Abstract
The main objective of this study was to optimize a culture media for low scale biomass production of Pleurotus spp. Future applications of this optimization will be implemented for “in situ” rice straw degradation, increase soil nutrients availability, and lower residue and rice culture management costs. Soil samples were taken from different points in six important rice production cities in Colombia. For carbon and nitrogen source selection a factorial 42 design was carried out. The Plackett-Burman design permitted to detect carbon, nitrogen and inducer effects on fungus growth (response variable for all designs). This optimization was carried out by a Box-Behnken design. Finally a re-optimization assay for glucose concentration was performed by means of a One Factor design. Only 4/33 (12 %) isolates showed and important laccase or manganese peroxidase activity compared to Pleurotus ostreatus (HPB/P3). We obtained an increased biomass production in Pleurotus spp. (T1.1.) with glucose, followed by rice husk. Rice straw was considered an inducing agent for lignin degradation. Glucose was a significant component with positive effects, whereas Tween 80 and pH had negative effects. On the contrary, rice husk, yeast extract and CaCl2 were not significant components for increase the biomass production. Final media composition consisted of glucose 25 g L−1, yeast extract 5 g L−1, Tween 80 0.38 % (v/v), Rice husk 10 g L−1, CaCl2 1 g L−1, and pH 4.88 ± 0.2. The Box-Behnken polynomial prediction resulted to be lower than the experimental validation of the model (6.59 vs. 6.91 Log10 CFU ml−1 respectively).
Keywords: Ligninolytic fungi, Rice straw, Response surface methodology, Plackett-Burman, Box-Behnken, Factorial design
Introduction
Rice is considered a staple food for more than 50 % of the world’s population and is second in the world’s harvested area after maize. In addition, rice production generates abundant by-products like rice husk and rice straw. Rice straw, accounts for 30 % of the total production and is estimated that approximately 525 million tons are discarded annually. Rice residues impede seedbed preparation and contribute to disease and weed problems. Therefore, open-field burns have been implemented. Such is the case for Colombia, where waste burning generated a great environmental impact, thus it has been restricted through the Decree 4296 of 2004 [1].
Proper management for the use of rice straw is a major world-wide concern [2]. Rice straw is made up of approximately 19 lignin, 44 % cellulose, hemicellulose 20.1, and 9.8 % non-digestible silica. All these components account for its low nutritive value. Hence, currently there are few options for rice straw due to its poor quality for forage, bioconversion, and engineering applications [3]. Fungi have been described to compost lignocellulosic waste. In order to address the issues related to rice straw waste, this study was undertaken to design a culture media for biomass production of an autoctonal ligninolytic fungus isolated from a rice crop. We attempt to employ this technique in future “in situ” rice straw degradation, in order to increase soil nutrients’ availability, and reduce costs related to residue and rice culture management.
Materials and Methods
Sampling
Six cities in Colombia’s main areas of rice production were selected for sampling. These two main areas cover 70 % of the annual rice production: Villavicencio and Acacias (Meta), Lerida and Armero (North of Tolima) Purificación, and Saldaña (South of Tolima). Representative samples with respect to the field size were taken (12 samples per city, approximately 1 kg each). Samples were taken at different points on a zig-zag sampling pattern, with a distance of 15–25 m between each sampling point [4].
Isolation and Ligninolytic Activity Screening
Ten grams of soil sample were mixed with 90 ml 0.85 % NaCl (w/v) and diluted to a 10−2 solution. Each dilution was seeded in lignin-agar (0.5 g L−1 K2HPO4, 0.2 g L−1 MgSO4 7H2O, 0.1 g L−1 NH4NO3, 0.1 g L−1 KCl2, 0.02 g L−1 FeSO4 7H2O, 0.05 g L−1 CaNO3 4H20, 2 g L−1 malt extract, 15 g L−1 agar–agar, 0.2 g L−1 KOH, 1 g L−1 alkaline lignin, 10 ml L−1 dioxane, 60 mg L−1 chloramphenicol, 30 mg L−1 penicillin G), [5]. In addition 0.4 ml L−1 guaiacol was added [6]. Petri dishes were incubated at 28 °C for 7 days. Brown halo colonies were selected. All procedures were carried-out in triplicate.
Ligninolytic Enzyme Activity
Colonies were cultured in wheat bran media (2 % (w/v) glucose, 0.5 % (w/v) peptone, 0.2 % (w/v) yeast extract, 0.0075 % (w/v) MgSO4 H2O, 175 g L−1 wheat bran) supplemented with Tween 80 (0.05 % v/v), guaiacol (0.2 mM) and MnSO4 (2 mM) as inducers of enzyme production [7]. Cultures were carried out in 50 ml flasks with 20 ml media at 30 °C. Flasks were shaken for 8 days at 120 rpm. Mycelium was discarded by centrifugation (2,000×g for 10 m at 25 °C). Crude extract was used for Manganese Peroxidase (E.C. 1.11.1.13) and Lacasse (E.C. 1.10.3.2) activity.
