Abstract
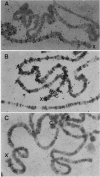
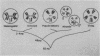
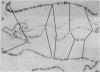
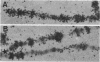
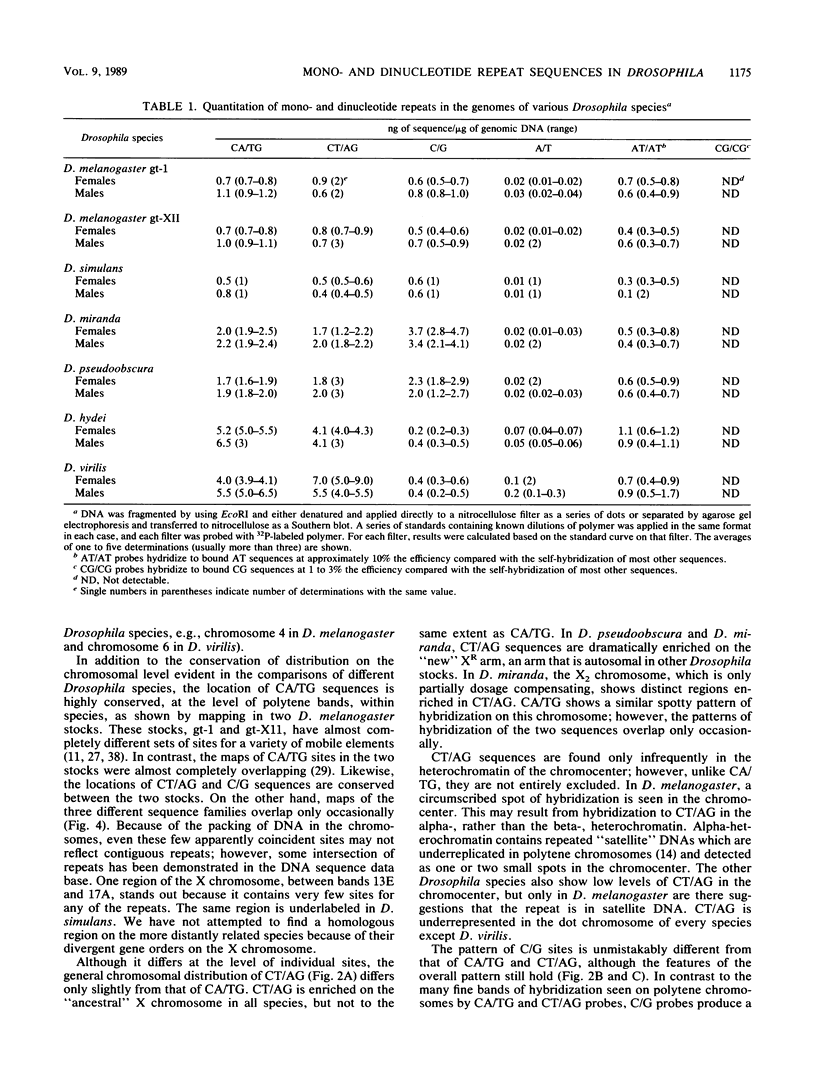
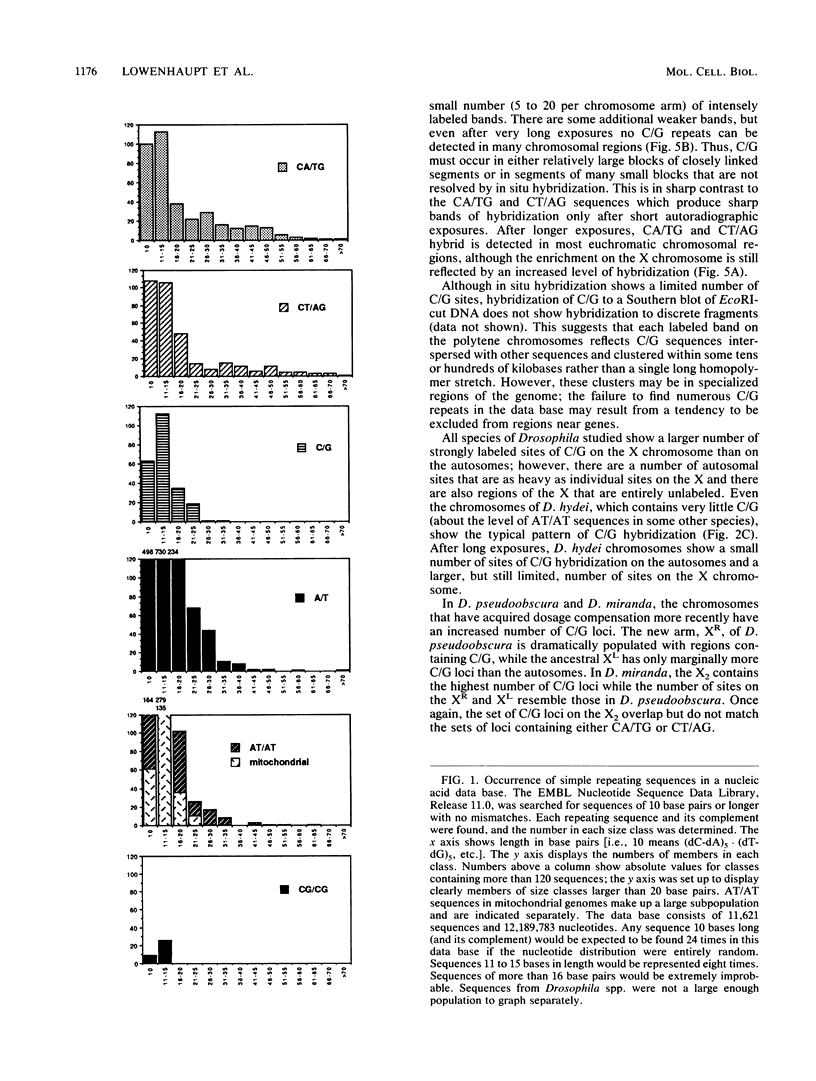
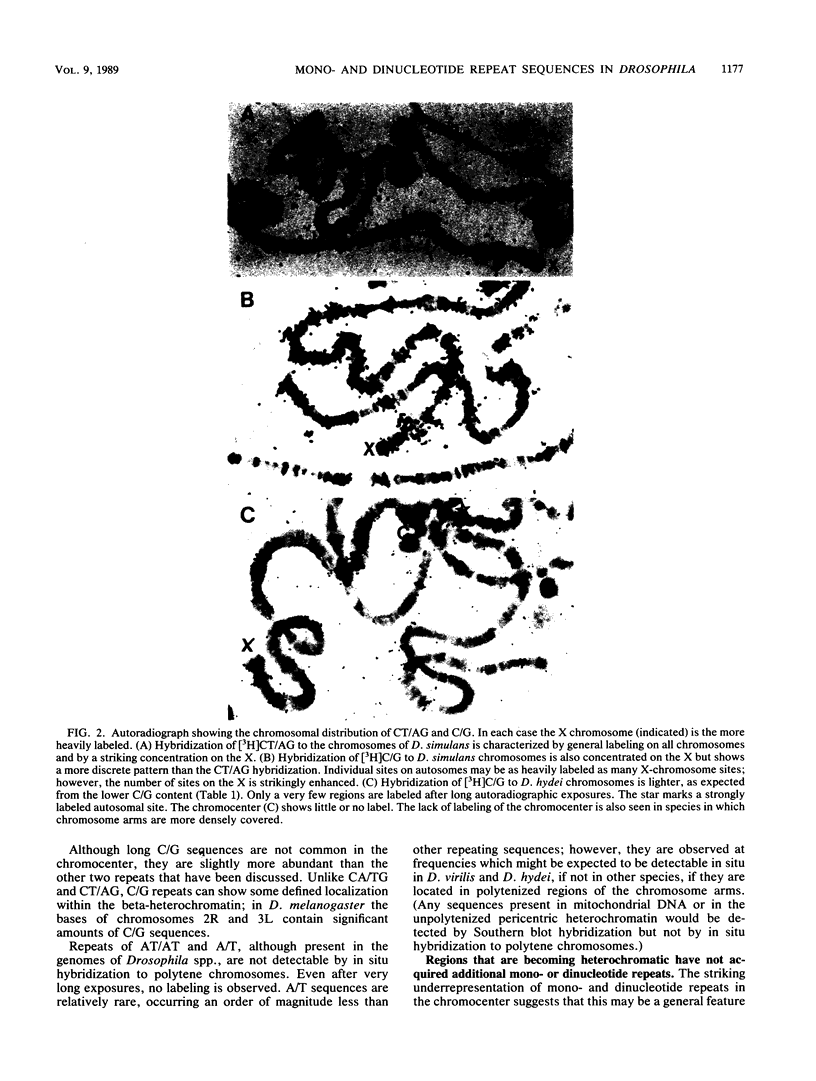
Long stretches of (dC-dA)n.(dT-dG)n, abbreviated CA/TG, have a distinctive distribution on Drosophila chromosomes (M.L. Pardue, K. Lowenhaupt, A. Rich, and A. Nordheim, EMBO J. 6:1781-1789, 1987). The distribution of CA/TG suggests a correlation with the overall transcriptional activity of chromosomal regions and with the ability to undergo meiotic recombination. These correlations are conserved among Drosophila species and may indicate one or more chromosomal functions. To test the generality of these findings, we analyzed the distribution of the rest of the six possible mono- and dinucleotide repeats (A/T, C/G, AT/AT, CA/TG, CT/AG, and CG/CG). All but CG/CG were present at significant levels in the genomes of the six Drosophila species studied; however, A/T levels were an order of magnitude lower than those of the other sequences. Data base analyses suggested that the same sequences are present in other eucaryotes. Like CA/TG, both CT/AG and C/G showed increased levels on dosage-compensating chromosomes; however, the individual sites clearly differed for each sequence. In contrast, A/T and AT/AT, although present in Drosophila DNA, could not be detected in situ in polytene chromosomes. We also used in situ hybridization to analyze the neo-Y chromosome of Drosophila miranda, an ancestral autosome that has become attached to the Y chromosome and is now partially heterochromatic. The neo-Y has acquired repeated DNA sequences; we found that the added sequences are as devoid of mono- and dinucleotide repeats as other heterochromatin. The distribution and function of these sequences are likely to result from both their repetitious nature and base contents.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abraham I., Lucchesi J. C. Dosage compensation of genes on the left and right arms of the X chromosome of Drosophila pseudoobscura and Drosophila willistoni. Genetics. 1974 Dec;78(4):1119–1126. doi: 10.1093/genetics/78.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker R. F. Binding of DNA to cellulose nitrate filters under denaturing conditions. Anal Biochem. 1977 Apr;78(2):569–571. doi: 10.1016/0003-2697(77)90119-1. [DOI] [PubMed] [Google Scholar]
