Abstract
In many biological systems, variability of the components can be expected to outrank statistical fluctuations in the shaping of self-organized patterns. In pioneering work in the late 1990s, it was hypothesized that a drift of cellular parameters (along a ‘developmental path’), together with differences in cell properties (‘desynchronization’ of cells on the developmental path) can establish self-organized spatio-temporal patterns (in their example, spiral waves of cAMP in a colony of Dictyostelium discoideum cells) starting from a homogeneous state. Here, we embed a generic model of an excitable medium, a lattice of diffusively coupled FitzHugh–Nagumo oscillators, into a developmental-path framework. In this minimal model of spiral wave generation, we can now study the predictability of spatio-temporal patterns from cell properties as a function of desynchronization (or ‘spread’) of cells along the developmental path and the drift speed of cell properties on the path. As a function of drift speed and desynchronization, we observe systematically different routes towards fully established patterns, as well as strikingly different correlations between cell properties and pattern features. We show that the predictability of spatio-temporal patterns from cell properties contains important information on the pattern formation process as well as on the underlying dynamical system.
Keywords: FitzHugh–Nagumo, developmental path, spatio-temporal pattern formation, variability, spiral waves, Dictyostelium discoideum
1. Introduction
1.1. Spatio-temporal pattern formation and biological variability
Spatio-temporal pattern formation is an essential component of studies of active media, in contexts encompassing various disparate fields of chemistry, physics and biology [1–3]. As self-sustaining structures, spiral waves are one of the most prominent manifestations of such patterns, and have been observed in several contexts in the realm of biological systems [4]. Perhaps the most notably studied case is the paradigmatic example of spiral waves in the life cycle of the social amoeba, Dictyostelium discoideum, when waves of cyclic adenosine monophosphate (cAMP) spread through a colony during starvation conditions, leading to the emergence of aggregation centres and thus triggering subsequent stages of the organisms' life cycle.
In biology, variability in the form of cell-to-cell differences (in some dynamically relevant parameter) is expected to contribute more significantly than stochastic noise to the heterogeneity in a biological system [5]. With the example of Dictyostelium in mind, such cell properties include the response to an incoming cAMP signal (‘excitability’), cell motility and cAMP receptor density (‘sensitivity’).
The potential importance of variability in biological pattern formation is one of the most striking differences between biological systems and those in physics or chemistry, where system components are essentially identical and random fluctuations determine the details of the self-organization process and the self-organized patterns. This has far-reaching implications for our understanding of biological systems, as, in biology, patterns are linked to function. The patterns frequently constitute a (precursor of a) collective systemic mode and are thus subject to evolutionary selection. Variability is one means of ‘implementing’ a single-element control on the collective mode. Sawai et al. [6] have convincingly made this case for Dictyostelium, relating the strength of a regulatory feedback loop to the spatial density of spiral wave patterns. By studying mutants in key components of feedback, the authors could vary this intrinsic parameter systematically and observe how the spatio-temporal patterns change accordingly. Their main finding is that wild-type feedback strength is optimized to yield maximal aggregation territory size. This allows for optimally sized basins of attraction for the consecutive aggregation process of the Dictyostelium cells in the formation of the multicellular stage capable of spore generation that completes the developmental cycle [6].
1.2. Theoretical investigations of biological variability
The fact that similar to noise, variability can induce patterns or trigger a transition from one pattern to another has been a focus of research in the 1990s and early 2000s. In the more theoretically oriented studies, our biologically motivated term ‘variability’ often appears as ‘diversity’, ‘disorder’ or ‘quenched noise’. In Glatt et al. [7], it is shown that variability can induce a phase transition in a system of coupled FitzHugh–Nagumo (FHN) oscillators from oscillatory behaviour to excitability. In Hütt et al. [8], the effect of variability on spatio-temporal patterns in a lattice of coupled biochemical oscillators has been investigated, showing that pattern complexity is highest at intermediate variability. These studies and others culminated in the phenomenon of diversity-induced resonance [9], where the response of an excitable or bistable system to an external subthreshold stimulus is optimal at intermediate levels of diversity. The work in Gassel et al. [10] extends the theoretical scope by exploring the interplay of additive and multiplicative variability in the context of diversity-induced resonance. In Tessone & Toral [11], a diversity-induced resonance was also observed in a model of opinion formation attributing a dynamical role to social diversity. In Marko [12], such a diversity-induced resonance was found in simulations of calcium dynamics, showing that waves propagate optimally at intermediate cell-to-cell variability, thus considering the effect of variability on spatio-temporal patterns. Relatedly, on a more theoretical level, in Glatt et al. [13], it is shown that parameter variability in a subexcitable network of FHN oscillators can induce pattern formation in the form of spiral waves of excitation.