For enzyme activity 800 μl of crude extract was supplemented with 100 μl of 20 mM 2,6-Dimethoxyphenol (DMP) and 100 μl buffer solution (sodium succinate, 25 mM, pH 4.5 ± 0.2 for laccase or sodium acetate, 100 mM pH 5 ± 0.2 for manganese peroxidase). Manganese peroxidase activity was performed in the presence of 100 mM manganese sulfate and 100 mM hydrogen peroxide. Activity was determined (enzyme units) by optical density (OD) at a wavelength of 468 nm (Spectronic 20 Genesys) before and after reaction time lapse (Eq. 1). An enzyme unit generated 1 mM of DMP oxidized per minute [8–10].
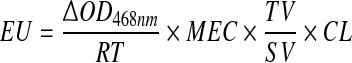 |
1 |
where ∆OD468nm is difference between optical densities measured before and after reaction. RT is reaction’s time lapse (3 min), MEC is the substrate’s molar extinction coefficient (49,600) [10]. TV is total reaction medium volume. SV is sample volume. CL is spectrophotometric cell length.
Culture Media and Optimization Conditions for Fungal Biomass Production
All tests were carried out in 250 ml flasks, with working volumes of 100 ml. All procedures were performed in triplicate.
Liquid Culture
Isolate was inoculated by puncturing the center of a PDA-agar Petri dish (Scharlau, Barcelona Spain), and incubated at 25 °C for 10 days. Saline medium (0.5 g L−1 KH2PO4, 0.2 g L−1 MgSO4 7H2O, 0.1 g L−1 NH4NO3, 0.1 g L−1 KCl, 0.02 g L−1 FeSO4 7H2O, 0.05 g L−1 CaNO3 4H2O, 0.06 g L−1 CuSO4) was used as base media for adding sources depending on the purpose of each experimental design. Inoculums were at a proportion of 3 agar discs (5 mm diameter × 4 mm depth) per 100 ml of culture media. Growing conditions were 25 °C, 150 rpm for 12 days. After incubation, the mycelia suspension was centrifuged (Sorvall) at 10,000×g during 15 min and washed twice with 100 ml saline solution (0.85 % (w/v) NaCl). The mycelium was resuspended in 0.85 % (w/v) NaCl solution.
Mycelia suspension was homogenized in a blade homogenizer for 3 min and diluted to a 10−4 solution. One hundred μl of homogenate was inoculated on PDA-agar supplemented with 0.5 % (w/v) chloramphenicol. Petri dishes were incubated at 25 °C for approximately 4–5 days to obtain a Log10 CFU ml−1 count.
Experimental Design
All design and result analysis were performed using Design Expert® V 8.05 2010 (Stat-Ease, Inc Minneapolis, MN USA).
Carbon and Nitrogen Source Selection
To determine the effect of the combination of 4 carbon and 4 nitrogen sources on fungus growth, a 42 factorial design was performed. Each variable consisted of four levels (20 g L−1 glucose, 20 g L−1 sucrose, 10 g L−1 rice straw and 10 g L−1 rice husk as the carbon source, and 10 g L−1 tryptone, 10 g L−1 yeast extract, 5 g L−1 NH2SO4 and 3 g L−1 NH4NO3 as the nitrogen source) for 16 assays. Sources and concentrations were selected based on optimal results reported in the literature [11, 12].
Effect of Carbon, Nitrogen and Inducers on Fungal Growth
To determine the effect of carbon, nitrogen, calcium, Tween 80 and pH changes on biomass production, a Plackett-Burman design [13], was performed in saline medium (Table 1).
Table 1.
Effect of carbon, nitrogen and inducers on fungal growth
| Variables | Units | Symbol code | Experimental values | |
|---|---|---|---|---|
| Lower (−1) | Higher (+1) | |||
| Glucose | g L−1 | A | 15 | 25 |
| Rice husk | g L−1 | B | 10 | 20 |
| Yeast extract | g L−1 | C | 5 | 15 |
| Tween 80 | % (v/v) | D | 0.05 | 0.15 |
| CaCl2 | g L−1 | E | 0.1 | 1.0 |
| pH | – | F | 4.5 ± 0.2 | 6.5 ± 0.2 |
Experimental variables at different levels used for the biomass production of ligninolytic fungus, using a Plackett-Burman design
Optimization of Carbon, Nitrogen and Selected Inducers
To optimize variables with the greatest effect on biomass production, obtained from the Plackett-Burman design, a Box-Behnken Design [14, 15] was used. Three variables were included in the design (Glucose, Tween 80 and pH). Each variable consisted of two levels (+1, −1). The base liquid media was the previously described saline solution in the liquid culture section.
Glucose Concentration Re-optimization Assay
To assay the effect of higher glucose concentration on Log10 CFU ml−1, a One Factor design was performed. For this experiment 5 levels were assayed (45, 59, 73, 86 and 100 g L−1) for the previously described saline solution. The other conditions were maintained as determined by the Box-Behnken design after 34 in-range optimization cycles.
