Abstract
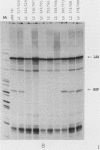
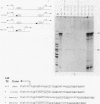
Human mitochondrial DNA contains two major promoters, one for transcription of each strand of the helix. Previous mapping and mutagenesis data have localized these regulatory elements and have suggested regions important to their function. In order to define, at high resolution, the sequences critical for accurate and efficient transcriptional initiation, a linker substitution analysis of the entire promoter region was performed. Each promoter was shown to consist of approximately 50 base pairs comprising two functionally distinct elements. These and previous data strongly support a mode of transcription initiation requiring minimal sequences surrounding the initiation sites that are likely interactive with core polymerase and upstream regulatory domains capable of binding a transcription factor that modulates the efficiency of transcription initiation. Furthermore, in at least one case, this upstream regulatory domain is capable of operating bidirectionally.
Full text
PDF
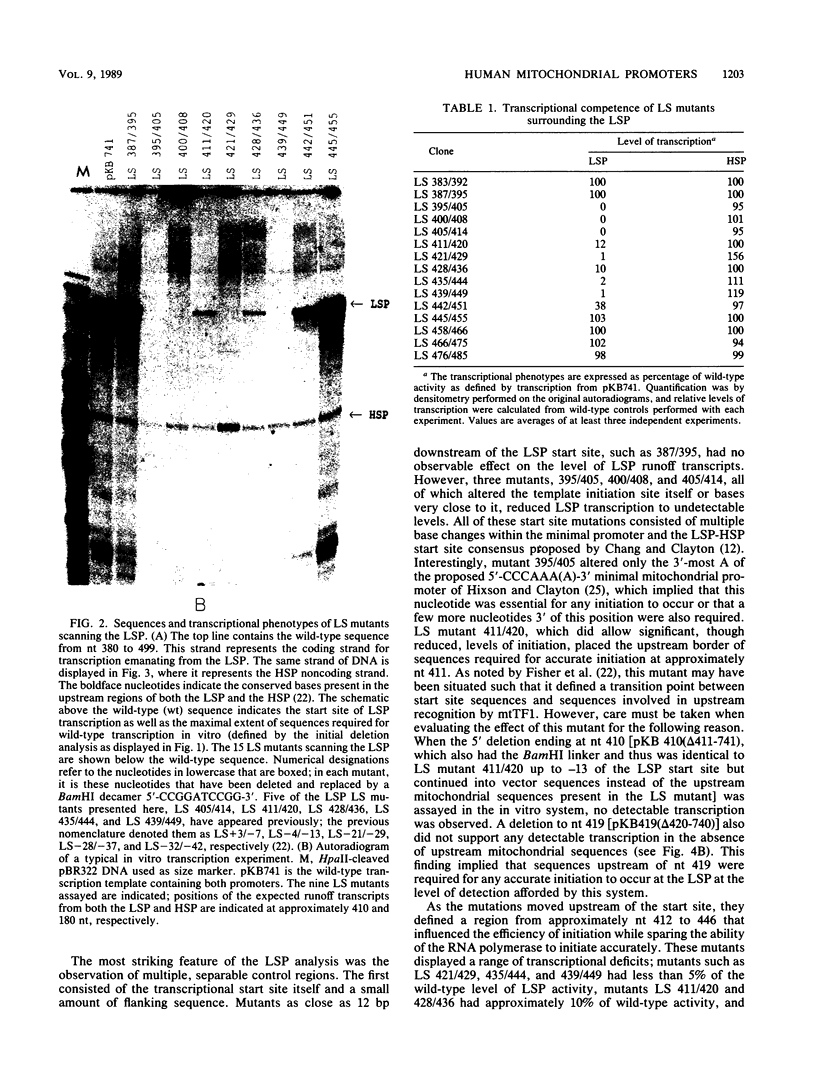
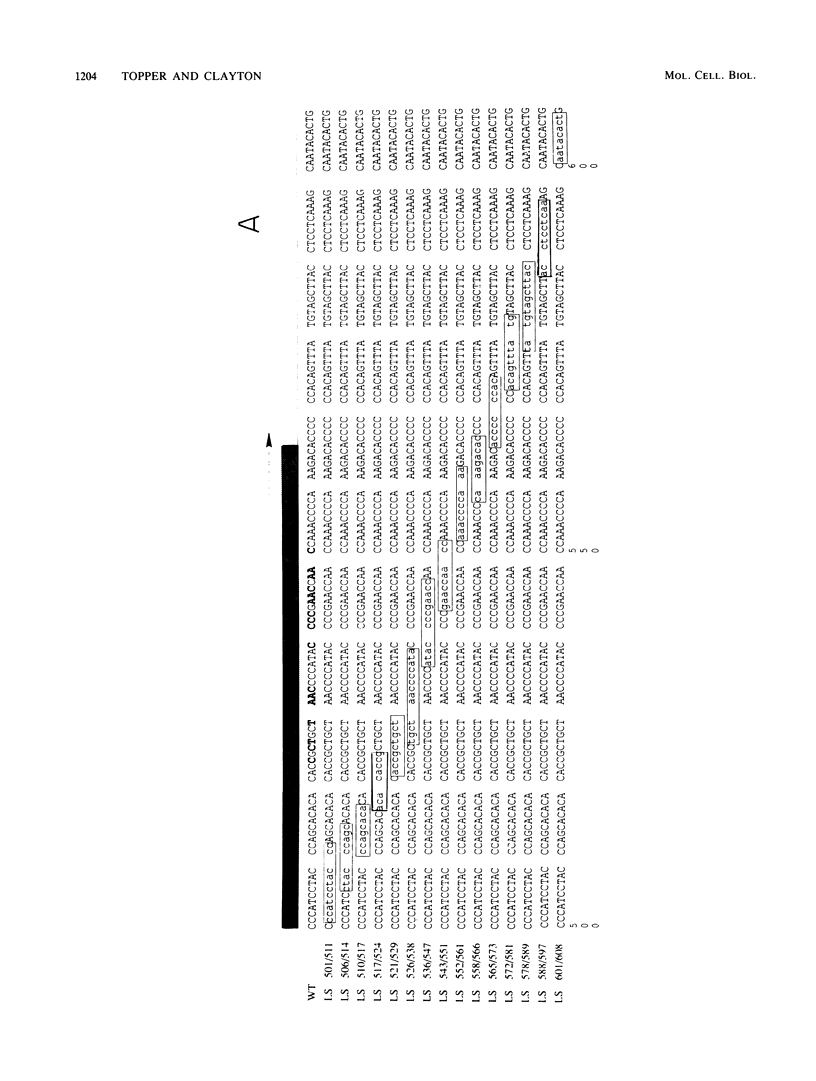
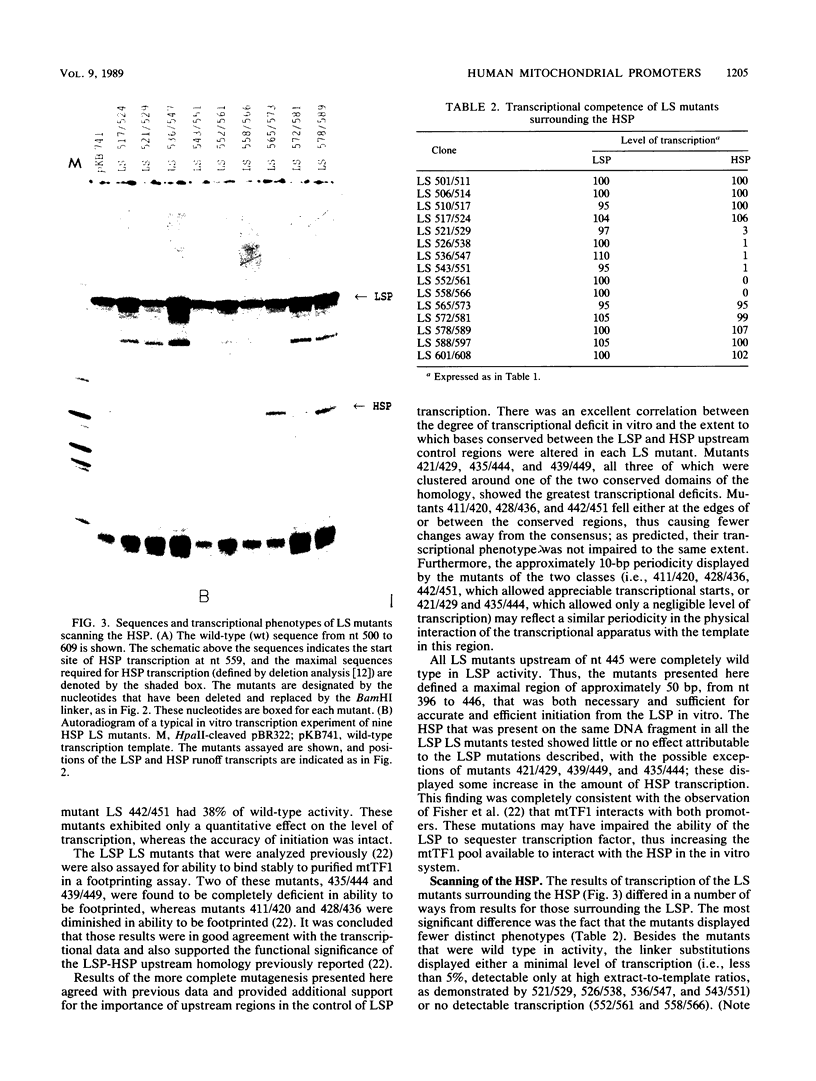

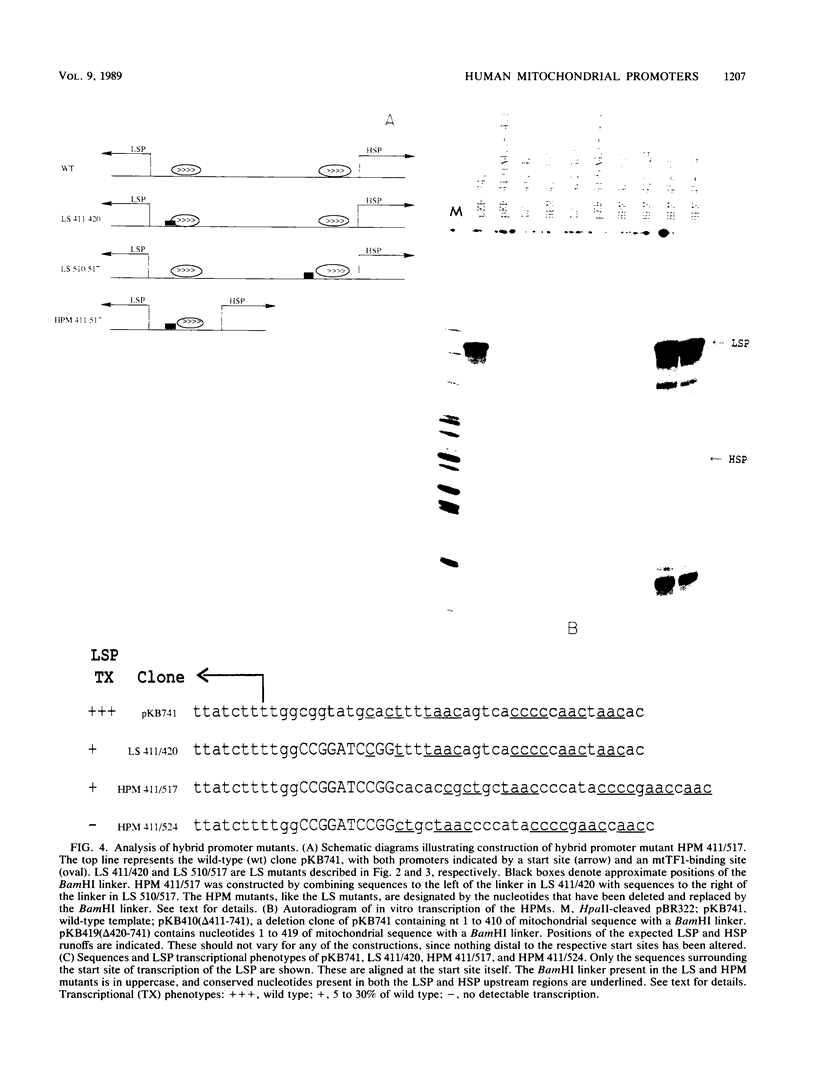




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison L. A., Moyle M., Shales M., Ingles C. J. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985 Sep;42(2):599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- Barrera-Saldana H., Takahashi K., Vigneron M., Wildeman A., Davidson I., Chambon P. All six GC-motifs of the SV40 early upstream element contribute to promoter activity in vivo and in vitro. EMBO J. 1985 Dec 30;4(13B):3839–3849. doi: 10.1002/j.1460-2075.1985.tb04156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggs J., Searles L. L., Greenleaf A. L. Structure of the eukaryotic transcription apparatus: features of the gene for the largest subunit of Drosophila RNA polymerase II. Cell. 1985 Sep;42(2):611–621. doi: 10.1016/0092-8674(85)90118-7. [DOI] [PubMed] [Google Scholar]
