Abstract
A gram positive, extreme haloalkaliphilic, radioresistant bacterium was isolated from mangrove region of Kerala (India) which was characterized as Exiguobacterium sp. HKG-126 using morphological, physiological, biochemical and molecular characterization. Present investigation was undertaken to examine Exiguobacterium sp. as a potential source of broad-spectrum antimicrobial activity and enhancement in this activity was observed due to cross-species/cross-genera induction and also in response to high dose of gamma (γ) irradiation. Individual studies on the antimicrobial activity of all the co-cultivated bacterial strains before and after mixed culture fermentation, showed excellent enhancement in antimicrobial activity of Exiguobacterium sp. against a variety of clinical pathogens. To the best of our knowledge, this is the first report showing existence of an extremely high radioresistant strain of (up to 15 kGy) Exiguobacterium sp.
Keywords: Exiguobacterium sp., Gamma irradiation, Cross-species induction, Extraction of secondary metabolites, Antimicrobial activity
Introduction
The production of antimicrobial compounds by microbial system is usually assayed under straight forward growth conditions that are very different from their natural environments. The primary role of any antimicrobial compound is to antagonize competitors [1–5]. Microbial interaction during mixed culture involves the production of some communicating chemicals known as quorum sensing signal molecules. When these molecules from one species or genus interact with those of another, they may stimulate or inhibit a number of physiological and biochemical activities including production of secondary metabolites like antimicrobial compounds. Such type of interaction is referred to as cross-species or cross-genera induction or inhibition [6, 7]. Thus, new cultivation strategies, particularly strategies mimicking the natural habitats of these microorganisms, can be used to enhance the production of secondary metabolites. We can greatly influence the productivity of such compounds by maximizing the types of samples collected, diversifying the isolation strategies, optimizing nutrients, temperature, pH, aeration, incubation time, exploiting talented strains by co-cultivation, and genetically modifying the strains by radiation exposure [5, 8–10]. Since these bacteria are able to flourish in extreme conditions, the bioactive secondary metabolites obtained from such microorganisms, have great structural and functional diversity, including antimicrobial compounds against clinical pathogens [11]. In the present study, we showed the enhancement of antimicrobial properties in response to high dose of gamma (γ) irradiation and cross-species induction of antimicrobial activity in presence of some pathogenic micro-organisms during mixed culture fermentation.
Materials and Methods
Sample Collection, Culture Conditions and Bacterial Identification
The bacterial strain was isolated from the Mangrove region of Kannamali, Kerala, India. Morphological, physiological and biochemical characterization of the isolated strains were performed as per Bergey’s manual of determinative bacteriology [12] and finally bacterial identification was done by using 16S rRNA sequence analysis as described earlier [13]. The temperature range, pH range and effect of salinity at 37 °C was determined at a temperature range of 10–50 °C, pH range between pH 5.0 and 12 and NaCl ranging from 0 to 14 %, respectively.
Gamma Irradiation/Extraction of Secondary Metabolites from Exiguobacterium sp
The radioresistant property of the Exiguobacterium sp. HKG-126 was evaluated by exposing them to gamma (γ) irradiation various dose levels of 5–15 kGy at 22 °C as described in previous studies [13, 14]. After molecular and biochemical characterization of the mutant population as compared to non-irradiated, shake flask fermentation of both, control strain (before irradiation) as well as gamma irradiated bacterial strains was done to retrieve bioactive molecule for evaluation of antimicrobial properties. The extraction of secondary metabolites was done by using Diaion HP-20 poly aromatic adsorbent resin from culture supernatant as described previously [15] followed by antimicrobial activity test.
Determination of Antimicrobial Activity and Minimum Inhibitory Concentration (MIC)
Antimicrobial activities of all strains were performed according to the method described previously [15, 16] in which gentamicin disc (10 μg/disc) and methanol were used as positive and negative controls, respectively. Minimum inhibitory concentration (MIC) of both extracts (0 and 15 kGy) on all sensitive test microorganisms was done by the twofold serial dilution method in 96-well microtiter plate and incubated at 37 °C overnight. Test organism without methanolic crude extract was used as negative control. As an indicator of bacterial growth, 50 μl of 0.2 mg/ml iodonitrotetrazolium chloride (INT) (Sigma–Aldrich) was added to the wells and incubated at 37 °C for 30 min. The lowest concentrations, which did not show any growth of tested microorganism was determined as MIC. All the assays were carried out in triplicates.