1.3. Relevance for Dictyostelium pattern formation
Beyond these works about the general impact of variability on dynamics, the influence of individual element properties on the macroscopic pattern development has received comparatively little attention. In particular, the question of how a spatial distribution of cell properties affects the emerging spatio-temporal patterns has, to the best of our knowledge, only been explored in references [14–17]. Some of these previous investigations discussed the prediction of spatio-temporal patterns from cell properties. For two models of Dictyostelium cAMP signalling, this has been conducted in Geberth et al. [15] (for the model from Levine et al. [18]) and in Geberth et al. [17] (for the model from Lauzeral et al. [19]).
Our motivation for this study is to design a minimal model representation guided by these previous works in order to better understand how cell properties are translated into pattern features in the course of the self-organization process.
Strong support for the predictability of patterns from cell properties arises from the observation that the direction and magnitude of a D. discoideum cell's response to a signal pulse is indeed a cell property, which is not randomly fluctuating but is preserved during the course of the cells' development [5] and therefore falls into the general scheme outlined within this study. In that work, the behaviour of single cells under periodic cAMP signals was analysed, leading to the observation that the characteristics of the gradient sensing response of an individual cell at a certain time point strongly correlate with that of the same cell at a later time point.
1.4. Developmental path model
The developmental path discussed in the following is a biologically motivated mechanism for translating variability into heterogeneity acting upon the patterns. Heterogeneity can be seen as a prerequisite for spiral wave formation (see [20] as well as the corresponding debate in [19,21]). The ‘developmental path’ concept was proposed by Lauzeral et al. [19], in the context of the model of D. discoideum pattern formation developed by Martiel & Goldbeter [22] (see [23]). In this ‘developmental path’, specified cell properties follow a defined trajectory over time, with this variation leading cells successively through quiescent, excitable, oscillatory and excitable regimes of dynamical behaviour. Desynchronization of the cells' properties on this path then provides the necessary cell-to-cell differences for spiral waves to form. This path incorporates variation in adenylate cyclase and phosphodiesterase, in correspondence to experimental observations. The developmental path concept thus provides a mechanism for the generation of variability from homogeneous initial conditions. This model was shown to successfully reproduce the sequence of macroscale patterning observed in experimental Dictyostelium colonies.
1.5. Outline of our investigation
In order to explore the generalizability of the developmental-path concept to a simple model of excitable dynamics and the mechanisms by which heterogeneity can result in spiral wave production, we construct a selection of analogous ‘developmental paths’ in a lattice of FHN oscillators. The parsimonious structure of the FHN model allows us to search for the generic principles of how cell properties constrain the emerging spatio-temporal patterns. In this minimal model of spiral wave generation, we can now study pattern predictability as a function of desynchronization (or ‘spread’) of cells along the developmental path and the drift speed on the developmental path. Focusing on four cases: high speed/high spread (++), high speed/low spread (+−), low speed/high spread (−+) and low speed/low spread (−−), we find very different sequences of events leading to established patterns, as well as strikingly different correlations between cell properties and pattern features. On these grounds, we advocate an event-based view of spatio-temporal patterns, where, over time, pattern types give rise to other pattern types in a predictable and distinct sequential manner depending on the properties of the developmental path, establishing causal links between the original information: the cell properties, and the later-stage asymptotic information: the distribution of spirals in the fully developed pattern.
2. Methods
2.1. Mathematical model
We chose the FHN model of neuronal excitation [24,25] as an established minimal model of excitable dynamics and a low-dimensional system of ordinary differential equations. The FHN equations for a set of spatially coupled oscillators forming a two-dimensional lattice with diffusion in one of the two dynamical variables uij are
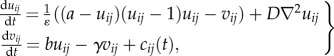 |
2.1 |
where the uij are the voltage-like variable at each lattice site, the vij denote a recovery variable, cij(t) represents the biological variability (in the dependence on i and j) and the shift of cell properties along a developmental path and D is the diffusion constant. Depending on the values of the governing parameters, each FHN element may be excitable, oscillatory or in a non-excitable steady state. In the following, the model parameters take the values a = −1, γ = b = 0.12, 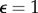 and D = 0.1. In equation (2.1), ∇2 represents the discretized two-dimensional Laplacian for diffusion in uij. We here use a five-point discretization:
and D = 0.1. In equation (2.1), ∇2 represents the discretized two-dimensional Laplacian for diffusion in uij. We here use a five-point discretization:
| 2.2 |
No explicit spatial scale is considered.
