Abstract
Objective: Interaction of genotype and environment results in a specific phenotype of the clinical course of asthma. TGFβ-1 gene belongs to the important group of genes involved in the regulation of proliferation, differentiation, adhesion, and migration of a variety of cell types. TGFβ-1 is inhibitory for B and T cells, as well as IgE production. In particular, it is engaged in inflammation of the bronchi and airway remodeling in asthma, which processes are critical in the pathogenesis of the disease. The aim of this study was to evaluate the correlation between the level of expression of TGFβ-1 and the severity of asthma. Methods: The study included 39 participants (20 healthy subjects and 19 patients with asthma). Each sample was analysed by using real time PCR. Results: There was statistical associations between the control group and the group of patients (p = 0,00007). It was demonstrated strong correlation between healthy and patients with severe asthma according GINA guidelines (p = 0,017). It was found the strong statistical correlation between healthy and patients with severe corticosteroid dependent asthma (p = 0,013). Correlations were observed between levels of asthma severity according to the ATS guidelines and controls. The influence of the level of TGFβ-1 mRNA expression and the severity of asthma (ATS) in the FEV1 (%) parameter value was found. Conclusion: It was found that an important role is played by TGFβ-1 in the pathogenesis of asthma.
Keywords: Asthma, inflammation, airway obstruction, transforming growth factor β-1, T regulatory cells
Introduction
The products of many genes are involved in the regulation and enhancement of the levels of cytokines, chemokines, and leukotrienes in bronchial asthma. The activated cells synthesize and release around 100 different substances [1] which mediate the complex inflammatory response in the airways [2]. These are responsible for the recruitment of other cells of the immune response, and cause bronchoconstriction, increased permeability and vasodilation, as well as an enhanced inflammatory reaction. Cytokines determine T cell differentiation towards Th2, increased production of IgE by B cells and prolongation of inflammatory cell survival [3].
A special cytokine seen in the regulation of the immune response in asthma is transforming growth factor-β (TGF-β), a member of the TGF-β superfamily [4], which consists of approximately 30 mammalian cytokines including different isoforms of TGF-β, Műllerian Inhibitory Substance (MIS), drosophila decapentaplegic gene, activins and inhibins, growth and differentiation factors, bone morphogenetic proteins (BMPs), glial-derived neurotrophic factor [5] and macrophage cytokine inhibitor 1 (MIC-1) [6]. These proteins are encoded by 28 genes and show similarities in regulatory functions [7].
TGF-β is produced by a number of inflammatory cells: eosinophils [8], neutrophils, mast cells, macrophages, monocytes, B- and T-cells [9]. This cytokine is also secreted by airway structural cells: smooth muscle cells, epithelial cells, endothelial cells and fibroblasts [10]. TGF-β has both paracrine and autocrine effects on nearby cells [5]. TGF-β mRNA transcription is regulated by a number of growth factors such as Nerve Growth Factor, transcription factors (eg AP-1) and oncogenes such as src, abl, ras, jun and fos [11]. The cytokine is formed from a propeptide removed from a crude amino-terminal fragment containing a LAP (latency associated protein) connected with a homodimer of mature TGF-β [12].
TGF-β has five isoforms, of which TGF-β1, TGF-β2 and TGF-β3 are encoded by different genes, but activate the same TGF-β receptors and stimulate similar path signals [13]. The TGF-β1 isoform has an immunosuppressive effect on T and B cells, and its deficiency results in an increased expression of autoimmune diseases [7]. This plays an important role in the complex pathogenesis of bronchial asthma, and overexpression of TGF-β1 mRNA and TGF-β protein significantly worsens the course of the disease [5], which affects the course of early recovery processes and airway remodeling [5]. TGF-β1 increases the synthesis of fibronectin [14], tenascin and antiproteases in lung epithelial cells [15], and decreases the expression of cathepsins [16].
TGF-β1 increases the secretion of IL-8 from the epithelial cells and airway smooth muscle cells [17], and increases the expression of adhesion molecules such as ICAM-1 on endothelial cells [18]. This cytokine enhances expression of RANTES and GM-CSF, which have a chemotactic effect on inflammatory cells: eosinophils, lymphocytes and monocytes [19]. It is worth noting that the expression of COX-2 and PGE2 is significantly increased by the action of TGF-β1 on the smooth muscle cells of the airways [20] and by doing so, TGF-β plays an important role in modulating the course of inflammation in the airways of patients with bronchial asthma. Hence, there is a need for molecular, biochemical, and clinical trials involving this multifunctional cytokine in the pathogenesis of airway hyperresponsiveness in obstructive disorders.