- Biswas T. K., Edwards J. C., Rabinowitz M., Getz G. S. Characterization of a yeast mitochondrial promoter by deletion mutagenesis. Proc Natl Acad Sci U S A. 1985 Apr;82(7):1954–1958. doi: 10.1073/pnas.82.7.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas T. K., Ticho B., Getz G. S. In vitro characterization of the yeast mitochondrial promoter using single-base substitution mutants. J Biol Chem. 1987 Oct 5;262(28):13690–13696. [PubMed] [Google Scholar]
- Bogenhagen D. F., Applegate E. F., Yoza B. K. Identification of a promoter for transcription of the heavy strand of human mtDNA: in vitro transcription and deletion mutagenesis. Cell. 1984 Apr;36(4):1105–1113. doi: 10.1016/0092-8674(84)90061-8. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Insdorf N. F. Purification of Xenopus laevis mitochondrial RNA polymerase and identification of a dissociable factor required for specific transcription. Mol Cell Biol. 1988 Jul;8(7):2910–2916. doi: 10.1128/mcb.8.7.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen D. F., Romanelli M. F. Template sequences required for transcription of Xenopus laevis mitochondrial DNA from two bidirectional promoters. Mol Cell Biol. 1988 Jul;8(7):2917–2924. doi: 10.1128/mcb.8.7.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen D. F., Yoza B. K. Accurate in vitro transcription of Xenopus laevis mitochondrial DNA from two bidirectional promoters. Mol Cell Biol. 1986 Jul;6(7):2543–2550. doi: 10.1128/mcb.6.7.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Carbon P., Murgo S., Ebel J. P., Krol A., Tebb G., Mattaj L. W. A common octamer motif binding protein is involved in the transcription of U6 snRNA by RNA polymerase III and U2 snRNA by RNA polymerase II. Cell. 1987 Oct 9;51(1):71–79. doi: 10.1016/0092-8674(87)90011-0. [DOI] [PubMed] [Google Scholar]