Results and Discussions
Screening and Ligninolytic Activity
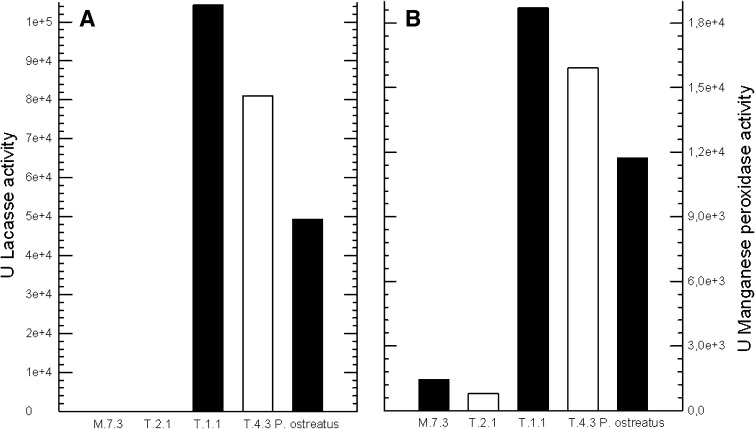
Thirty-three fungi with presumptive ligninolytic activity were isolated. In solid culture only 4/33 (12 %) displayed an important activity. Therefore, they were subsequently evaluated in liquid culture. Isolates were evaluated under the same conditions as the control Pleurotus ostreatus (HPB/P3). T.4.3 and T.1.1 isolates had a higher activity compared to control (Fig. 1). On the contrary, activity for strains M.7.3 and T.2.1 was very low.
Fig. 1.
Laccase (a) and manganese peroxidase (b) activity
Isolate T1.1 was selected for culture conditions optimization. It was molecularly identified as Pleurotus spp., following the methodology described by White et al. (data not shown) [16]. This result is not surprising, since most white-rot fungi degrade lignin via oxidation by secreting three enzymes: lignin peroxidase (EC 1.11.1.14), manganese peroxidase (MnP) (EC 1.11.1.13) and laccase (EC 1.10.3.2) [17]. Most species of Pleurotus produce manganese peroxidase and laccase, with high expression levels, in particular for laccase [11, 18].
Selection of Carbon and Nitrogen Sources
Nitrogen and carbon are both important for fungi growth. Nitrogen is essential for amino acid and nucleic acid synthesis. Carbon has an even greater importance, since it is in higher proportion compared to other elements. It is necessary for energy production and synthesis of various cell-wall lipids [19].
The Factorial Design indicated that the applied model was significant (Prob > F = 0.0095). This result suggests that at least one variable affects biomass production with a 95 % confidence interval (Table 2).
Table 2.
Carbon and nitrogen sources selection
| Source | Sum of squares | df | Mean square | F value | Prob > F |
|---|---|---|---|---|---|
| Factorial model | 144.02 | 15 | 9.60 | 3.44 | 0.0095* |
| A: Carbon source | 15.84 | 3 | 5.28 | 1.89 | 0.1712 |
| B: Nitrogen source | 95.20 | 3 | 31.73 | 11.39 | 0.0003* |
| AB | 32.98 | 9 | 3.66 | 1.31 | 0.3031 |
| Pure error | 44.60 | 16 | 2.79 | ||
| Cor. total | 188.62 | 31 |
ANOVA results for selection of carbon and nitrogen sources in a factorial design. Results are presented in Log10 CFU ml−1
R2 = 0.7636, Adj. R2 = 0.5419, Adeq. precision = 5, 540
*Significant at 95 %
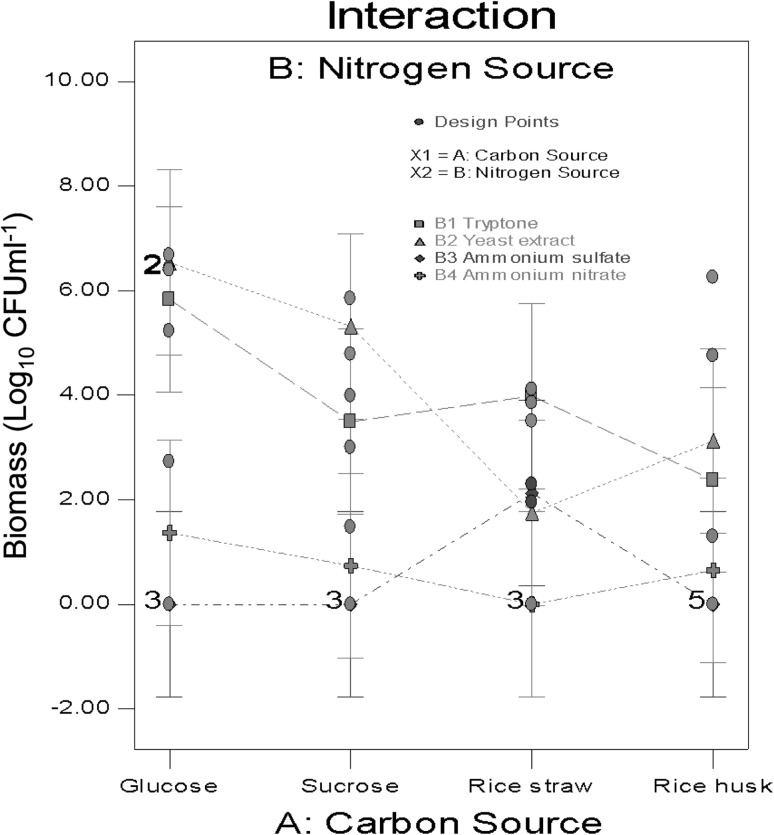
An increased biomass production was obtained using glucose (Fig. 2), followed by sucrose as substrates. Glucose and fructose dimer is easier to hydrolyse compared to rice waste (cellulose, hemicellulose 57–69 %, lignin 4.07 %, and silica 10–18 %). To degrade these substrates ligninolytic and cellulolytic enzymes are required [20, 21]. Although we worked with a ligninolytic white-rot fungus capable of degrading complex substrates, we accelerated fungal growth by using more degradable carbohydrates.