The aim of the study is an evaluation of the relationship between the level of expression of TGF-β1 mRNA and the disease phenotype in a Polish population of patients with bronchial asthma.
Materials and methods
The study was approved by the local ethics committee (Consent of Research Review Board at the Medical University of Lodz, Poland, No RNN/133/09/KE). At the commencement of the study, participants were invited to attend voluntarily. Before enrollment, written informed consent was obtained from every patient.
The study was conducted on a group of 19 patients with bronchial asthma. Asthma diagnosis was established according to GINA (The Global Initiative for Asthma) and ATS (American Thoracic Society) recommendations, based on clinical asthma symptoms and a lung function test. The level of asthma severity and control was determined on the basis of ATS/GINA report guidelines. In order to determine the nature of their glucocorticoid resistance, as well as establish whether genetic factors are primary or secondary, all the participants underwent both objective and subjective examinations. Apart from a subjective examination, structuralized anamnesis was performed and a number of other factors were examined: gender, obesity, tobacco smoking, duration of bronchial asthma, allergy to house dust mites, animal fur, mould spores, cockroach allergens, and hypersensitivity to non-steroid anti-inflammatory drugs (NSAIDs). The results of pulmonary function tests and allergological tests were obtained from the individual medical records of the patients. If there were no results of spirometry or allergological tests available, such examinations were additionally performed during the recruitment visit.
The exclusion criteria included subjects suffering from clinically significant exacerbations, or who were using such drugs as rifampicin, phenobarbital, phenytoin or ephedrine which might induce resistance to glucocorticoids; subjects with signs of viral infections, either generalized, or affecting the respiratory tract; subjects failing to comply with the recommendations of their doctor. The control arm included a group of 20 healthy adults who met the following criteria: no history or symptoms of either bronchial asthma, or other pulmonary diseases, no history or symptoms of allergy, no history or symptoms of atopic dermatitis, no history or signs of hypersensitivity to aspirin, negative results of skin tests for 12 common allergens, no first-degree relatives with bronchial asthma or atopic disorders [21].
According to the standards developed by the Polish Society for Pulmonary Diseases, the best result of three spirometry measurements was selected for the analysis of obstructive disorders and disease severity. The correlation analysis took into consideration FEV1 (forced expiratory volume) expressed in liters, FEV1% (A/N% - percentage ratio of the measured to expected value) expressed as a percentage of the expected value and the FEV1% FVC index (FEV1 to FVC ratio - forced vital capacity) expressed as absolute numbers. Spirometry tests were conducted in the Outpatient Department according to ERS (European Respiratory Society) / ATS standards, and allergological tests according to the EAACI guidelines (European Academy of Allergy and Clinical Immunology) [22].
The level of asthma control was assessed with the Asthma Control Test, which is clear and easy for patients. The Asthma Control Test (ACT™) consists of five questions. It was developed by Nathan et al. in cooperation with general practitioners and specialists in diagnostics and therapy of asthma. Bronchial asthma control level was calculated on the basis of the following patient ACT scores: 00 - 19 points - no asthma control, 20 - 24 points - partially controlled asthma and 25 points - well-controlled asthma.
The study included 39 participants: 20 healthy subjects and 19 patients with asthma. The control group comprised 12 women and 8 men: mean age 45.31 years, minimum age 23 years, maximum age 71 years, SE±17.11. The average FEV1 value (%) was 93.73. SE±8.7 and average FEV1 value (L) was 2.90. SE±0.9.
The case group comprised 16 women and 3 men: mean age 50.04 years, minimum age 24 years, maximum age 72 years, SE±16.2. The average FEV1 value (%) was 74.19, SE±20.06 and the average FEV1 value (L) was 2.20, SE±0.9.
Mild asthma occurred in 15.79% of patients, and moderate and severe in 52.63% and 31.58%, respectively. The average ACTTM value in patients was 19.6 points, SE±4.9. The minimum value of ACTTM was 9 points and maximum 25 points. A detailed discussion of the parameters of descriptive statistics for the study groups are presented in Table 1.