- Chang D. D., Clayton D. A. Identification of primary transcriptional start sites of mouse mitochondrial DNA: accurate in vitro initiation of both heavy- and light-strand transcripts. Mol Cell Biol. 1986 May;6(5):1446–1453. doi: 10.1128/mcb.6.5.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D. D., Clayton D. A. Precise assignment of the heavy-strand promoter of mouse mitochondrial DNA: cognate start sites are not required for transcriptional initiation. Mol Cell Biol. 1986 Sep;6(9):3262–3267. doi: 10.1128/mcb.6.9.3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D. D., Clayton D. A. Precise assignment of the light-strand promoter of mouse mitochondrial DNA: a functional promoter consists of multiple upstream domains. Mol Cell Biol. 1986 Sep;6(9):3253–3261. doi: 10.1128/mcb.6.9.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D. D., Clayton D. A. Precise identification of individual promoters for transcription of each strand of human mitochondrial DNA. Cell. 1984 Mar;36(3):635–643. doi: 10.1016/0092-8674(84)90343-x. [DOI] [PubMed] [Google Scholar]
- Chang D. D., Hixson J. E., Clayton D. A. Minor transcription initiation events indicate that both human mitochondrial promoters function bidirectionally. Mol Cell Biol. 1986 Jan;6(1):294–301. doi: 10.1128/mcb.6.1.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clary D. O., Wolstenholme D. R. The mitochondrial DNA molecular of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol. 1985;22(3):252–271. doi: 10.1007/BF02099755. [DOI] [PubMed] [Google Scholar]
- Clayton D. A. Transcription of the mammalian mitochondrial genome. Annu Rev Biochem. 1984;53:573–594. doi: 10.1146/annurev.bi.53.070184.003041. [DOI] [PubMed] [Google Scholar]
- Fisher R. P., Clayton D. A. A transcription factor required for promoter recognition by human mitochondrial RNA polymerase. Accurate initiation at the heavy- and light-strand promoters dissected and reconstituted in vitro. J Biol Chem. 1985 Sep 15;260(20):11330–11338. [PubMed] [Google Scholar]