Fig. 2.
Effects of carbon a nitrogen sources on ligninolytic fungi, Pleurotus spp. (T1.1), cultured for 12 days. aCarbon sources glucose (20 g L−1), sucrose (20 g L−1), rice straw (10 g L−1) and rice husk (10 g L−1). bNitrogen sources tryptone (10 g L−1), yeast extract (10 g L−1), NH2SO4 (5 g L−1) and NH2NO3 (5 g L−1)
Rice husk culture medium CFU ml−1 results were comparable to those obtained from glucose with yeast extract as the organic nitrogen source (Fig. 2). Henceforth, rice husk was considered the second carbon source for biomass production. In addition, the fungi’s inducing capacity for lignin degradation was assessed during culture and prior to growth with rice straw. To evaluate the effect of Pleurotus spp. (T1.1) on biomass production a Plackett-Burman design was carried-out, yeast extract was selected as the organic nitrogen source. A combination of glucose/rice husk was employed as a carbon source to accelerate cell growth and stimulate ligninolytic capacity [20].
Carbon, Nitrogen, and Inducer Effect on Fungal Growth
Plackett-Burman design results (Table 3) determined that component combination used in experiment number 8 (in bold) generated the highest biomass concentration (Log10 CFU ml−1). The model was significant at 95 % confidence interval (Table 4) and demonstrated through F that there was only a 4.69 % chance that a “Model F-Value” this large could occur due to noise. Furthermore, results from the Plackett-Burman design established an Adequate Precision measure (signal to noise ratio) greater than 4. Our ratio of 6.694 was an adequate indicator; confirming that the design space could be navigated. The model revealed glucose, Tween 80, and pH were significant components (95 %) for T1.1. growth (Table 4). In addition, these results pointed out which component had a positive effect (glucose) or a negative effect (Tween 80 and pH) on growth.
Table 3.
Carbon, nitrogen, and inducer effect on fungal growth
| Run order | Factors combinations | Response | |||||
|---|---|---|---|---|---|---|---|
| A: Glucose | B: Rice husk | C: Yeast extract | D: Tween 80 | E: CaCl2 | F: pH ± 0.2 | Log10 CFU ml−1 | |
| 1 | 25 (+1) | 20 (+1) | 15 (+1) | 0.5 (−1) | 1 (+1) | 6.5 (+1) | 5.7655 |
| 2 | 15 (−1) | 10 (−1) | 5 (−1) | 1.5 (+1) | 1 (+1) | 6.5 (+1) | 0.6505 |
| 3 | 25 (+1) | 10 (−1) | 15 (+1) | 0.5 (−1) | 0.1 (−1) | 4.5 (−1) | 6.0636 |
| 4 | 15 (−1) | 10 (−1) | 15 (+1) | 1.5 (+1) | 1 (+1) | 4.5 (−1) | NG |
| 5 | 15 (−1) | 10 (−1) | 5 (−1) | 0.5 (−1) | 0.1 (−1) | 4.5 (−1) | 5.1818 |
| 6 | 15 (−1) | 20 (+1)) | 5 (−1) | 0.5 (−1) | 0.1 (−1) | 6.5 (+1) | NG |
| 7 | 15 (−1) | 20 (+1) | 15 (+1) | 1.5 (+1) | 0.1 (−1) | 6.5 (+1) | NG |
| 8 | 25 (+1) | 10 (−1) | 5 (−1) | 0.5 (−1) | 1 (+1) | 6.5 (+1) | 7.0792 |
| 9 | 15 (−1) | 20 (+1) | 15 (+1) | 0.5 (−1) | 1 (+1) | 4.5 (−1) | 5.5640 |
| 10 | 25 (+1) | 20 (+1) | 5 (−1) | 1.5 (+1) | 1 (+1) | 4.5 (−1) | 6.3580 |
| 11 | 25 (+1) | 10 (−1) | 15 (+1) | 1.5 (+1) | 0.1 (−1) | 6.5 (+1) | NG |
| 12 | 25 (+1) | 20 (+1) | 5 (−1) | 1.5 (+1) | 0.1 (−1) | 4.5 (−1) | 5.6531 |
NG no growth was observed; for modeling results effect of these values were feeding as 0.000. In bold the highest results
Table 4.
