed] [Google Scholar]
- Goldschmidt B, El-Jaick KB, Souza LM, Carvalho ECQ, Moura VLS, Benevides Filho IM. Cryptorchidism associated with 78, XY/79, XXY mosaicism in dog. Israel J Vet Med. 2001;56:56–58. [Google Scholar]
- Moreno-Garcia M, Miranda EB. Chromosomal anomalies in cryptorchidism and hypospadias. J Urol. 2002;168(5):2170–2172. doi: 10.1016/S0022-5347(05)64346-7. discussion 2172. [DOI] [PubMed] [Google Scholar]
- Prabhakara K, Angalena R, Ramadevi AR. Familial (9;11)(p22;p15.5)pat translocation and XX sex reversal in a phenotypic boy with cryptorchidism and delayed development. Genetic counseling (Geneva, Switzerland) 2004;15(1):37–41. [PubMed] [Google Scholar]
- Brito L, Sertich PL, Durkin K, Chowdhary BP, Turner RM, Greene LM, McDonnell S. Autosomic 27 Trisomy in a Standardbred Colt. J Equine Vet Sci. 2008;28(7):431–436. [Google Scholar]
- Tartaglia N, Davis S, Hench A, Nimishakavi S, Beauregard R, Reynolds A, Fenton L, Albrecht L, Ross J, Visootsak J. A new look at XXYY syndrome: medical and psychological features. Am J Med Genet A. 2008;146A(12):1509–1522. doi: 10.1002/ajmg.a.32366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veken LT, Dieleman MM, Douben H, van de Brug JC, van de Graaf R, Hoogeboom AJ, Poddighe PJ, de Klein A. Low grade mosaic for a complex supernumerary ring chromosome 18 in an adult patient with multiple congenital anomalies. Mol Cytogenet. 2010;3:13. doi: 10.1186/1755-8166-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyazov DM, Nawaz Z, Justice AN, Toriello HV, Martin CL, Adam MP. Genotype/phenotype correlations in two patients with 12q subtelomere deletions. Am J Med Genet A. 2007;143A(22):2700–2705. doi: 10.1002/ajmg.a.32005. [DOI] [PubMed] [Google Scholar]
- Melis D, Genesio R, Boemio P, Del Giudice E, Cappuccio G, Mormile A, Ronga V, Conti A, Imperati F, Nitsch L. Clinical description of a patient carrying the smallest reported deletion involving 10p14 region. Am J Med Genet A. 2012;158A(4):832–835. doi: 10.1002/ajmg.a.34133. [DOI] [PubMed] [Google Scholar]
- Tannour-Louet M, Han S, Corbett ST, Louet JF, Yatsenko S, Meyers L, Shaw CA, Kang SH, Cheung SW, Lamb DJ. Identification of de novo copy number variants associated with human disorders of sexual development. PLoS One. 2010;5(10):e15392. doi: 10.1371/journal.pone.0015392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltonen L, Perola M, Naukkarinen J, Palotie A. Lessons from studying monogenic disease for common disease. Hum Mol Genet. 2006;15:R67–R74. doi: 10.1093/hmg/ddl060. Spec No 1. [DOI] [PubMed] [Google Scholar]
- Gianotten J, van der Veen F, Alders M, Leschot NJ, Tanck MW, Land JA, Kremer JA, Hoefsloot LH, Mannens MM, Lombardi MP. Chromosomal region 11p15 is associated with male factor subfertility. Mol Hum Reprod. 2003;9(10):587–592. doi: 10.1093/molehr/gag081. [DOI] [PubMed] [Google Scholar]
- Klonisch T, Fowler PA, Hombach-Klonisch S. Molecular and genetic regulation of testis descent and external genitalia development. Dev Biol. 2004;270(1):1–18. doi: 10.1016/j.ydbio.2004.02.018. [DOI] [PubMed] [Google Scholar]
- Kreidberg JA, Sariola H, Loring JM, Maeda M, Pelletier J, Housman D, Jaenisch R. WT-1 IS REQUIRED FOR EARLY KIDNEY DEVELOPMENT. Cell. 1993;74(4):679–691. doi: 10.1016/0092-8674(93)90515-r. [DOI] [PubMed] [Google Scholar]
- Ikadai H, Ajisawa C, Taya K, Imamichi T. Suprainguinal ectopic scrota of TS inbred rats. J Reprod Fertil. 1988;84(2):701–707. doi: 10.1530/jrf.0.0840701. [DOI] [PubMed] [Google Scholar]
- Li X, Nokkala E, Yan W, Streng T, Saarinen N, Warri A, Huhtaniemi I, Santti R, Makela S, Poutanen M. Altered structure and function of reproductive organs in transgenic male mice overexpressing human aromatase. Endocrinology. 2001;142(6):2435–2442. doi: 10.1210/endo.142.6.8211. [DOI] [PubMed] [Google Scholar]