- Fisher R. P., Clayton D. A. Purification and characterization of human mitochondrial transcription factor 1. Mol Cell Biol. 1988 Aug;8(8):3496–3509. doi: 10.1128/mcb.8.8.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. P., Topper J. N., Clayton D. A. Promoter selection in human mitochondria involves binding of a transcription factor to orientation-independent upstream regulatory elements. Cell. 1987 Jul 17;50(2):247–258. doi: 10.1016/0092-8674(87)90220-0. [DOI] [PubMed] [Google Scholar]
- Gidoni D., Kadonaga J. T., Barrera-Saldaña H., Takahashi K., Chambon P., Tjian R. Bidirectional SV40 transcription mediated by tandem Sp1 binding interactions. Science. 1985 Nov 1;230(4725):511–517. doi: 10.1126/science.2996137. [DOI] [PubMed] [Google Scholar]
- Haltiner M., Kempe T., Tjian R. A novel strategy for constructing clustered point mutations. Nucleic Acids Res. 1985 Feb 11;13(3):1015–1025. doi: 10.1093/nar/13.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hixson J. E., Clayton D. A. Initiation of transcription from each of the two human mitochondrial promoters requires unique nucleotides at the transcriptional start sites. Proc Natl Acad Sci U S A. 1985 May;82(9):2660–2664. doi: 10.1073/pnas.82.9.2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kownin P., Bateman E., Paule M. R. Eukaryotic RNA polymerase I promoter binding is directed by protein contacts with transcription initiation factor and is DNA sequence-independent. Cell. 1987 Aug 28;50(5):693–699. doi: 10.1016/0092-8674(87)90327-8. [DOI] [PubMed] [Google Scholar]
- Lassar A. B., Martin P. L., Roeder R. G. Transcription of class III genes: formation of preinitiation complexes. Science. 1983 Nov 18;222(4625):740–748. doi: 10.1126/science.6356356. [DOI] [PubMed] [Google Scholar]
- Lennard A. C., Egly J. M. The bidirectional upstream element of the adenovirus-2 major late promoter binds a single monomeric molecule of the upstream factor. EMBO J. 1987 Oct;6(10):3027–3034. doi: 10.1002/j.1460-2075.1987.tb02608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters B. S., Stohl L. L., Clayton D. A. Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell. 1987 Oct 9;51(1):89–99. doi: 10.1016/0092-8674(87)90013-4. [DOI] [PubMed] [Google Scholar]