Carbon, nitrogen, and inducer effect on fungal growth
| Source | Coefficient | Standard error | Sum of squares | df | Mean square | F-value | Prob > F |
|---|---|---|---|---|---|---|---|
| Factorial model | 87.76 | 6 | 14.63 | 5.31 | 0.0435* | ||
| A: Glucose | 1.63 | 0.48 | 31.76 | 1 | 31.76 | 11.54 | 0.0193*a |
| B: Rice husk | 0.36 | 0.48 | 1.59 | 1 | 1.59 | 0.58 | 0.4817a |
| C: Yeast extract | −0.63 | 0.48 | 4.72 | 1 | 4.72 | 1.72 | 0.2471b |
| D: Tween 80 | −1.42 | 0.48 | 24.06 | 1 | 24.06 | 8.74 | 0.0316*b |
| E: CaCl2 | 0.71 | 0.48 | 6.05 | 1 | 6.05 | 2.20 | 0.1984a |
| F: pH | −1.28 | 0.48 | 19.57 | 1 | 19.57 | 7.11 | 0.0445*b |
| Residual | 13.76 | 5 | 2.75 | ||||
| Cor. Total | 101.52 | 11 |
Plackett-Burman. Estimated coefficient, standard error and corresponding F and P values for biomass production (Log10 CFU ml−1) of Pleurotus spp. (T1.1) for six experimented variables
R2 = 0.8644, Adj. R2 = 0.7018, Adeq. Precision = 6.694
*Significant at 95 %
aPositive effect
bNegative effect
The results obtained in the Plackett-Burman design were consistent. Tween 80 is an anionic surfactant with amphipathic properties that coat the lignocellulosic substrate, increasing the adhesion of ligninolytic enzymes. It also reduces surface tension and increases solubility and mobility, allowing biodegradation of insoluble organic compounds [22]. Nonetheless, Brar et al. [23] have reported that excess addition of Tween 80 produces a toxic reaction to the microorganism and reduces biomass production. Likewise, pH plays a decisive role in the degradation of different lignocellulosic materials. According to Liu et al. [12] the maximum laccase activity for some fungi can be achieved at pH 4 ± 0.2. When the pH is less than 2.5 ± 0.2 or greater than 5.0 ± 0.2 laccase enzyme can only reach 40 % of its activity. Considering the results obtained by using the Plackett-Burman design, we proceeded to optimize concentration levels through a Box-Behnken design.
Carbon, Nitrogen and Selected Inducer Optimization
By using the Box-Behnken design it was determined that rice husk, yeast extract, and CaCl2 had no significant effect on biomass production. Consequently, we maintained the concentrations of these components as in experiment number 8 (Plackett-Burman Design, Table 3), only glucose, Tween 80 and pH levels were adjusted (Table 5).
Table 5.
Carbon, nitrogen and selected inducer optimization
| Run order | Point type | Experimental values | Log10 CFU ml−1 | |||
|---|---|---|---|---|---|---|
| Glucose (g L−1) | Tween 80 (ml L−1) | pH ± 0.2 | Observed | Predicted | ||
| 1 | Fact | 25 (−1) | 0.25 (0) | 4.00 (−1) | 5.99 | 6.57 |
| 2 | Fact | 35 (0) | 0.00 (−1) | 6.00 (+1) | 0.00 | 1.18 |
| 3 | Fact | 35 (0) | 0.00 (−1) | 6.00 (+1) | 6.03 | 5.94 |
| 4 | Center | 35 (0) | 0.25 (0) | 5.00 (0) | 6.66 | 6.12 |
| 5 | Fact | 45 (+1) | 0.50 (+1) | 5.00 (0) | 5.67 | 6.17 |
| 6 | Fact | 45 (+1) | 0.00 (−1) | 5.00 (0) | 5.60 | 5.00 |
| 7 | Fact | 45 (+1) | 0.25 (0) | 4.00 (−1) | 6.17 | 6.85 |
| 8 | Fact | 35 (0) | 0.50 (+1) | 4.00 (−1) | 5.98 | 4.80 |
| 9 | Fact | 25 (−1) | 0.25 (0) | 6.00 (+1) | 6.03 | 5.94 |
| 10 | Fact | 45 (+1) | 0.25 (0) | 6.00 (+1) | 5.90 | 5.31 |
| 11 | Fact | 35 (0) | 0.50 (+1) | 6.00 (+1) | 6.03 | 6.12 |
| 12 | Fact | 25 (−1) | 0.50 (+1) | 5.00 (0) | 5.84 | 6.44 |
| 13 | Center | 35 (0) | 0.25 (0) | 5.00 (0) | 5.82 | 6.12 |
| 14 | Fact | 25 (−1) | 0.00 (−1) | 5.00 (0) | 4.31 | 3.81 |
| 15 | Center | 35 (0) | 0.25 (0) | 5.00 (0) | 5.88 | 6.12 |
Box-Behnken. Experiments and results obtained from the response variable (Log10 CFU ml−1)
aThe observed values of Log10 CFU ml−1 were the mean values of triplicates. Highest values (results) in bold
Glucose levels utilized for the Box-Behnken (Table 5) were determined after evaluating the results obtained in the Plackett-Burman design (Table 4). Accordingly, level +1 in the Plackett-Burman design was set to −1 level in the Box-Behnken design (Table 5). Levels 0 and +1 of the Box-Behnken design were established by increasing the concentration in intervals of 10 g L−1. Since Tween 80 had a negative effect determined by the Plackett-Burman design (Table 4), the −1 value in the Plackett-Burman design (Table 3) was set to +1 in the Box-Behnken design. Thus, levels 0 and −1 in the Box-Behnken design were established in decreases of 0.25 ml L−1 (Table 5). The pH factor levels tested in the Box-Behnken design (Table 5) were selected decreasing the levels tested in the Plackett-Burman by 0.5 ± 0.2 (Table 3), since pH had a negative effect on growth (Table 4).
