Early metagenetic surveys of micro-eukaryotic diversity revealed numerous phylotypes that seemed unrelated to any described eukaryotic taxa. It has been proposed that some of them may represent novel kingdom-level taxonomic diversity in eukaryotes. However, detailed analysis of DNA sequences retrieved from environmental samples and assigned to these putative new kingdoms showed that most of them were undetected chimeras or incorrectly placed fast-evolving phylotypes [1]. Since then, the number of new eukaryotic lineages revealed by environmental surveys grew exponentially, but the question of how many of them deserve the highest taxonomic level has remained.
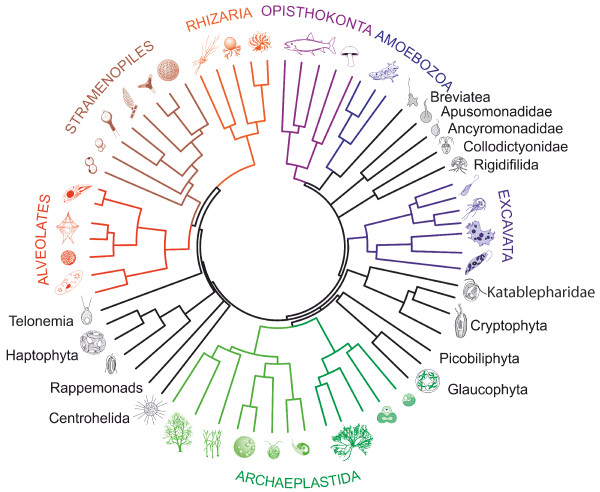
Over the past few years, reconstruction of deep eukaryote phylogeny focused on grouping various eukaryotic lineages into large monophyletic assemblages. Advances in phylogenomic studies led to the formation of four to seven supergroups (Figure 1). The classical multicellular kingdoms of animals and fungi are placed in the supergroup Opisthokonta, while green plants together with red algae form a supergroup of Archaeplastida. All other supergroups are composed of typically unicellular eukaryotes: Amoebozoa comprise the lobose amoebae and slime molds, Stramenopiles include among others diatoms, kelps, and oomycetes (previously classified as fungi), Alveolata are composed of ciliates, dinoflagellates and parasitic apicomplexans (among which the causative agents of malaria and toxoplasmosis), Rhizaria are dominated by amoeboid protists, such as foraminifera, radiolarians or filose amoebae, and heterotrophic flagellates with filose pseudopodia, and Excavata group together euglenozoans, heterolobosean amoebae and some amitochondriate parasitic phyla (such as diplomonads and parabasalids). However, with an increasing amount of phylogenetic and metagenetic data available for larger taxon sampling of eukaryote diversity, there was growing evidence that not all lineages could be placed inside the established supergroups. More than ten lineages were considered as of uncertain placement and left as incertae sedis in the recently revised eukaryote classification [2].
Figure 1.
Deep phylogeny of eukaryotes showing the position of small eukaryotic lineages that branch outside the seven supergroups (modified after Burki et al. [12]; drawings S Chraiti).
What are these hypothetical novel eukaryotic micro-kingdoms? First of all, many of them are not really new. For example, the genus Collodictyon was described 150 years ago but the diversity and evolutionary importance of Collodictyonidae have been investigated only recently [3]. The unclassified eukaryotic lineages represent a large variety of forms and modes of life, from free-swimming or gliding biflagellated cells to amoeboflagellates or filopodia-bearing amoeboid cells. Some lineages have cells covered with mineralized scales or spicules (Haptophyta, Centrohelida) or organic thecae (Apusomonadidae, Rigidifilida). Few lineages are phototrophs and possess chloroplasts acquired as a result of the secondary symbiosis (Cryptophyta, some Haptophyta). Most of them are mixotrophs or heterotrophs, often bacterivorous, living in marine or freshwater environments. Only one lineage (Breviatea) is amitochondriate and lives in anaerobic conditions, but interestingly no group comprises parasitic species. With the exception of Haptophytes and Cryptophytes, all other lineages are represented by very few described species, although their environmental diversity may be quite large. For example, the genus Telonema, represented by only two described morphospecies, has seen its diversity explode with more than 20 phylotypes revealed by environmental study [4].