Table 1.
Detailed statistics for groups of patients with regard to epidemiological data, level of expression of TGF-β1 (2-ΔΔCT) and pulmonary function
| Parameter | HEALTHY | ||||
|
| |||||
| Mean | Median | Minimum | Maximum | Standard deviation | |
|
| |||||
| 2-ΔΔCT | 12946.76 | 9946.68 | 337.79 | 40342.10 | 11127.84 |
| Age (years) | 45.31 | 50.00 | 23.00 | 71.00 | 17.11 |
| FEV1 (L) | 2.89 | 2.69 | 1.66 | 4.70 | 0.84 |
| FEV1 (%) | 93.73 | 93.00 | 67.00 | 128.00 | 8.70 |
| FVC (L) | 2.90 | 3.53 | 2.03 | 6.40 | 0.90 |
| FVC (%) | 98.33 | 100.00 | 74.00 | 150.00 | 17.89 |
| FEV1 % FVC | 78.75 | 76.45 | 65.97 | 95.20 | 7.61 |
| FEV1 % FVC (%) | 98.57 | 98.00 | 84.00 | 115.00 | 8.00 |
|
| |||||
| Parameter | PATIENTS | ||||
|
| |||||
| Mean | Median | Minimum | Maximum | Standard deviation | |
|
| |||||
| 2-ΔΔCT | 29520.42 | 17620.90 | 2946.93 | 114500.90 | 33398.41 |
| Age (years) | 50.04 | 49.00 | 24.00 | 72.00 | 16.20 |
| FEV1 (L) | 2.07 | 2.13 | 0.37 | 4.20 | 0.86 |
| FEV1 (%) | 74.19 | 79.00 | 19.00 | 105.00 | 20.06 |
| FVC (L) | 2.20 | 2.88 | 1.11 | 6.00 | 0.90 |
| FVC (%) | 90.67 | 93.00 | 46.00 | 115.00 | 15.20 |
| FEV1 % FVC | 68.26 | 70.680 | 33.3300 | 91.0 | 14.71 |
| FEV1 % FVC (%) | 84.95 | 89.000 | 44.0000 | 113.0 | 17.30 |
Detailed statistics for groups of patients with regard to epidemiological data, level of expression of TGF-β1 (2-ΔΔCT) and pulmonary function.
An amount of 10 μg total RNA was extracted from the peripheral blood lymphocytes using TRI Reagent® Solution (Ambion, NY, USA) according to the standard acid-guanidinium-phenol-chloroform method [23]. The extracted RNA was analyzed by agarose gel electrophoresis and only cases with preserved 28S, 18S and 5S ribosomal RNA bands, indicating good RNA quality, were used in the study. Total RNA was digested with DNase (GIBCO) at room temperature for 15 min. The amount of purified RNA was determined using spectrophotometry at 260 nm in a Nanodrop analyser (ND-100; Nanodrop Technologies, Wilmington, DE, USA). The purity was verified according to the ratio of 260/280 nm measurements, such that values between 1.8 and 2.1 indicated that the quality of the RNA obtained was optimal and suitable for the quantitative real-time polymerase chain reaction (qRT-PCR).
Reverse transcription of 1μg RNA was performed using an AccuScript PfuUltraII RT-PCR kit (Agilent Technologies, CA, USA). The cDNA was subjected to real-time quantitative PCR using gene-specific primers (5’-GCC TCC GGA GGG TGT CAG TG-3’ and 5’-GTG TCT GCC TCC TGA CCC TTC-3’ SIGMA-ALDRICH, Germany) for TGFβ-1 and GAPDH (5’-AGC CAC ATC GCT CAG ACA -3’ and 5’-GCC CAA TAC GAC CAA ATC C-3’ IBB PAN, Polska) using a Brilliant II SYBR Green QRT-PCR Master Mix Kit (Stratagene, CA, USA). Amplification was performed using the normal 2 steps and a standard thermal profile. Primer annealing temperature was 63°C, and primer annealing time was 20 seconds. An Agilent Technologies Stratagene Mx3000P was used for the PCR reaction. For each sample, the CT values were calculated with the help of Mx-Pro software. The RT-PCR amplification of the TGFβ-1 gene was compared to that of the GAPDH reference gene. Real-time PCR data was automatically calculated with the data analysis module. The results were analyzed according to the 2-ΔΔCT method [24,25]. Validation of PCR efficiency was performed with a standard curve.