After 5 days of culture the Box-Behnken, reported a significant quadratic model (85 %). Predicted values compared to model results are shown in Table 5. The quadratic model explained the mathematical relationship between the independent variable and the dependent response by Eq. 2.
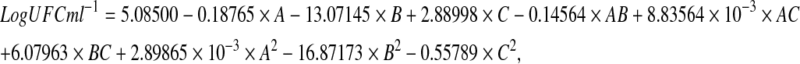 |
2 |
where A is glucose concentration, B is concentration of Tween 80, C is pH. ANOVA results (Table 6) indicated the predictability of the model at 85 % confidence interval and a determination coefficient (R2) of 0.8269. These results display a reliable equation, confirming that the model is suitable for predicting within the range of the chosen variables (Fig. 3I–III). After 34 in-range factors optimization cycles navigating through the design surface, the following combination was selected: glucose (43 g L−1), Tween 80 (0.19 % v/v), and pH (4.02 ± 0.2), (Fig. 3IV).
Table 6.
Box-Behnken design analysis of variance (ANOVA) for response surface regression model
| Source | Sum of squares | df | Mean square | F value | Prob > F |
|---|---|---|---|---|---|
| Quadratic model | 28.90 | 9 | 3.21 | 2.65 | 0.1475*** |
| A: Glucose | 0.42 | 1 | 0.42 | 0.35 | 0.5795 |
| B: Tween 80 | 7.20 | 1 | 7.20 | 5.95 | 0.0588* |
| C: pH | 5.91 | 1 | 5.91 | 4.89 | 0.0780** |
| AB | 0.53 | 1 | 0.53 | 0.44 | 0.5372 |
| AC | 0.031 | 1 | 0.031 | 0.026 | 0.8787 |
| BC | 9.24 | 1 | 9.24 | 7.64 | 0.0397* |
| A2 | 0.31 | 1 | 0.31 | 0.26 | 0.6341 |
| B2 | 4.11 | 1 | 4.11 | 3.39 | 0.1248 |
| C2 | 1.15 | 1 | 1.15 | 0.95 | 0.3745 |
| Pure error | 0.43 | 2 | 0.22 | ||
| Cor. total | 34.95 | 14 |
R2 = 0.8269, Adj. R2 = 0.5153, Adeq. Precision = 6.318
*Significant at 95 %
**Significant at 90 %
***Significant at 85 %
Fig. 3.
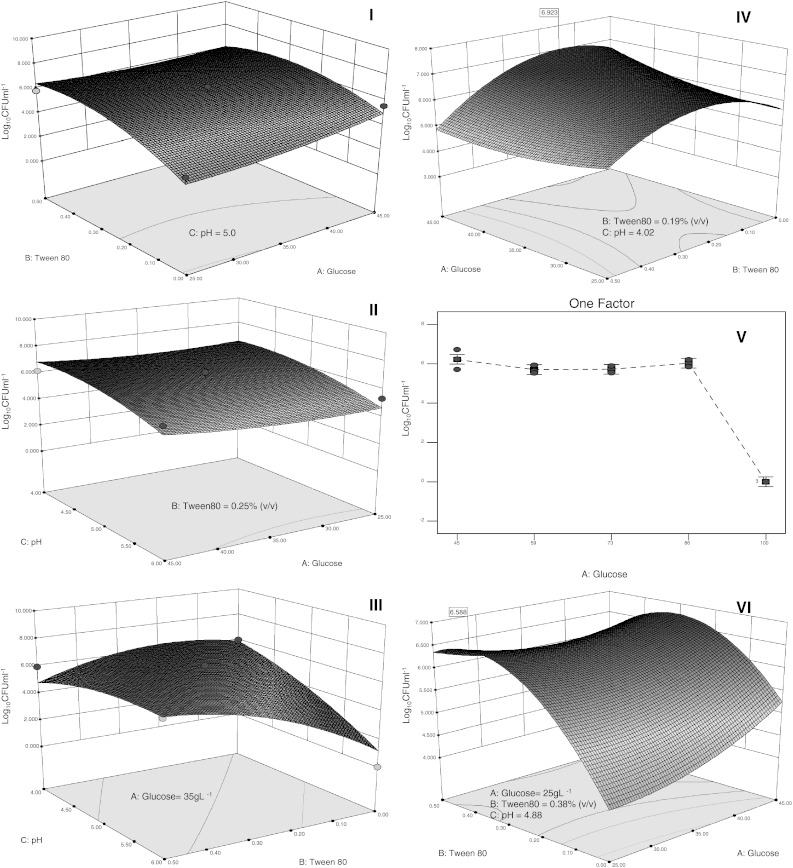
Response surface methodology (RSM), (I–VI) and Factorial Design (V). Box-Behnken showing the effects of: I Glucose-Tween 80; II Glucose-pH and III pH-Tween 80 on Log10 CFUml−1. IV In range optimization of glucose, Tween 80 and pH. V One Factor Design effect of Glucose on Log10 CFU ml−1 maintaining the range optimization of Tween 80 and pH. VI Box-Behnken optimization minimizing the glucose concentration and maintaining Tween 80 and pH within the established range
As evidenced by the Plackett-Burman- and the Box-Behnken-design glucose concentration could be further increased. We inferred that an increase in glucose concentration would result in an augmented biomass production. On account of these results a One Factor Design was performed to determine the highest glucose concentration needed to increase biomass production.