- Caron P, Imbeaud S, Bennet A, Plantavid M, Camerino G, Rochiccioli P. Combined hypothalamic-pituitary-gonadal defect in a hypogonadic man with a novel mutation in the DAX-1 gene. J Clin Endocrinol Metab. 1999;84(10):3563–3569. doi: 10.1210/jcem.84.10.6030. [DOI] [PubMed] [Google Scholar]
- Donaldson KM, Tong SY, Washburn T, Lubahn DB, Eddy EM, Hutson JM, Korach KS. Morphometric study of the gubernaculum in male estrogen receptor mutant mice. J Androl. 1996;17(2):91–95. [PubMed] [Google Scholar]
- Gorlov IP, Kamat A, Bogatcheva NV, Jones E, Lamb DJ, Truong A, Bishop CE, McElreavey K, Agoulnik AI. Mutations of the GREAT gene cause cryptorchidism. Hum Mol Genet. 2002;11(19):2309–2318. doi: 10.1093/hmg/11.19.2309. [DOI] [PubMed] [Google Scholar]
- Canto P, Escudero I, Soderlund D, Nishimura E, Carranza-Lira S, Gutierrez J, Nava A, Mendez JP. A novel mutation of the insulin-like 3 gene in patients with cryptorchidism. J Hum Genet. 2003;48(2):86–90. doi: 10.1007/s100380300012. [DOI] [PubMed] [Google Scholar]
- Ferlin A, Garolla A, Bettella A, Bartoloni L, Vinanzi C, Roverato A, Foresta C. Androgen receptor gene CAG and GGC repeat lengths in cryptorchidism. Eur J Endocrinol. 2005;152(3):419–425. doi: 10.1530/eje.1.01860. [DOI] [PubMed] [Google Scholar]
- Yoshida R, Fukami M, Sasagawa I, Hasegawa T, Kamatani N, Ogata T. Association of cryptorchidism with a specific haplotype of the estrogen receptor alpha gene: implication for the susceptibility to estrogenic environmental endocrine disruptors. J Clin Endocrinol Metab. 2005;90(8):4716–4721. doi: 10.1210/jc.2005-0211. [DOI] [PubMed] [Google Scholar]
- Ferlin A, Bogatcheva NV, Gianesello L, Pepe A, Vinanzi C, Agoulnik AI, Foresta C. Insulin-like factor 3 gene mutations in testicular dysgenesis syndrome: clinical and functional characterization. Mol Hum Reprod. 2006;12(6):401–406. doi: 10.1093/molehr/gal043. [DOI] [PubMed] [Google Scholar]
- Silva-Ramos M, Oliveira JM, Cabeda JM, Reis A, Soares J, Pimenta A. The CAG repeat within the androgen receptor gene and its relationship to cryptorchidism. Int Braz J Urol. 2006;32(3):330–334. doi: 10.1590/s1677-55382006000300014. discussion 335. [DOI] [PubMed] [Google Scholar]
- Wada Y, Okada M, Fukami M, Sasagawa I, Ogata T. Association of cryptorchidism with Gly146Ala polymorphism in the gene for steroidogenic factor-1. Fertil Steril. 2006;85(3):787–790. doi: 10.1016/j.fertnstert.2005.09.016. [DOI] [PubMed] [Google Scholar]
- Bogatcheva NV, Ferlin A, Feng S, Truong A, Gianesello L, Foresta C, Agoulnik AI. T222P mutation of the insulin-like 3 hormone receptor LGR8 is associated with testicular maldescent and hinders receptor expression on the cell surface membrane. Am J Physiol Endocrinol Metab. 2007;292(1):E138–E144. doi: 10.1152/ajpendo.00228.2006. [DOI] [PubMed] [Google Scholar]
- El Houate B, Rouba H, Sibai H, Barakat A, Chafik A, Chadli el B, Imken L, Bogatcheva NV, Feng S, Agoulnik AI. Novel mutations involving the INSL3 gene associated with cryptorchidism. J Urol. 2007;177(5):1947–1951. doi: 10.1016/j.juro.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Yamazawa K, Wada Y, Sasagawa I, Aoki K, Ueoka K, Ogata T. Mutation and polymorphism analyses of INSL3 and LGR8/GREAT in 62 Japanese patients with cryptorchidism. Horm Res. 2007;67(2):73–76. doi: 10.1159/000096089. [DOI] [PubMed] [Google Scholar]
- Wang Y, Barthold J, Figueroa E, Gonzalez R, Noh PH, Wang M, Manson J. Analysis of five single nucleotide polymorphisms in the ESR1 gene in cryptorchidism. Birth Defects Res A Clin Mol Teratol. 2008;82(6):482–485. doi: 10.1002/bdra.20458. [DOI] [PubMed] [Google Scholar]