- Braaten D. C., Thomas J. R., Little R. D., Dickson K. R., Goldberg I., Schlessinger D., Ciccodicola A., D'Urso M. Locations and contexts of sequences that hybridize to poly(dG-dT).(dC-dA) in mammalian ribosomal DNAs and two X-linked genes. Nucleic Acids Res. 1988 Feb 11;16(3):865–881. doi: 10.1093/nar/16.3.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock P., Miller J., Botchan M. Effects of poly[d(pGpT).d(pApC)] and poly[d(pCpG).d(pCpG)] repeats on homologous recombination in somatic cells. Mol Cell Biol. 1986 Nov;6(11):3948–3953. doi: 10.1128/mcb.6.11.3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor C. R., Efstratiadis A. Possible structures of homopurine-homopyrimidine S1-hypersensitive sites. Nucleic Acids Res. 1984 Nov 12;12(21):8059–8072. doi: 10.1093/nar/12.21.8059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chino M, Kikkawa H. Mutants and Crossing over in the Dot-like Chromosome of DROSOPHILA VIRILIS. Genetics. 1933 Mar;18(2):111–116. doi: 10.1093/genetics/18.2.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clary D. O., Wolstenholme D. R. The mitochondrial DNA molecular of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol. 1985;22(3):252–271. doi: 10.1007/BF02099755. [DOI] [PubMed] [Google Scholar]
- Dawid I. B., Long E. O., DiNocera P. P., Pardue M. L. Ribosomal insertion-like elements in Drosophila melanogaster are interspersed with mobile sequences. Cell. 1981 Aug;25(2):399–408. doi: 10.1016/0092-8674(81)90058-1. [DOI] [PubMed] [Google Scholar]
- Dowsett A. P., Young M. W. Differing levels of dispersed repetitive DNA among closely related species of Drosophila. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4570–4574. doi: 10.1073/pnas.79.15.4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauron C. M., Wolstenholme D. R. Intraspecific diversity of nucleotide sequences within the adenine + thymine-rich region of mitochondrial DNA molecules of Drosophila mauritiana, Drosophila melanogaster and Drosophila simulans. Nucleic Acids Res. 1980 Nov 25;8(22):5391–5410. doi: 10.1093/nar/8.22.5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall J. G., Cohen E. H., Polan M. L. Reptitive DNA sequences in drosophila. Chromosoma. 1971;33(3):319–344. doi: 10.1007/BF00284948. [DOI] [PubMed] [Google Scholar]