Glucose Concentration Re-optimization Assay
The One Factor Design revealed that glucose concentrations between 45 and 86 g L−1 had no increasing effect on Log10 CFU ml−1. When 100 gL−1 was used, a substrate repression effect was evident (Fig. 3V). These results demonstrated that glucose concentration was not a feasible strategy. Thence, considering glucose’s cost, we ran again 34 optimization cycles using the previous Box-Behnken results with a modification; that consisted in lowering glucose concentration.
Based on highest desirability (figure not shown) and highest biomass production, (Log10 CFU ml−1), (Fig 3VI) the predicted conditions to produce Pleurotus spp. (T1.1) were as follow: glucose (25 g L−1), yeast extract (5 g L−1), Tween 80 (0.38 % v/v), rice husk (10 g L−1), CaCl2 (1 g L−1) and pH (4.88 ± 0.2). A predictive value of 6.588 Log10 CFU ml−1 was obtained when the levels of each factor in the polynomial resulting from the Box-Behnken design was replaced. Once the final design was validated at the laboratory, the value attained was 6.91 Log10 CFU ml−1, which exceeded the prediction.
Conclusions
Pleurotus spp. (T1.1) was isolated from a rice crop, and a feasible culture media was optimized for biomass production. Enzyme activities for manganese peroxidase and lacasse were higher compared with the results obtained with Pleurotus ostreatus (HPB/P3). The Box-Behnken polynomial prediction value compared to the experimental validation model was lower. Finally, biological assays using Pleurotus spp. T1.1 have been performed to degrade rice straw under the conditions determined in this work. We have obtained very encouraging preliminary results (unpublished data).
Acknowledgments
This project was funded by the “Ministerio de Agricultura y Desarrollo Rural” (2007B6423.161) with the approval of the “Vicerrectoría Académica” (001518) at the Pontificia Universidad Javeriana, Bogotá, D.C. Colombia. The authors thank María Lucía Gutiérrez for helpful contribution with English editing of the manuscript.
References
- 1.Ministerio de Ambiente Vivienda y Desarrollo Territorial (2004) Decreto 4296. In, p http://www.presidencia.gov.co/prensa_new/decretoslinea/2004/diciembre/2020/dec4296201204.pdf. Accessed 20 Aug 2011
- 2.Kérouanton A, Marault M, Petit L, Grout J, Dao TT, et al. Evaluation of a multiplex PCR assay as an alternative method for Listeria monocytogenes serotyping. J Microbiol Methods. 2010;80:134–137. doi: 10.1016/j.mimet.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 3.Devevre OC, Horwáth WR. Decomposition of rice straw and microbial carbon use efficiency under different soil temperatures and moistures. Soil Biol Biochem. 2000;32:1773–1785. doi: 10.1016/S0038-0717(00)00096-1. [DOI] [Google Scholar]
- 4.Alef K, Nannipieri P, editors. Nutrients, sterilization, aerobic and anaerobic culture techniques. San Diego: Academic Press; 1995. [Google Scholar]
- 5.Thorn R, Adinarayana R, Harris D, Paul A. Isolation of saprophytic basidiomycetes from soil. Appl Environ Microbiol. 1996;62:4288–4292. doi: 10.1128/aem.62.11.4288-4292.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiiskinen LL, Ratto M, Kruus K. Screening for novel laccase-producing microbes. J Appl Microbiol. 2004;97:640–646. doi: 10.1111/j.1365-2672.2004.02348.x. [DOI] [PubMed] [Google Scholar]
- 7.Martínez-Salgado M, Pedroza-Rodríguez A, Rodríguez-Vásquez R, Rosas-Acosta J. Efecto de la glucosa y nitrato de amonio sobre las enzimas ligninolíticas producidas por Trametes versicolor inmovilizado en espuma y la decoloración de un efluente papelero en un biorreactor de lecho fluidizado. Univ Scient. 2005;10:27–36. [Google Scholar]
- 8.Schmutzer M, Schwanninger M, Fackler K, Messner K, Gradinger C. Comparison of methods to evaluate the potential of fungal growth on decay of spruce wood after short-time treatment. Int Biodeterior Biodegrad. 2007;61:319–324. doi: 10.1016/j.ibiod.2007.09.005. [DOI] [Google Scholar]