HPLC Analysis
The final analysis of bioactive compounds from 0 to 15 kGy gamma irradiated Exiguobacterium sp. HKG-126, was carried out using high performance liquid chromatography (HPLC) column (Waters 600 controller, USA) equipped with a waters 2996 photodiode array (PDA) and waters 2475 multi-wavelength fluorescence detector. The column used was SunFire C18 reverse phase column (4.6 mm interior diameter × 150 mm long) with a particle size of 5 μm. HPLC of 0.22 μ filtered sample (20 μl) was performed at room temperature with isocratic CH3OH:H2O (40:60) containing 0.1 % (v/v) tri-fluoro acetic acid as a mobile phase over a 20 min period.
Mixed Culture Fermentation of Microbial Strains
Four bacterial isolates: Arthrobacter sp. HKG 115 [15], Bacillus sonorensis HKG 102 [11], Micrococcus luteus and Staphylococcus aureus were randomly selected from diverse environmental sources, such as high altitude region of Leh, Himalaya region, sediment sample from Laccadive Sea, India and clinically isolated strains from patient samples, for mixed culture fermentation. Above selected isolates along with Exiguobacterium sp. HKG-126 were grouped in two separate batches and fermented at 37 °C for 5 days in HiMedia nutrient broth. Individually grown bacterial cultures in nutrient broth were taken as control. After 5 days of fermentation, all retrieved bacterial strains are preliminary identified by morphological and biochemical tests and confirmation was done by using 16S rRNA sequencing. All the strains before and after mixed culture fermentation were processed for the extraction of secondary metabolites followed by antimicrobial activity test, as described earlier.
Result and Discussion
Isolation, Identification and Characterization of Bacterial Strains
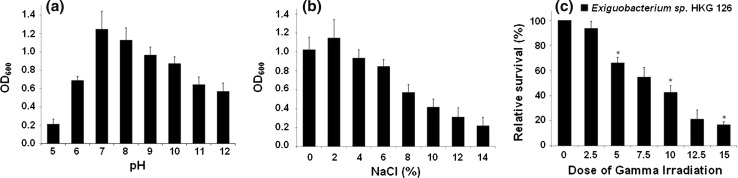
We have screened a unique radiation resistant bacterial strain, isolated from mangrove region of Kannamali, Kerala (India) which was identified as Exiguobacterium sp. through morphological, biochemical (Table 1) and molecular characterization. The 16S rDNA sequence of strain was submitted as Exiguobacterium sp. HKG-126 to NCBI GenBank under accession no. HM012684. Significant growth of this bacterium was observed at temperature 10–40 °C, pH level 6–12 and in media containing 0–14 % NaCl (Fig. 1a, b), and it was found to withstand extremely high dose of gamma radiation up to 15 kGy (Fig. 1c).
Table 1.
The morphological, physiological and biochemical characteristics of Exiguobacterium sp. HKG-126
| Test | Result | Test | Result |
|---|---|---|---|
| Shape | Rod shaped | Casein hydrolysis | Positive |
| Gram reaction | Gram positive | Starch hydrolysis | Positive |
| Colonies | Circular (1–5 mm in diameter) | β-galactosidase | Positive |
| Colour | Orange coloured | Acid phosphatase activities | Positive |
| NaCl tolerance | Up to 14 % | Lipid hydrolysis | Negative |
| Growth temperature | 10-40 °C (optimum 37 °C) | Indole test | Negative |
| Catalase test | Positive | Voges Proskauer test | Negative |
| Methyl red test | Positive | H2S production | Negative |
| ONPG test | Positive | Reduction of NO3− to NO2− | Negative |
Fig. 1.