- Gross D. S., Garrard W. T. The ubiquitous potential Z-forming sequence of eucaryotes, (dT-dG)n . (dC-dA)n, is not detectable in the genomes of eubacteria, archaebacteria, or mitochondria. Mol Cell Biol. 1986 Aug;6(8):3010–3013. doi: 10.1128/mcb.6.8.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H., Petrino M. G., Kakunaga T. A novel repeated element with Z-DNA-forming potential is widely found in evolutionarily diverse eukaryotic genomes. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6465–6469. doi: 10.1073/pnas.79.21.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D., Morgan A. R. Unique structures formed by pyrimidine-purine DNAs which may be four-stranded. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1637–1641. doi: 10.1073/pnas.75.4.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Larsen A., Weintraub H. An altered DNA conformation detected by S1 nuclease occurs at specific regions in active chick globin chromatin. Cell. 1982 Jun;29(2):609–622. doi: 10.1016/0092-8674(82)90177-5. [DOI] [PubMed] [Google Scholar]
- Macknight R H. The Sex-Determining Mechanism of Drosophila Miranda. Genetics. 1939 Mar;24(2):180–201. doi: 10.1093/genetics/24.2.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A. S., Chatterjee S. N. Chromosomal basis of dosage compensation in Drosophila VIII. Faster replication and hyperactivity of both arms of the X-chromosome in males of Drosophila pseudoobscura and their possible significance. Chromosoma. 1975 Nov 24;53(2):91–105. doi: 10.1007/BF00333038. [DOI] [PubMed] [Google Scholar]
- Nordheim A., Pardue M. L., Weiner L. M., Lowenhaupt K., Scholten P., Möller A., Rich A., Stollar B. D. Analysis of Z-DNA in fixed polytene chromosomes with monoclonal antibodies that show base sequence-dependent selectivity in reactions with supercoiled plasmids and polynucleotides. J Biol Chem. 1986 Jan 5;261(1):468–476. [PubMed] [Google Scholar]