- 9.Rothschild N, Novotný C, Šašek V, Dosoretz CG. Ligninolytic enzymes of the fungus Irpex lacteus (Polyporus tulipiferae): isolation and characterization of lignin peroxidase. Enzyme Microb Technol. 2002;31:627–633. doi: 10.1016/S0141-0229(02)00171-0. [DOI] [Google Scholar]
- 10.Šnajdr J, Valášková V, Merhautová V, Cajthaml T, Baldrian P. Activity and spatial distribution of lignocellulose-degrading enzymes during forest soil colonization by saprotrophic basidiomycetes. Enzyme Microb Technol. 2008;43:186–192. doi: 10.1016/j.enzmictec.2007.11.008. [DOI] [Google Scholar]
- 11.Bettin F, Montanari Q, Calloni R, Gaio TA, Silveira MM, et al. Production of laccases in submerged process by Pleurotus sajor-caju PS-2001 in relation to carbon and organic nitrogen sources, antifoams and Tween 80. J Ind Microbiol Biotechnol. 2009;36:1–9. doi: 10.1007/s10295-008-0463-1. [DOI] [PubMed] [Google Scholar]
- 12.Liu L, Lin Z, Zheng T, Lin L, Zheng C, et al. Fermentation optimization and characterization of the laccase from Pleurotus ostreatus strain 10969. Enzyme Microb Technol. 2009;44:426–433. doi: 10.1016/j.enzmictec.2009.02.008. [DOI] [Google Scholar]
- 13.Chauhan K, Trivedi U, Patel KC. Statistical screening of medium components by Plackett-Burman design for lactic acid production by Lactobacillus sp. KCP01. Bioresour Technol. 2007;98:98–103. doi: 10.1016/j.biortech.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 14.Martendal E, Budziak D, Carasek E. Application of fractional factorial experimental and Box-Behnken designs for optimization of single-drop microextraction of 2,4,6-trichloroanisole and 2,4,6-tribromoanisole from wine samples. J Chromatograph A. 2007;1148:131–136. doi: 10.1016/j.chroma.2007.02.079. [DOI] [PubMed] [Google Scholar]
- 15.Duque-Jamaica R, Arévalo-Galvis A, Poutou-Piñales RA, Trespalacios-Rangel AA. Sequential statistical improvement of the liquid cultivation of Helicobacter pylori. Helicobacter. 2010;15:303–312. doi: 10.1111/j.1523-5378.2010.00763.x. [DOI] [PubMed] [Google Scholar]
- 16.White TJ, Bruns T, Lee S, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 17.Wong DWS. Structure and action mechanism of ligninolytic enzymes. Appl Biochem Biotechnol. 2009;157:174–209. doi: 10.1007/s12010-008-8279-z. [DOI] [PubMed] [Google Scholar]
- 18.Tinoco R, Acevedo A, Galindo E, Serrano-Carreón L. Increasing Pleurotus ostreatus laccase production by culture medium optimization and copper/lignin synergistic induction. J Ind Microbiol Biotechnol. 2011;38:531–540. doi: 10.1007/s10295-010-0797-3. [DOI] [PubMed] [Google Scholar]
- 19.Palmieri G, Giardina P, Bianco C, Fontanella B, Sannia G. Copper induction of laccase isoenzymes in the ligninolytic fungus Pleurotus ostreatus. Appl Environ Microbiol. 2000;66:920–924. doi: 10.1128/AEM.66.3.920-924.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh Arora D, Kumar Sharma R. Ligninolytic fungal laccases and their biotechnological applications. Appl Biochem Biotechnol. 2010;160:1760–1788. doi: 10.1007/s12010-009-8676-y. [DOI] [PubMed] [Google Scholar]
- 21.Membrillo I, Sánchez C, Meneses M, Favela E, Loera O. Effect of substrate particle size and additional nitrogen source on production of lignocellulolytic enzymes by Pleurotus ostreatus strains. Bioresour Technol. 2008;99:7842–7847. doi: 10.1016/j.biortech.2008.01.083. [DOI] [PubMed] [Google Scholar]
- 22.Zhou J, Jiang W, Ding J, Zhang X, Gao S. Effect of Tween 80 and b-cyclodextrin on degradation of decabromodiphenyl ether (BDE-209) by White Rot Fungi. Chemosphere. 2007;70:172–177. doi: 10.1016/j.chemosphere.2007.06.036. [DOI] [PubMed] [Google Scholar]
- 23.Brar SK, Verma M, Barnabé S, Tyagi RD, Valéro JR, et al. Impact of Tween 80 during Bacillus thuringiensis fermentation of wastewater sludges. Proc Biochem. 2005;40:2695–2705. doi: 10.1016/j.procbio.2004.12.003. [DOI] [Google Scholar]