- Harris RM, Finlayson C, Weiss J, Fisher L, Hurley L, Barrett T, Emge D, Bathgate RA, Agoulnik AI, Jameson JL. A missense mutation in LRR8 of RXFP2 is associated with cryptorchidism. Mamm Genome. 2010;21(9–10):442–449. doi: 10.1007/s00335-010-9291-5. [DOI] [PubMed] [Google Scholar]
- Tang KF, Zheng JZ, Xing JP. Molecular analysis of SNP12 in estrogen receptor alpha gene in hypospadiac or cryptorchid patients from Northwestern China. Urol Int. 2011;87(3):359–362. doi: 10.1159/000330902. [DOI] [PubMed] [Google Scholar]
- Feng S, Ferlin A, Truong A, Bathgate R, Wade JD, Corbett S, Han S, Tannour-Louet M, Lamb DJ, Foresta C. INSL3/RXFP2 signaling in testicular descent. Ann N Y Acad Sci. 2009;1160:197–204. doi: 10.1111/j.1749-6632.2009.03841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgaard MD, Weinhold N, Edsgard D, Silver JD, Pers TH, Nielsen JE, Jorgensen N, Juul A, Gerds TA, Giwercman A. A genome-wide association study of men with symptoms of testicular dysgenesis syndrome and its network biology interpretation. J Med Genet. 2012;49(1):58–65. doi: 10.1136/jmedgenet-2011-100174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolon TF, Wiener JS, Lewitton M, Roth DR, Gonzales ET Jr, Lamb DJ. Analysis of homeobox gene HOXA10 mutations in cryptorchidism. J Urol. 1999;161(1):275–280. [PubMed] [Google Scholar]
- Teles MG, Trarbach EB, Noel SD, Guerra-Junior G, Jorge A, Beneduzzi D, Bianco SD, Mukherjee A, Baptista MT, Costa EM. A novel homozygous splice acceptor site mutation of KISS1R in two siblings with normosmic isolated hypogonadotropic hypogonadism. Eur J Endocrinol. 2010;163(1):29–34. doi: 10.1530/EJE-10-0012. [DOI] [PubMed] [Google Scholar]
- Williams GA, Ott TL, Michal JJ, Gaskins CT, Wright RW Jr, Daniels TF, Jiang Z. Development of a model for mapping cryptorchidism in sheep and initial evidence for association of INSL3 with the defect. Anim Genet. 2007;38(2):189–191. doi: 10.1111/j.1365-2052.2007.01585.x. [DOI] [PubMed] [Google Scholar]
- Cassata R, Iannuzzi A, Parma P, De Lorenzi L, Peretti V, Perucatti A, Iannuzzi L, Di Meo GP. Clinical, cytogenetic and molecular evaluation in a dog with bilateral cryptorchidism and hypospadias. Cytogenet Genome Res. 2008;120(1–2):140–143. doi: 10.1159/000118753. [DOI] [PubMed] [Google Scholar]
- Zhao X, Du ZQ, Rothschild MF. An association study of 20 candidate genes with cryptorchidism in Siberian Husky dogs. J Anim Breed Genet. 2010;127(4):327–331. doi: 10.1111/j.1439-0388.2010.00859.x. [DOI] [PubMed] [Google Scholar]
- Galan JJ, Guarducci E, Nuti F, Gonzalez A, Ruiz M, Ruiz A, Krausz C. Molecular analysis of estrogen receptor alpha gene AGATA haplotype and SNP12 in European populations: potential protective effect for cryptorchidism and lack of association with male infertility. Hum Reprod. 2007;22(2):444–449. doi: 10.1093/humrep/del391. [DOI] [PubMed] [Google Scholar]
- Pathirana IN, Tanaka K, Kawate N, Tsuji M, Kida K, Hatoya S, Inaba T, Tamada H. Analysis of single nucleotide polymorphisms in the 3′ region of the estrogen receptor 1 gene in normal and cryptorchid Miniature Dachshunds and Chihuahuas. J Reprod Dev. 2010;56(4):405–410. doi: 10.1262/jrd.09-195t. [DOI] [PubMed] [Google Scholar]
- Lo Giacco D, Ars E, Bassas L, Galan JJ, Rajmil O, Ruiz P, Caffaratti J, Guarducci E, Ruiz-Castane E, Krausz C. ESR1 promoter polymorphism is not associated with nonsyndromic cryptorchidism. Fertil Steril. 2011;95(1):369–371. doi: 10.1016/j.fertnstert.2010.08.051. 371 e361-362. [DOI] [PubMed] [Google Scholar]
- Krausz C, Quintana-Murci L, Fellous M, Siffroi JP, McElreavey K. Absence of mutations involving the INSL3 gene in human idiopathic cryptorchidism. Mol Hum Reprod. 2000;6(4):298–302. doi: 10.1093/molehr/6.4.298. [DOI] [PubMed] [Google Scholar]