- Pardue M. L., Dawid I. B. Chromosomal locations of two DNA segments that flank ribosomal insertion-like sequences in Drosophila: flanking sequences are mobile elements. Chromosoma. 1981;83(1):29–43. doi: 10.1007/BF00286014. [DOI] [PubMed] [Google Scholar]
- Pardue M. L., Fostel J. M., Cech T. R. DNA-protein interactions in the Drosophila virilis mitochondrial chromosome. Nucleic Acids Res. 1984 Feb 24;12(4):1991–1999. doi: 10.1093/nar/12.4.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardue M. L., Lowenhaupt K., Rich A., Nordheim A. (dC-dA)n.(dG-dT)n sequences have evolutionarily conserved chromosomal locations in Drosophila with implications for roles in chromosome structure and function. EMBO J. 1987 Jun;6(6):1781–1789. doi: 10.1002/j.1460-2075.1987.tb02431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulleyblank D. E., Haniford D. B., Morgan A. R. A structural basis for S1 nuclease sensitivity of double-stranded DNA. Cell. 1985 Aug;42(1):271–280. doi: 10.1016/s0092-8674(85)80122-7. [DOI] [PubMed] [Google Scholar]
- Steinemann M. Multiple sex chromosomes in Drosophila miranda: a system to study the degeneration of a chromosome. Chromosoma. 1982;86(1):59–76. doi: 10.1007/BF00330730. [DOI] [PubMed] [Google Scholar]
- Strobel E., Pelling C., Arnheim N. Incomplete dosage compensation in an evolving Drosophila sex chromosome. Proc Natl Acad Sci U S A. 1978 Feb;75(2):931–935. doi: 10.1073/pnas.75.2.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treco D., Arnheim N. The evolutionarily conserved repetitive sequence d(TG.AC)n promotes reciprocal exchange and generates unusual recombinant tetrads during yeast meiosis. Mol Cell Biol. 1986 Nov;6(11):3934–3947. doi: 10.1128/mcb.6.11.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young B. S., Pession A., Traverse K. L., French C., Pardue M. L. Telomere regions in Drosophila share complex DNA sequences with pericentric heterochromatin. Cell. 1983 Aug;34(1):85–94. doi: 10.1016/0092-8674(83)90138-1. [DOI] [PubMed] [Google Scholar]
- Young M. W., Schwartz H. E. Nomadic gene families in Drosophila. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):629–640. doi: 10.1101/sqb.1981.045.01.081. [DOI] [PubMed] [Google Scholar]