Metagenetic studies played a key role in discovery and assessment of the diversity of these putative micro-kingdoms. Two lineages (Picobiliphytes, Rappemonads) are known exclusively from environmental sequence data [5,6]. Very little is known about their morphology and cell characteristics and in some cases even the features that seemed to be well established, like the presence of chloroplasts in Picobiliphytes, have been questioned based on a single-cell genomic study [7]. Compared to some other planktonic groups revealed by metagenetics, such as marine stramenopiles (MAST) or marine alveolates (MALV) [8], the environmental diversity of novel lineages seems relatively modest, though some of them have been found to be quite diverse when specifically targeted [9].
The phylogenetic position of these lineages remained an unresolved conundrum, in spite of genomic and/or transcriptomic data available for most of the groups that include cultivable species. A new supergroup, called Hacrobia or CCTH has been created for some of these orphan lineages [10,11], but later analyses did not support this hypothetical grouping [12]. An up-to-date phylogenomic tree (Figure 1) shows 13 independent lineages branching in three paraphyletic assemblages placed at the base of SAR assemblage grouping Stramenopiles, Alveolata and Rhizaria (Telonema, Haptophytes, Rappemonads, Centrohelids), next to Archaeplastida (Glaucophytes, Picobiliphytes, Cryptophytes, Katablepharids), and close to Amoebozoa (Breviatea, Collodictyon, Rigidifilida, Apusomonads, and Ancyromonads). Phylogenetic relations between these lineages and the adjacent supergroups are not well established. Some of them, like Breviatea and Glaucophyta are included in Amoebozoa and Archaeplastida, respectively. However, support for these groupings is usually weak and there is no general consensus about their classification.
One of the most striking characteristics of these orphan lineages and the reason why it is so difficult to place them in the eukaryotic tree is their extreme genetic divergence. High genetic distances separating them from other supergroups suggest that they represent the deepest eukaryotic lineages. Identifying and characterizing these deep lineages is essential for understanding the early evolution of eukaryotes. However, their number and diversity is difficult to assess. It has been assumed that many new deep lineages will be revealed by next-generation sequencing surveys of environmental diversity but their identification using short and standardized sequence tags [13] might not be straightforward. The single-cell genomic approach [7] may represent a much more efficient way to unveil the diversity of these unique lineages and establish their phylogenetic position.
Our view on the dawn of eukaryote evolution has profoundly changed as a result of phylogenetic and metagenetic studies. The traditional oversimplified classification of unicellular eukaryotes based on their mode of locomotion or capacity to photosynthesize has been replaced by a phylogenetically robust system of large, monophyletic eukaryotic supergroups that originated more than 1 billion years ago. Still, relatively little is known about the radiation of numerous small lineages that preceded the divergence of these supergroups. The challenge of future eukaryotic research is to describe the vastness of this radiation and to determine the ecological and evolutionary importance of the new micro-kingdoms.
Note
This article is part of the BMC Biology tenth anniversary series. Other articles in this series can be found at http://www.biomedcentral.com/bmcbiol/series/tenthanniversary.