2.2. Developmental paths
We explored a wide range of candidates for developmental paths and then selected the class of (one-dimensional) paths incorporating temporal variation in the parameter cij(t). This set-up proved most reliable in producing an asymptotic pattern of fully developed spiral waves, while the reduction to a one-dimensional variation allows translation of the original concept to a truly minimal system. The parameter cij(t) determines the point at which the linear  -nullcline crosses the cubic
-nullcline crosses the cubic  -nullcline (figure 1): varying cij(t) alters the excitability of the corresponding cells. The parameters are chosen so that only one fixed point exists.
-nullcline (figure 1): varying cij(t) alters the excitability of the corresponding cells. The parameters are chosen so that only one fixed point exists.
Figure 1.
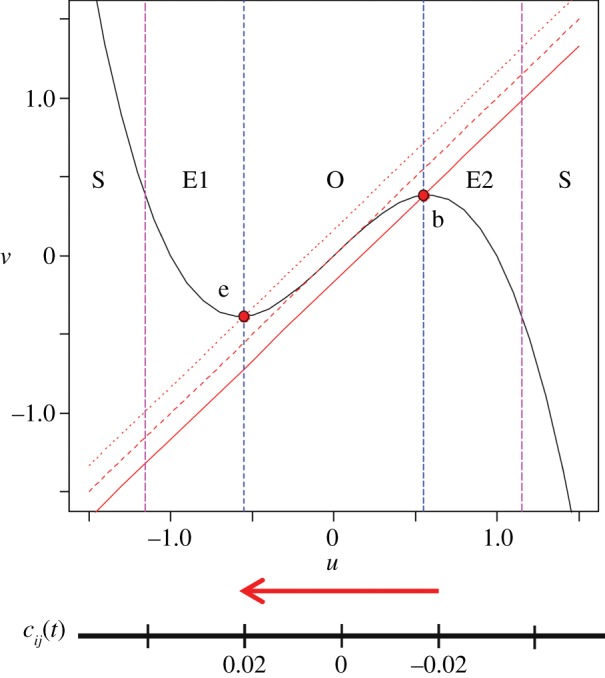
Nullclines of the single FitzHugh–Nagumo oscillator at the beginning (b), midpoint and end (e) of the developmental path. The dynamical regimes are non-excitable steady state (S), excitable (E1 and E2) and oscillatory behaviour (O). The one-dimensional path in cij(t) shown below the x-axis involves variation of cij(t) between−0.02 (b) and 0.02 (e), causing a shift in the linear nullcline. (Online version in colour.)
The parameter c varies with time in a sigmoidal fashion, similar to the parametrization in Lauzeral et al. [19]:
| 2.3 |
where t is the ‘global time’ from equation (2.1). In the FHN model, the value of c0 determines the dynamical regimes encompassed by the path. By setting the value of c0 to 0.02, the developmental paths were restricted to the oscillatory regime, as the minimal system in which sustained spiral waves appeared viable. Hence, the lattice values of cij(t) vary between approximately −0.02 at the start of the developmental paths and 0.02 at the end. At the chosen parameter values, a value of 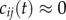 leads to the maximal oscillatory frequency.
leads to the maximal oscillatory frequency.
Each cell receives its own time offset Δtij on this path, thus implementing a distribution of cell properties over the lattice. Following the scheme from Lauzeral et al. [19], this time offset is randomly drawn from an exponential distribution:
| 2.4 |
where Δ is a desynchronization parameter, determining the spread of time offsets Δtij.
A simple comparison of dimensions between our construct and the set-up from Lauzeral et al. [19] illustrates the minimal character of our construct, combining the FHN model with a developmental path, which translates a pattern of cell-to-cell differences into a heterogeneous, drifting input for the pattern formation process: the model is two-dimensional (compared with the three-dimensional model in Lauzeral et al. [19]), it has five parameters (compared with 15 in Lauzeral et al. [19]) and we use a one-dimensional developmental path (compared with the two-dimensional paths in Lauzeral et al. [19]).