Characterization of Exiguobacterium sp. HKG 126; a, b Effect of pH and NaCl (%) on the growth rate of Exiguobacterium sp. HKG 126, respectively; c Relative survival (%) of Exiguobacterium sp. HKG 126 (unirradiated control and irradiated strains) at different doses of gamma radiation. All growth experiments were performed in triplicates and the values are mean ± standard deviation of three independent experiments
Enhancement of Antimicrobial Activity After Exposing them to Gamma Irradiation
As demonstrated by measuring zone of inhibition, methanol extracts showed excellent antibacterial activity against Dietzia sp., Staphylococcus aureus, Bacillus cereus, Micrococcus flavus and Pseudomonas aeruginosa (Table 2). MIC was calculated only for two conditions (0 kGy: control and 15 kGy: irradiated), which were supposed to be more significant in irradiation experiment. It was observed that the lowest MIC was demonstrated against Micrococcus flavus, and Dietzia sp. (Table 2). The MIC for the antibacterial, extracted from the 15 kGy supernatant extract was less than that of 0 kGy supernatant extract of Exiguobacterium sp. which indicates that the antimicrobial agents from 15 kGy supernatant extract was more active than from 0 kGy extract. Earlier, it was believed that enhancement in antimicrobial activity occurs at only lower dose (up to 3 kGy) and highly decreased at higher dose level (>5 kGy), but interestingly, in this report, it was observed at both, lower and higher dose levels (up to 15 kGy). Thus, alteration in antimicrobial properties in response to γ-irradiation was found to be dose-dependent, i.e., the higher the dose used, higher the alteration in properties was observed (Table 2).
Table 2.
Inhibition zones (mm) and minimum inhibitory concentration (MIC) against several pathogenic microbial test strains, caused by methanolic extracts of Exiguobacterium sp. (EC)
| Microbial test strains | Zone of inhibition (mm)a | MIC (mg/ml) | |||||
|---|---|---|---|---|---|---|---|
| ECb | EC-5c | EC-10c | EC-15c | Gentd | ECe | EC-15e | |
| Dietzia sp. K44 (MTCC 7402) | 8.66 | 10.33 | 12.33 | 16.0 | 23.66 | 3.67 | 1.83 |
| Micrococcus flavus (NCIM 2378) | 9.66 | 9.66 | 11.0 | 14.33 | 24.33 | 3.33 | 2.33 |
| Bacillus subtilis (NCIM-2063) | – | – | 7.33 | 9.0 | 29.0 | – | 3.67 |
| Escherichia coli (NCIM 2739) | 7.0 | 7.33 | 8.66 | 11.66 | 29.66 | 4.17 | 2.83 |
| Pseudomonas aeroginosa (NCIM 2053) | 9.0 | 8.33 | 9.66 | 12.0 | 29.0 | 3.67 | 2.83 |
| Staphylococcus aureus | 9.33 | 9.0 | 9.33 | 11.33 | 22.0 | 3.67 | 3.17 |
| Bacillus cereus | 8.33 | 8.66 | 10.66 | 12.33 | 21.33 | 3.83 | 2.83 |
| Salmonella typhi | – | 6.33 | 7.33 | 11.33 | 21.33 | – | 3.17 |
| Micrococcus luteus | 6.33 | 6.33 | 7.66 | 11.66 | 16.33 | – | 2.83 |
All the values are mean of three independent experiments and the values of standard deviation were in the range of 5–10 %
aDiameter of zone of inhibition (mm), including disc diameter of 6 mm
bMethanol extract (100 μg/disc) from Exiguobacterium sp. before treatment with gamma irradiation
cMethanol extract (100 μg/disc) from Exiguobacterium sp. after exposing them to 5 kGy, 10 kGy and 15 kGy gamma irradiation, respectively
dGentamicin (10 μg/disc) as a positive control
eMIC value (mg/ml) of methanol extract from Exiguobacterium sp. before and after 15 kGy gamma irradiation exposure
HPLC Chromatogram Based Analysis