The T-test for independent samples and multiple regression tests were used to evaluate the correlation between the variables. An advanced linear model and a general regression model were used to analyze the data. The level of significance was p = 0.05. Statistical analysis was performed with the help of a licensed version of STATISTICA (StatSoft, Inc. 2011. STATISTICA data analysis software system, version 10. Licence number AXAP202E504303AR-A). The genotyping was performed by two investigators who were unaware of the phenotypes.
Results
Significant relationships were found between the control group and the group of patients (p = 0.00007. Standard error 3527.0). According to GINA, a strong correlation was demonstrated between healthy subjects and those with severe asthma (p = 0.017. Controls SD±9119.98. Cases SD±26154.20), and no similar associations were observed between healthy subjects and non-severe asthma patients. A strong correlation was found between healthy subjects and those with severe corticosteroid dependent asthma (p = 0.013. Controls SD±9119.98. Cases SD± 28794.42). There were no associations found concerning TGFβ-1 mRNA expression between the groups of patients with non-severe asthma and severe asthma.
The relationship between TGFβ-1 mRNA expression levels seen in controls and asthma patients was also evaluated according to the ATS criteria. Statistically significant relationships between the control group and the degrees of severity of asthma graded according to the American Thoracic Society, is given in Table 2.
Table 2.
Correlation of expression levels of TGF-1 mRNA in groups of patients with severe refractory asthma, according to the ATS, with those of the control group
| ATS Criteria: | Level of expression TGFβ-1 mRNA | ||
|---|---|---|---|
|
| |||
| ATS 0 (No) - unfulfilled | b | Standard error | p |
| ATS 1 (Yes) - fulfilled | |||
| Control group vs. Asthma ATS 0 | 0.15 | 0.03 | 0.001 |
| Control group vs. Asthma ATS 1 | -0.01 | 0.003 | 0.04 |
| Asthma ATS 0 vs. Asthma ATS 1 | -0.03 | 0.01 | 0.23 |
Correlation of expression levels of TGF-1 mRNA in groups of patients with severe refractory asthma, according to the ATS, with those of the control group.
The correlation between the degree of asthma control according to ACTTM, and expression of TGF-1 mRNA was statistically insignificant (p > 0.05).
The relationship between the pulmonary function tests and the levels of expression of TGF-1 mRNA was evaluated. A correlation was found only for FEV1 (%); other spirometric variables did not correlate significantly with ΔΔCT. Correlations in multivariate analyses for FEV1 (%) were found in patients with asthma as described by the ATS criteria, but not in those described by GINA. A graphical interpretation of the expression of ΔΔCT TGF-1 mRNA against FEV1 in patients with severe asthma, refractory to treatment according to the ATS, is given in the scatterplot in Figure 1.
Figure 1.
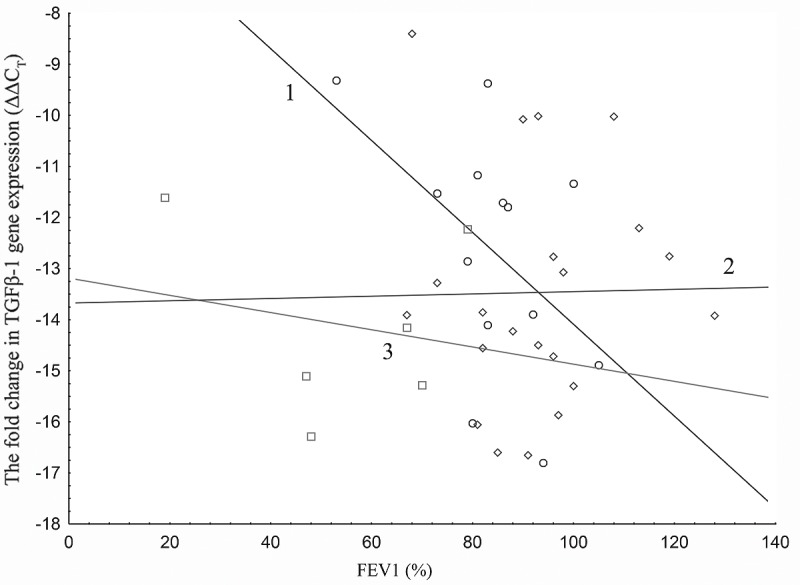
Line 1 - the ΔΔCT of TGF-1 mRNA expression in asthma patients does not meet the diagnostic criteria for severe asthma refractory to treatment, according to the ATS FEV1 value (%). Line 2 - the ΔΔCT of TGF-1 mRNA expression for the control group Line 3 - the ΔΔCT of TGF-1 mRNA expression in patients with asthma meets the diagnostic criteria for severe asthma refractory to treatment according to the ATS FEV1 value (%). The ΔΔCT of TGF-1 mRNA expression in patients with asthma (according to ATS) with regard to the level of airway obstruction is based on FEV1 (%) values.