References
- Berney C, Fahrni J, Pawlowski J. How many novel eukaryotic "kingdoms"? Pitfalls and limitations of environmental DNA surveys. BMC Biol. 2004;2:13. doi: 10.1186/1741-7007-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adl SM, Simpson AG, Lane CE, Lukeš J, Bass D, Bowser SS, Brown MW, Burki F, Dunthorn M, Hampl V, Heiss A, Hoppenrath M, Lara E, Le Gall L, Lynn DH, McManus H, Mitchell EA, Mozley-Stanridge SE, Parfrey LW, Pawlowski J, Rueckert S, Shadwick L, Schoch CL, Smirnov A, Spiegel FW. The revised classification of eukaryotes. J Eukaryot Microbiol. 2012;59:429–493. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Burki F, Bråte J, Keeling PJ, Klaveness D, Shalchian-Tabrizi K. Collodictyon--an ancient lineage in the tree of eukaryotes. Mol Biol Evol. 2012;29:1557–1568. doi: 10.1093/molbev/mss001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bråte J, Klaveness D, Rygh T, Jakobsen KS, Shalchian-Tabrizi K. Telonemia-specific environmental 18S rDNA PCR reveals unknown diversity and multiple marine-freshwater colonizations. BMC Microbiol. 2010;10:168. doi: 10.1186/1471-2180-10-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Not F, Valentin K, Romari K, Lovejoy C, Massana R, Töbe K, Vaulot D, Medlin LK. Picobiliphytes: a marine picoplanktonic algal group with unknown affinities to other eukaryotes. Science. 2007;315:253–255. doi: 10.1126/science.1136264. [DOI] [PubMed] [Google Scholar]
- Kim E, Harrison JW, Sudek S, Jones MD, Wilcox HM, Richards TA, Worden AZ, Archibald JM. Newly identified and diverse plastid-bearing branch on the eukaryotic tree of life. Proc Natl Acad Sci USA. 2011;108:1496–1500. doi: 10.1073/pnas.1013337108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HS, Price DC, Stepanauskas R, Rajah VD, Sieracki ME, Wilson WH, Yang EC, Duffy S, Bhattacharya D. Single-cell genomics reveals organismal interactions in uncultivated marine protists. Science. 2011;332:714–717. doi: 10.1126/science.1203163. [DOI] [PubMed] [Google Scholar]
- Logares R, Audic S, Santini S, Pernice MC, de Vargas C, Massana R. Diversity patterns and activity of uncultured marine heterotrophic flagellates unveiled with pyrosequencing. ISME J. 2012;6:1823–1833. doi: 10.1038/ismej.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Probert I, Uitz J, Claustre H, Aris-Brosou S, Frada M, Not F, de Vargas C. Extreme diversity in noncalcifying haptophytes explains a major pigment paradox in open oceans. Proc Natl Acad Sci USA. 2009;106:12803–12808. doi: 10.1073/pnas.0905841106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto N, Chantangsi C, Horák A, Leander BS, Keeling PJ. Molecular phylogeny and description of the novel katablepharid Roombia truncata gen. et sp. nov., and establishment of the Hacrobia taxon nov. PLoS One. 2009;4:e7080. doi: 10.1371/journal.pone.0007080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki F, Inagaki Y, Brate J, Archibald JM, Keeling PJ, Cavalier-Smith T, Horak A, Sakaguchi M, Hashimoto T, Klaveness D, Jakobsen KS, Pawlowski J, Shalchian-Tabrizi K. Early evolution of eukaryotes: two enigmatic heterotrophic groups are related to photosynthetic chromalveolates. Genome Biol Evol. 2009;1:231–238. doi: 10.1093/gbe/evp022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki F, Okamoto N, Pombert JF, Keeling PJ. The evolutionary history of haptophytes and cryptophytes: phylogenomic evidence for separate origins. Proc Biol Sci. 2012;279:2246–2254. doi: 10.1098/rspb.2011.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski J, Audic S, Adl S, Bass D, Belbahri L, Berney C, Bowser SS, Cepicka I, Decelle J, Dunthorn M, Fiore-Donno AM, Gile GH, Holzmann M, Jahn R, Jirků M, Keeling PJ, Kostka M, Kudryavtsev A, Lara E, Lukeš J, Mann DG, Mitchell EA, Nitsche F, Romeralo M, Saunders GW, Simpson AG, Smirnov AV, Spouge JL, Stern RF, Stoeck T. et al. CBOL Protist Working Group: Barcoding Eukaryotic Richness beyond the Animal, Plant, and Fungal Kingdoms. PLoS Biol. 2012;10:e1001419. doi: 10.1371/journal.pbio.1001419. [DOI] [PMC free article] [PubMed] [Google Scholar]