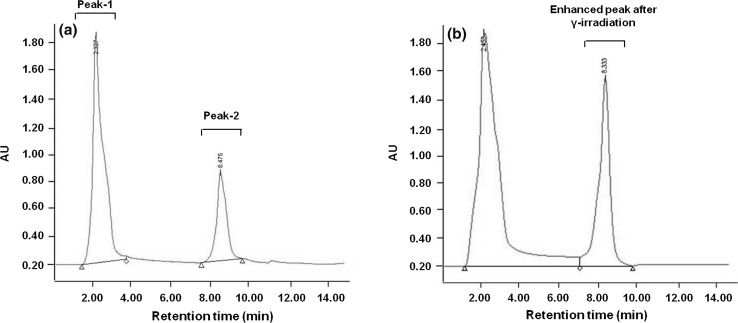
Enhancement in one chromatographically distinct peak (peak 2) out of two chromatographically distinct peaks (peak 1 and peak 2) was observed in crude methanol extracts of Exiguobacterium sp. HKG-126, after exposing to 15 kGy γ-radiations (Fig. 2). It might be possible that the enhanced antimicrobial activity in γ-irradiated strain was due to the compound showing enhanced peak and not any new compound, because of similar retention time of induced peak. Although the HPLC profile is one of the basic evidences to identify antimicrobial compounds, similarities in the HPLC chromatogram might explain that compounds produced by strains may have similar structure. By comparing HPLC chromatogram of known antibiotic compounds to that of active compounds in supernatant extract might explain the ability of the Exiguobacterium sp. to produce those compounds.
Fig. 2.
HPLC chromatogram of crude methanol extracts from Exiguobacterium sp. HKG 126 showing enhanced peak after gamma irradiation; a unirradiated control; b 15 kGy gamma irradiated strains
Enhancement of Antimicrobial Activity During Mixed Culture Fermentation
In order to find more novel structures and to enhance antibiotic production from microbial systems, new ways of screening for these compounds must be applied. Knowledge concerning mixed culture fermentation, especially with regards to antimicrobial production, could be an attractive research challenge from ecological relationship and industrial points of view. To test, the effect of mixed culture fermentation on such type of enhancement in antimicrobial activity as observed during γ-irradiation exposure, we adopted co-cultivation strategy for the production and enhancement of new antibiotics. Interestingly, we observed an enhancement in antimicrobial activity of Exiguobacterium sp. during and after mixed culture fermentation (Table 3a, b). It is possible that Exiguobacterium sp. might be able to sense the presence of any foreign species, possibly via some communication molecules secreted into the medium by the inducer strain. The antimicrobial properties of lyophilized extracts was tested before and after frozen or heated (−80 to +60 °C) conditions and was found stable under different pH (5–9) and temperature conditions and data were reproducible after incubation in conditions provided.
Table 3.
Determination of the antibacterial activity of crude methanolic extracts from different bacterial strains isolated before and after mixed culture fermentation and their combinations by measuring zone of inhibition (mm) and minimum inhibitory concentration (MIC), against several pathogenic microbial test strains
| Microbial test strains | Zone of inhibition (mm)a | MIC (mg/ml) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Before mix culture fermentationb | During mixedm culture fermentation | Positive Standard | Before fermentation | ||||||
| ECb | ABb1 | MLb1 | BSb2 | SAb2 | Comb-1m1 | Comb-2m2 | Gentg | ECb | |
| (a) | |||||||||
| Dietzia sp. K44 (MTCC 7402) | 8.66 | 10.33 | 8.66 | – | 7.66 | 12.33 | 9.33 | 23.66 | 3.67 |
| Micrococcus flavus (NCIM 2378) | 9.66 | 16.66 | – | – | – | 17.0 | 10.66 | 24.33 | 3.33 |
| Bacillus subtilis (NCIM-2063) | – | 19.66 | – | – | – | 21.66 | 7.66 | 29.0 | – |
| Escherichia coli (NCIM 2739) | 7.0 | 11.0 | – | – | – | 15.33 | 7.66 | 29.66 | 4.17 |