Table 3 shows the relationship between the multivariate analysis of FEV1 (%): the levels of expression of TGF-1 mRNA and the severity of asthma according to ATS.
Table 3.
There was no correlation between TGF-1 mRNA expression and age, gender, number of allergens or smoking (p > 0.05)
| Parameter | Statistics | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| FEV1 (%) Param. | FEV1 (%) ±SE | FEV1 (%) t | FEV1 (%) p | -95.00% Confidence limit | +95.00%Confidence limit | |
| ΔΔCT | -3.46 | 1.59 | -2.17 | 0.04 | -6.88 | -0.04 |
| Asthma strongly refactory to treatment according to ATS-No | 10.86 | 4.73 | 2.29 | 0.03 | 0.71 | 21.02 |
| Asthma strongly refactory to treatment according to ATS-Yes | 9.39 | 4.67 | 2.00 | 0.06 | -0.63 | 19.41 |
| Sex | -8.92 | 4.85 | -1.83 | 0.08 | -19.34 | 1.48 |
Multivariate analysis of variables affecting the pulmonary functional parameter FEV1 (%).
There was no correlation between TGF-1 mRNA expression and age, gender, number of allergens or smoking (p > 0.05).
Discussion
TGF-1 is a cytokine which plays an important role in influencing the differentiation of precursor and T-helper cells, modulating airway inflammation in the course of obstructive diseases. Thus, the level of expression of the marker should correlate significantly with either the severity of asthma and the degree of airway narrowing, and the level of disease control according to the ACTTM test. This pilot study is a prelude to a cross-regional study of patients with asthma, the aim of which is to determine the expression levels of TGF-1 mRNA at different stages of the disease, according to both international GINA and ATS guidelines, and the phenotypic factors correlated with it.
The study found significantly higher levels of expression of TGF-1 mRNA in patients compared with the control population, as well as between patients with severe asthma according to GINA, and the control group. Similar observations have been found in patients with severe steroid dependent asthma who have higher levels of TGF-1 mRNA expression compared with the healthy population. No significantly different expression levels were seen between patients with severe and non-severe asthma. No association was found between mild persistent asthma (N = 3), moderate asthma (n = 10) and the control group, which may be due to the insufficient number of observations. In addition, multivariate analysis did not find a statistically significant correlation between TGF-1 mRNA level and lung function parameters of GINA-graded asthma severity.
Patients were graded based on the degree of asthma severity according to the ATS criteria. TGFβ-1 mRNA levels were measured both in patients with severe asthma refractive to treatment, and in those not fulfilling this criterion. However, these two populations did not differ from each other with regard to the level of cytokine expression. In multivariate analysis, it was found that the parameter FEV1 (%) significantly correlated with the level of expression of TGF-1 (ΔΔCT) and asthma severity as defined by the ATS. A multifactorial model showed the severity of asthma only correlated with mRNA expression in patients who had non-severe, refractory asthma. There was no such relevance for patients with severe asthma resistant to treatment. This is probably due to the low number of patients included in this subgroup, as prior analysis has shown that the more severe form of the disease, the higher the levels of TGF-1 mRNA expression in patients.
It should be noted that the criteria for severity of illness, determined arbitrarily, may not always be a sufficient tool to categorize the stage of the disease. This is due to the fact that asthma may well be a heterogeneous collection of different clinical syndromes: chronic airway inflammation and reversible airway obstruction bronchitis. Grading the disease according to the GINA and ATS criteria are therefore temporary measures. Without a doubt, the multidimensional approach to evaluating asthma best reflects its clinical properties. Because of the central role of TGF-β in the pathogenesis of asthma, this protein may serve as potential for new forms of targeted therapy, although the evaluation of its usefulness in this area requires further investigation.