- McClure W. R. Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Kingsbury R. Transcriptional control signals of a eukaryotic protein-coding gene. Science. 1982 Jul 23;217(4557):316–324. doi: 10.1126/science.6283634. [DOI] [PubMed] [Google Scholar]
- Murphy S., Di Liegro C., Melli M. The in vitro transcription of the 7SK RNA gene by RNA polymerase III is dependent only on the presence of an upstream promoter. Cell. 1987 Oct 9;51(1):81–87. doi: 10.1016/0092-8674(87)90012-2. [DOI] [PubMed] [Google Scholar]
- Pieler T., Hamm J., Roeder R. G. The 5S gene internal control region is composed of three distinct sequence elements, organized as two functional domains with variable spacing. Cell. 1987 Jan 16;48(1):91–100. doi: 10.1016/0092-8674(87)90359-x. [DOI] [PubMed] [Google Scholar]
- Schinkel A. H., Koerkamp M. J., Touw E. P., Tabak H. F. Specificity factor of yeast mitochondrial RNA polymerase. Purification and interaction with core RNA polymerase. J Biol Chem. 1987 Sep 15;262(26):12785–12791. [PubMed] [Google Scholar]
- Sollner-Webb B., Tower J. Transcription of cloned eukaryotic ribosomal RNA genes. Annu Rev Biochem. 1986;55:801–830. doi: 10.1146/annurev.bi.55.070186.004101. [DOI] [PubMed] [Google Scholar]
- Sommerville J. RNA polymerase I promoters and transcription factors. Nature. 1984 Jul 19;310(5974):189–190. doi: 10.1038/310189a0. [DOI] [PubMed] [Google Scholar]
- Walberg M. W., Clayton D. A. In vitro transcription of human mitochondrial DNA. Identification of specific light strand transcripts from the displacement loop region. J Biol Chem. 1983 Jan 25;258(2):1268–1275. [PubMed] [Google Scholar]
- de Vries E., van Driel W., van den Heuvel S. J., van der Vliet P. C. Contactpoint analysis of the HeLa nuclear factor I recognition site reveals symmetrical binding at one side of the DNA helix. EMBO J. 1987 Jan;6(1):161–168. doi: 10.1002/j.1460-2075.1987.tb04734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]