| Pseudomonas aeroginosa (NCIM 2053) | 9.0 | 8.66 | – | 8.33 | – | 8.33 | 9.0 | 29.0 | 3.67 |
| Staphylococcus aureus | 9.33 | 8.33 | – | – | – | 13.66 | 11.33 | 22.0 | 3.67 |
| Bacillus cereus | 8.33 | 9.33 | – | – | – | 12.0 | 10.33 | 21.33 | 3.83 |
| Salmonella typhi | – | 12.0 | 6.33 | – | 7.33 | 13.66 | 7.66 | 21.33 | – |
| Micrococcus luteus | 6.33 | 7.66 | – | 7.66 | 8.0 | 8.33 | 10.0 | 16.33 | – |
| Microbial test strains | Zone of inhibition (mm)a | MIC (mg/ml) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| After mix culture fermentationf | Positive standard | After fermentation | |||||||
| ECf1 | ECf2 | ABf1 | MLf1 | BSf2 | SAf2 | Gentg | ECf1 | ECf2 | |
| (b) | |||||||||
| Dietzia sp. K44 (MTCC 7402) | 8.33 | 10.66 | 11.66 | – | 6.33 | 8.0 | 23.66 | 3.83 | 3.17 |
| Micrococcus flavus (NCIM 2378) | 13.66 | 10.66 | 16.66 | – | – | – | 24.33 | 2.33 | 3.17 |
| Bacillus subtilis (NCIM-2063) | 8.66 | 7.33 | 19.33 | – | – | – | 29.0 | 3.67 | 4.17 |
| Escherichia coli (NCIM 2739) | 8.66 | 7.66 | 13.33 | – | 6.66 | 6.33 | 29.66 | 3.67 | 3.83 |
| Pseudomonas aeroginosa (NCIM 2053) | 8.66 | 10.66 | 8.66 | – | 8.66 | – | 29.0 | 3.67 | 3.17 |
| Staphylococcus aureus | 12.0 | 10.33 | 9.66 | – | – | – | 22.0 | 2.83 | 3.17 |
| Bacillus cereus | 8.33 | 9.66 | 8.66 | – | – | – | 21.33 | 3.83 | 3.33 |
| Salmonella typhi | 7.33 | – | 12.0 | – | – | 7.66 | 21.33 | 4.17 | – |
| Micrococcus luteus | 7.33 | 8.66 | 7.66 | – | 8.33 | 8.33 | 16.33 | 4.17 | 3.67 |
All the values are mean of three independent experiments and the values of standard deviation were in the range of 5–10 %
aDiameter of zone of inhibition (mm) including disc diameter of 6 mm showing antibacterial activity of methanol extracts (100 μg/disc) from bacterial supernatant of, EC, Exiguobacterium sp.; AB, Arthrobacter sp.; ML, Micrococcus luteus; BS, Bacillus sonorensis; SA, Staphylococcus aureus
m1Comb-1 corresponds to mixed bacterial cultures of bEC, b1AB and b1ML
m2Comb-2 corresponds to mixed bacterial cultures of bEC, b2BS and b2SA
f1Purified bacterial culture after mixed culture fermentation of combination-1
f2Purified bacterial culture after mixed culture fermentation of combination-2
gGentamicin (10 μg/disc) as a positive control
In case of f1ML, it was not characterized and identified after mixed culture fermentation. The reason behind them supposed to be killing of f1ML during mixed culture due to antimicrobial agents produced from other co-cultivated strains like EC and AB. Keeping these in mind, it is tempting to speculate that induction and enhancement in antimicrobial activity does not occur in all microbial strains and it depends on the pathogen used. A reason for this could be that induction by communication molecules affects only certain types of microbial population by means of quorum sensing (QS) and this response may be a defence mechanism which has evolved to deter or kill a potential competitor. There are several reports which have shown the production of such type of communication molecules from bacterial organisms such as Edwardsiella ictaluri [17]. Besides to these QS molecules, production of some other molecules which can degrades these QS signals by inhibiting the production of QS signal molecules or degradation of the AHLs, also has been reported from bacterial organisms, called quorum quenching (QQ) [18–21].