Conclusions
The observed correlation indicates the relationship between the level of expression of TGFβ-1, and the prevalence of asthma and the severity of its course. It was found that an important role is played by TGFβ-1 in the pathogenesis of asthma, particularly severe asthma. The relationship between TGFβ-1 mRNA expression and the degree of airway obstruction expressed by spirometry, as well as the degree of asthma severity, was found to have statistical and clinical significance. The study should be continued and the results be verified on a larger number of subjects.
Acknowledgements
The Authors would like to express their gratitude to Dr. Damian Tworek for help with patient recruitment and blood collection; to Joanna Molinska for help with administration; to Beata Małachowska for her help with the statistical analysis; Lastly, we offer our thanks to all of those who supported us in any way during the completion of the project.
This study was supported by the Grant of Clinic of Internal Medicine, Asthma and Allergies, II Chair of Internal Medicine, Medical University of Lodz, Nr 503/1-095-03/503-01. Beata Małachowska was supported by the TEAM project financed from the Innovative Economy Operational Program and coordinated by the Foundation for Polish Science.
The study was approved by the local ethics committee (Consent of Research Review Board at the Medical University of Lodz, Poland, No RNN/133/09/KE).
Conflict of interest statement
We declare no conflict of interest.
References
- 1.Droszcz W. Astma. Warszawa: Wydawnictwo Lekarskie PZWL Press; 2007. [Google Scholar]
- 2.Vercelli D. Discovering susceptibility genes for asthma and allergy. Nat Rev Immunol. 2008;8:169–182. doi: 10.1038/nri2257. [DOI] [PubMed] [Google Scholar]
- 3.Fal A. Alergia, choroby alergiczne, astma. Kraków: Medycyna Praktyczna Press; 2010. [Google Scholar]
- 4.Radwan-Oczko M, Boratyńska M, Banasik M, Filipowski H. Transforming growth factor beta 1 (TGF-ß1) in renal transplant recipients with and without gingival hyperplasia. Czas Stomat. 2005;2:95–101. [Google Scholar]
- 5.Duvernelle C, Freund V, Frossard N. Transforming growth factor-beta and its role in asthma. Pulm Pharmacol Ther. 2003;16:181–196. doi: 10.1016/S1094-5539(03)00051-8. [DOI] [PubMed] [Google Scholar]
- 6.Attisano L, Wrana JL. Signal transduction by the TGF-beta superfamily. Science. 2002;296:1646–1647. doi: 10.1126/science.1071809. [DOI] [PubMed] [Google Scholar]
- 7.Stępień-Wyrobiec O, Hrycek A, Wyrobiec G. Transforming growth factor beta (TGF-beta): Its structure, function, and role in the pathogenesis of systemic lupus erythematosus. Postepy Hig Med Dosw (Online) 2008;62:688–693. [PubMed] [Google Scholar]
- 8.Ohno I, Nitta Y, Yamauchi K, Hoshi H, Honma M, Woolley K, O’Byrne P, Tamura G, Jordana M, Shirato K. Transforming growth factor beta 1 (TGF beta 1) gene expression by eosinophils in asthmatic airway inflammation. Am J Respir Cell Mol Biol. 1996;15:404–409. doi: 10.1165/ajrcmb.15.3.8810646. [DOI] [PubMed] [Google Scholar]
- 9.Magnan A, Retornaz F, Tsicopoulos A, Brisse J, Van Pee D, Gosset P, Chamlian A, Tonnel AB, Vervloet D. Altered compartmentalization of transforming growth factor-beta in asthmatic airways. Clin Exp Allergy. 1997;27:389–395. [PubMed] [Google Scholar]
- 10.Magnan A, Frachon I, Rain B, Peuchmaur M, Monti G, Lenot B, Fattal M, Simonneau G, Galanaud P, Emilie D. Transforming growth factor beta in normal human lung: preferential location in bronchial epithelial cells. Thorax. 1994;49:789–792. doi: 10.1136/thx.49.8.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim SJ, Romeo D, Yoo YD, Park K. Transforming growth factorbeta: expression in normal and pathological conditions. Horm Res. 1994;42:5–8. doi: 10.1159/000184136. [DOI] [PubMed] [Google Scholar]