- Takahashi I, Takahashi T, Komatsu M, Matsuda J, Takada G. Ala/Thr60 variant of the Leydig insulin-like hormone is not associated with cryptorchidism in the Japanese population. Pediatr Int. 2001;43(3):256–258. doi: 10.1046/j.1442-200x.2001.01390.x. [DOI] [PubMed] [Google Scholar]
- Baker LA, Nef S, Nguyen MT, Stapleton R, Nordenskjold A, Pohl H, Parada LF. The insulin-3 gene: lack of a genetic basis for human cryptorchidism. J Urol. 2002;167(6):2534–2537. [PubMed] [Google Scholar]
- Bertini V, Bertelloni S, Valetto A, Lala R, Foresta C, Simi P. Homeobox HOXA10 gene analysis in cryptorchidism. J Pediatr Endocrinol Metab. 2004;17(1):41–45. doi: 10.1515/jpem.2004.17.1.41. [DOI] [PubMed] [Google Scholar]
- Nuti F, Marinari E, Erdei E, El-Hamshari M, Echavarria MG, Ars E, Balercia G, Merksz M, Giachini C, Shaeer KZ. The leucine-rich repeat-containing G protein-coupled receptor 8 gene T222P mutation does not cause cryptorchidism. J Clin Endocrinol Metab. 2008;93(3):1072–1076. doi: 10.1210/jc.2007-1993. [DOI] [PubMed] [Google Scholar]
- Simoni M, Tuttelmann F, Michel C, Bockenfeld Y, Nieschlag E, Gromoll J. Polymorphisms of the luteinizing hormone/chorionic gonadotropin receptor gene: association with maldescended testes and male infertility. Pharmacogenet Genomics. 2008;18(3):193–200. doi: 10.1097/FPC.0b013e3282f4e98c. [DOI] [PubMed] [Google Scholar]
- Kunej T, Zorn B, Peterlin B. Y chromosome microdeletions in infertile men with cryptorchidism. Fertil Steril. 2003;79(Suppl 3):1559–1565. doi: 10.1016/s0015-0282(03)00375-3. [DOI] [PubMed] [Google Scholar]
- Bor P, Hindkjaer J, Kolvraa S, Rossen P, von der Maase H, Jorgensen TM, Sorensen VT, Eiberg H, Ingerslev HJ. Screening for Y microdeletions in men with testicular cancer and undescended testis. J Assist Reprod Genet. 2006;23(1):41–45. doi: 10.1007/s10815-005-9001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurbuz N, Ozbay B, Aras B, Tasci AI. Do microdeletions in the AZF region of the Y chromosome accompany cryptorchidism in Turkish children? Int Urol Nephrol. 2008;40(3):577–581. doi: 10.1007/s11255-007-9318-3. [DOI] [PubMed] [Google Scholar]
- Hejmej A, Gorazd M, Kosiniak-Kamysz K, Wiszniewska B, Sadowska J, Bilinska B. Expression of aromatase and oestrogen receptors in reproductive tissues of the stallion and a single cryptorchid visualised by means of immunohistochemistry. Domest Anim Endocrinol. 2005;29(3):534–547. doi: 10.1016/j.domaniend.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Nguyen MT, Delaney DP, Kolon TF. Gene expression alterations in cryptorchid males using spermatozoal microarray analysis. Fertil Steril. 2009;92(1):182–187. doi: 10.1016/j.fertnstert.2008.05.043. [DOI] [PubMed] [Google Scholar]
- Hutson JM, Watts LM, Farmer PJ. Congenital undescended testes in neonatal pigs and the effect of exogenous calcitonin gene-related peptide. J Urol. 1998;159(3):1025–1028. [PubMed] [Google Scholar]
- Zuccarello D, Morini E, Douzgou S, Ferlin A, Pizzuti A, Salpietro DC, Foresta C, Dallapiccola B. Preliminary data suggest that mutations in the CgRP pathway are not involved in human sporadic cryptorchidism. J Endocrinol Invest. 2004;27(8):760–764. doi: 10.1007/BF03347519. [DOI] [PubMed] [Google Scholar]
- Bertola DR, Pereira AC, Passetti F, de Oliveira PS, Messiaen L, Gelb BD, Kim CA, Krieger JE. Neurofibromatosis-Noonan syndrome: molecular evidence of the concurrence of both disorders in a patient. Am J Med Genet A. 2005;136(3):242–245. doi: 10.1002/ajmg.a.30813. [DOI] [PubMed] [Google Scholar]
- Digilio MC, Lepri F, Baban A, Dentici ML, Versacci P, Capolino R, Ferese R, De Luca A, Tartaglia M, Marino B. RASopathies: Clinical Diagnosis in the First Year of Life. Molecular syndromology. 2011;1(6):282–289. doi: 10.1159/000331266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AE, Araki T, Swanson KD, Montgomery KT, Schiripo TA, Joshi VA, Li L, Yassin Y, Tamburino AM, Neel BG. Germline gain-of-function mutations in SOS1 cause Noonan syndrome. Nat Genet. 2007;39(1):70–74. doi: 10.1038/ng1926. [DOI] [PubMed] [Google Scholar]