Natural genetic transformation is also believed to be the essential mechanism for exploiting innate microbial capacity in bacterial populations, during mixed culture fermentation via horizontal gene transfer. In an early report, microbial sequencing studies and result of genome analyses indicates that the biosynthetic potential of microbial strains is much greater than that observed by traditional natural product isolation [22–24]. Therefore, with countless possible microbial combinations and increasingly sophisticated chemical isolation and structure determination methods, the potential for mixed fermentation in natural product drug discovery seems quite promising. Some of these independent studies involving particular approaches like co-cultivation and radiation exposure strategies to enhance antimicrobial activity support the findings reported here and these findings have important implications for the discovery of novel antimicrobial compounds from Exiguobacterium sp. HKG-126 and may allow the development of new methods for screening novel compounds active against multi-drug-resistant bacteria. In addition, the effect of mixed cultures of bacterial population on antimicrobial activity could be investigated as this would give a more in-depth picture of ecological relationships. Encouraged by these findings, further attempts are being directed towards the characterization of the mechanism involved; it is difficult to predict whether this approach will serve as a general model for producing other unusual antibiotics. In future, we are focusing on the purification of these compounds from crude extracts using HPLC and determining the exact composition and structure elucidation of compound responsible for antimicrobial property using analytical study.
Acknowledgments
The authors are grateful to Dr. Rajesh Gokhle, director of CSIR-IGIB for providing infrastructural facilities and financial assistance (HKG 61). R. Pathak would also like to acknowledge University Grants Commission (UGC), India, for providing Senior Research Fellowship.
References
- 1.Demain AL. Induction of microbial secondary metabolism. Int Microbiol. 1998;1:259–264. [PubMed] [Google Scholar]
- 2.Mearns SA, Bregu M, Boyd KG, Burgess JG. Cross-species induction and enhancement of antimicrobial activity produced by epibiotic bacteria from marine algae and invertebrates, after exposure to terrestrial bacteria. Lett Appl Microbiol. 1998;27:142–146. doi: 10.1046/j.1472-765X.1998.00416.x. [DOI] [PubMed] [Google Scholar]
- 3.Slattery M, Rajbhandari I, Wesson K. Competition mediated antibiotic induction in the marine bacterium Streptomyces tenjimariensis. Microb Ecol. 2001;41:90–96. doi: 10.1007/s002480000084. [DOI] [PubMed] [Google Scholar]
- 4.Oh DC, Kauffman CA, Jensen PR, Fenical W. Induced production of emericellamides A and B from the marine-derived fungus Emericella sp. in competing co-culture. J Nat Prod. 2007;70:515–520. doi: 10.1021/np060381f. [DOI] [PubMed] [Google Scholar]
- 5.Pettit RK. Mixed fermentation for natural product drug discovery. Appl Microbiol Biotechnol. 2009;83:19–25. doi: 10.1007/s00253-009-1916-9. [DOI] [PubMed] [Google Scholar]
- 6.Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 7.Dusane DH, Matkar P, Venugopalan VP, Kumar AR, Zinjarde SS. Cross-species induction of antimicrobial compounds, biosurfactants and quorum-sensing inhibitors in tropical marine epibiotic bacteria by pathogens and biofouling microorganisms. Curr Microbiol. 2011;62:974–980. doi: 10.1007/s00284-010-9812-1. [DOI] [PubMed] [Google Scholar]
- 8.Donadio S, Monciardini P, Alduina R, Mazza P, Chiocchini C, Cavaletti L, Sosio M, Puglia AM. Microbial technologies for the discovery of novel bioactive metabolites. J Biotechnol. 2002;99:187–198. doi: 10.1016/S0168-1656(02)00209-2. [DOI] [PubMed] [Google Scholar]