- 12.Munger JS, Harpel JG, Giancotti FG, Rifkin DB. Interactions between growth factors and integrins: latent forms of transforming growth factor-beta are ligands for the integrin alphavbeta1. Mol Biol Cell. 1998;9:2627–2638. doi: 10.1091/mbc.9.9.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bierie B, Moses HL. TGFbeta: the molecular Jekyll and Hyde of cancer. Nat Rev Cancer. 2006;6:506–520. doi: 10.1038/nrc1926. [DOI] [PubMed] [Google Scholar]
- 14.Romberger DJ, Beckmann JD, Claassen L, Ertl RF, Rennard SI. Modulation of fibronectin production of bovine bronchial epithelial cells by transforming growth factor-beta. Am J Respir Cell Mol Biol. 1992;7:149–155. doi: 10.1165/ajrcmb/7.2.149. [DOI] [PubMed] [Google Scholar]
- 15.Linnala A, Kinnula V, Laitinen LA, Lehto VP, Virtanen I. Transforming growth factor-beta regulates the expression of fibronectin and tenascin in BEAS 2B human bronchial epithelial cells. Am J Respir Cell Mol Biol. 1995;13:578–585. doi: 10.1165/ajrcmb.13.5.7576694. [DOI] [PubMed] [Google Scholar]
- 16.Gerber A, Wille A, Welte T, Ansorge S, Buhling F. Interleukin-6 and transforming growth factor beta 1 control expression of cathepsins B ans L in human lung epithelial cells. J Interferon Cytokine Res. 2001;21:11–19. doi: 10.1089/107999001459114. [DOI] [PubMed] [Google Scholar]
- 17.Kumar NM, Rabadi NH, Sigurdson LS, Schunemann HJ, Lwebuga-Mukasa JS. Induction of interleukin-1 and interleukin-8 mRNAs and proteins by TGF beta 1 in rat lung alveolar epithelial cells. J Cell Physiol. 1996;169:186–199. doi: 10.1002/(SICI)1097-4652(199610)169:1<186::AID-JCP19>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki Y, Tanigaki T, Heimer D, Mang W, Ross WG, Murphy GA, Sakai A, Sussman HH, Vu TH, Raffin TA. TGF-beta 1 causes increased endothelial ICAM-1 expression and lung injury. J Appl Physiol. 1994;77:1281–1287. doi: 10.1152/jappl.1994.77.3.1281. [DOI] [PubMed] [Google Scholar]
- 19.Jagels MA, Hugli TE. Mixed effects of TGF-beta on human airway epithelial-cell chemokine responses. Immunopharmacology. 2000;48:17–26. doi: 10.1016/s0162-3109(99)00190-3. [DOI] [PubMed] [Google Scholar]
- 20.Fong CY, Pang L, Holland E, Knox AJ. TGF-beta1 stimulates IL-8 release, COX-2 expression, and PGE(2) release in human airway smooth muscle cells. Am J Physiol Lung Cell Mol Physiol. 2000;279:L201–207. doi: 10.1152/ajplung.2000.279.1.L201. [DOI] [PubMed] [Google Scholar]
- 21.Pietras T, Panek M, Tworek D, Oszajca K, Wujcik R, Górski P, Kuna P, Szemraj J. The Bcl I single nucleotide polymorphism of the human glucocorticoid receptor gene h-GR/NR3C1 promoter in patients with bronchial asthma: pilot study. Mol Biol Rep. 2011;38:3953–3958. doi: 10.1007/s11033-010-0512-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panek M, Pietras T, Antczak A, Fabijan A, Przemęcka M, Górski P, Kuna P, Szemraj J. The N363S and I559N single nucleotide polymorphisms of the h-GR/NR3C1 gene in patients with bronchial asthma. Int J Mol Med. 2012;30:142–150. doi: 10.3892/ijmm.2012.956. [DOI] [PubMed] [Google Scholar]
- 23.Chomczynski P, Sacchi N. Single - step method of RNA isolation by acid guanidinium thiocyanate- phenol - chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 24.Winer J, Jung CK, Shackel I, Williams PM. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem. 1999;270:41–49. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]