- Razzaque MA, Nishizawa T, Komoike Y, Yagi H, Furutani M, Amo R, Kamisago M, Momma K, Katayama H, Nakagawa M. Germline gain-of-function mutations in RAF1 cause Noonan syndrome. Nat Genet. 2007;39(8):1013–1017. doi: 10.1038/ng2078. [DOI] [PubMed] [Google Scholar]
- Cannistraci CV, Ravasi T, Montevecchi FM, Ideker T, Alessio M. Nonlinear dimension reduction and clustering by Minimum Curvilinearity unfold neuropathic pain and tissue embryological classes. Bioinformatics. 2010;26(18):i531–i539. doi: 10.1093/bioinformatics/btq376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navlakha S, Kingsford C. The power of protein interaction networks for associating genes with diseases. Bioinformatics (Oxford, England) 2010;26:1057–1063. doi: 10.1093/bioinformatics/btq076. England. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzhetsky A, Wajngurt D, Park N, Zheng T. Probing genetic overlap among complex human phenotypes. P Natl Acad Sci USA. 2007;104(28):11694–11699. doi: 10.1073/pnas.0704820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Jiang R, Zhang MQ, Li S. Network-based global inference of human disease genes. Mol Syst Biol. 2008;4:189. doi: 10.1038/msb.2008.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Liu Q, Jiang R. Align human interactome with phenome to identify causative genes and networks underlying disease families. Bioinformatics. 2009;25(1):98–104. doi: 10.1093/bioinformatics/btn593. [DOI] [PubMed] [Google Scholar]
- Loscalzo J, Barabasi AL. Systems biology and the future of medicine. Wiley Interdiscip Rev Syst Biol Med. 2011;3(6):619–627. doi: 10.1002/wsbm.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oti M, Brunner HG. The modular nature of genetic diseases. Clin Genet. 2007;71(1):1–11. doi: 10.1111/j.1399-0004.2006.00708.x. [DOI] [PubMed] [Google Scholar]
- Ogorevc J, Dovc P, Kunej T. Polymorphisms in microRNA targets: a source of new molecular markers for male reproduction. Asian J Androl. 2011;13(3):505–508. doi: 10.1038/aja.2011.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunej T, Skok DJ, Horvat S, Dovc P, Jiang Z. The glypican 3-hosted murine mir717 gene: sequence conservation, seed region polymorphisms and putative targets. Int J Biol Sci. 2010;6(7):769–772. doi: 10.7150/ijbs.6.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Chromosomal abnormalities and CNVs associated with cryptorchidism.
Selected clinical syndromes that feature CO in their clinical picture.
Transgenic and knock-out murine models that display cryptorchid phenotype.
Genes tested for association with CO.
Genes with expression patterns associated with CO. Genes with expression patterns associated with CO in rat (adapted from [7]) and genomic location of their human orthologs.
The literature collected candidate genes involved in multiple (four or more) CO-associated pathways.
Forty-three network-predicted CO-associated candidate genes.
Protein network information matrix for candidate genes. Protein network information matrix for candidate genes involved in at least two pathways significant for literature-collected and network-predicted candidate genes. The matrix consists of 50 rows (each row corresponds to a different gene involved in at least two pathways) and 12 columns (each column corresponds to a different pathway), where 0 indicates that the gene is not present in a pathway and 1 indicates that the gene is present in it.
STRING network data: list of protein network interactions present in STRING with high confidence score.