- 9.Knight V, Sanglier JJ, DiTullio D, Braccili S, Bonner P, Waters J, Hughes D, Zhang L. Diversifying microbial natural products for drug discovery. Appl Microbiol Biotechnol. 2003;62:446–458. doi: 10.1007/s00253-003-1381-9. [DOI] [PubMed] [Google Scholar]
- 10.Genilloud O, Gonzalez I, Salazar O, Martin J, Tormo JR, Vicente F. Current approaches to exploit actinomycetes as a source of novel natural products. J Ind Microbiol Biotechnol. 2011;38:375–389. doi: 10.1007/s10295-010-0882-7. [DOI] [PubMed] [Google Scholar]
- 11.Rakesh OD, Pathak R, Dhaker AS, Arora R, Kumar R, Rajaram R, Gautam HK. Isolation, characterization and bioactivity of deep sea bacteria with special reference to induction of antibacterial and antioxidant metabolites following gamma irradiation. Can J Pure Appl Sci. 2011;5:1363–1370. [Google Scholar]
- 12.Bergey DH, Holt JG, Kreig NR, Sneath PHA. Bergey’s manual of determinative bacteriology. 9. Baltimore: Williams and Wilkins; 1994. [Google Scholar]
- 13.Gupta AK, Pathak R, Singh B, Gautam H, Kumar R, Kumar R, Arora R, Gautam HK. Proteomic analysis of global changes in protein expression during exposure of gamma radiation in Bacillus sp. HKG-112 isolated from saline soil. J Microbiol Biotechnol. 2011;21:574–581. [PubMed] [Google Scholar]
- 14.Beaume N, Pathak R, Yadav VK, Kota S, Misra HS, Gautam HK, Chowdhury S. Genome-wide study predicts promoter-G4 DNA motifs regulate selective functions in bacteria: radioresistance of D. radiodurans involves G4 DNA-mediated regulation. Nucleic Acids Res. 2013;41(1):76–89. doi: 10.1093/nar/gks1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pathak R, Singh R, Singh A, Gautam H, Dhaker AS, Kumar R, Arora R, Gautam HK. Assessment of antibacterial and free radical scavenging activity in psychrophilic Arthrobacter sp. Pharmacologyonline. 2011;1:344–355. [Google Scholar]
- 16.Mahato M, Arora V, Pathak R, Gautam HK, Sharma AK. Fabrication of nanostructures through molecular self-assembly of small amphiphilic glyco-dehydropeptides. Mol Biosyst. 2012;8:1742–1749. doi: 10.1039/c2mb25023c. [DOI] [PubMed] [Google Scholar]
- 17.Yang Q, Han Y, Tinh NTN, Hien NT, Bossier P. Detection of quorum sensing signal molecules in edwardsiella ictaluri Ei-151. Indian J Microbiol. 2012;52(4):581–586. doi: 10.1007/s12088-012-0312-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalia VC, Raju SC, Purohit HJ. Genomic analysis reveals versatile organisms for quorum quenching enzymes: acyl-homoserine lactone-acylase and lactonase. Open Microbiol J. 2011;5:1–13. doi: 10.2174/1874285801105010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalia VC, Purohit HJ. Quenching the quorum sensing system: potential antibacterial drug targets. Crit Rev Microbiol. 2011;37(2):121–140. doi: 10.3109/1040841X.2010.532479. [DOI] [PubMed] [Google Scholar]
- 20.Huma N, Shankar P, Kushwah J, Bhushan A, Joshi J, Mukherjee T, Raju S, Purohit HJ, Kalia VC. Diversity and polymorphism in AHL-lactonase gene (aiiA) of Bacillus. J Microbiol Biotechnol. 2011;21(10):1001–1011. doi: 10.4014/jmb.1105.05056. [DOI] [PubMed] [Google Scholar]
- 21.Kalia VC (2013) Quorum sensing inhibitors: an overview. Biotechnol Adv. doi:10.1016/j.biotechadv.2012.10.004 [DOI] [PubMed]
- 22.Maiden MC. Horizontal genetic exchange, evolution, and spread of antibiotic resistance in bacteria. Clin Infect Dis. 1998;27:S12–S20. doi: 10.1086/514917. [DOI] [PubMed] [Google Scholar]
- 23.Udwary DW, Zeigler L, Asolkar RN, Singan V, Lapidus A, Fenical W, Jensen PR, Moore BS. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc Natl Acad Sci USA. 2007;104:10376–10381. doi: 10.1073/pnas.0700962104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lal S, Cheema S, Kalia VC. Phylogeny vs genome reshuffling: horizontal gene transfer. Indian J Microbiol. 2008;48:228–242. doi: 10.1007/s12088-008-0034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]