The parameter drift in cij(t) (referred to as c from here), together with the desynchronization of cells across this drift, provides a method of generating the heterogeneity in excitability needed to trigger the pattern formation process. In the following, we will explore how this heterogeneity influences the emerging spatio-temporal patterns. Our two main questions are: (i) how much information does the distribution of cell properties contain about the asymptotic patterns (pattern predictability) and (ii) how do the patterns vary with the parameters of the developmental path?
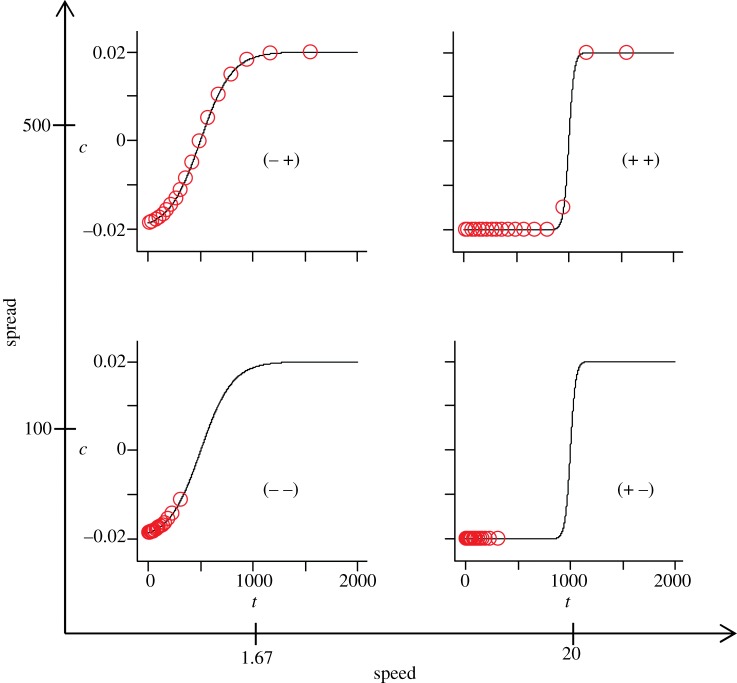
In order to characterize the properties of the developmental path route to spiral formation, we systematically vary two features: the parameter Δ, which controls the spread of cells on the developmental path, and the shape of the path, determined by the function c(t) (figure 2). This function varies with the parameters tc and Tc, which determine the initial slope of the path and the general steepness of the slope: we designate these two features as ‘spread’ and ‘speed’, respectively. Four paths are then selected as representing distinct features of interest in terms of the mechanisms observed en route to pattern formation.
Figure 2.
The four developmental path case studies with the starting distribution of the property c across the FHN lattice elements for each path, arranged in a matrix with differing values of ‘spread’ (the desynchronization parameter Δ, y-axis) and ‘speed’, the ratio of initial slope to general slope steepness (tc /Tc ). Each descriptor of the form e.g. ( −+) refers to low (−) or high (+) values of the parameters ‘speed’ and ‘spread’, respectively. One circle represents a group of 5% of the elements. (Online version in colour.)
2.3. Numerical simulation and data analysis
We use a custom-developed software suite to recognize the locations of spiral wave tips and target wave origins, described in Geberth & Hütt [26]. Spiral wave tips are identified with the phase singularity method from Bray et al. [27], then this signal is filtered to remove double tip recognition artefacts, whereas target wave origins are detected by fitting a three-dimensional cone to waves in the space–time cube (see [26] for details). Our resulting space–time event plots, combined with observations of the development of the field u, allow insights into the behaviour of the system along the different developmental paths. These are inspected for 50 different sets of time offsets for each path.
In order to assess the statistical relationships between cell properties and spiral and target wave distributions, and thus gain an understanding of the underlying mechanisms, 100 developmental runs with different time offset distributions for each path are simulated, with the initial values of u and v randomly selected between −0.01 and 0.01. The correlation coefficients corresponding to the correlations between the varying cell property parameter c, the locations of target wave origins and the positions of spiral wave tips are computed, together with mean values of the time-averaged spatial densities of spiral wave tips and target wave origins. To approximate the relationships between signals on different spatial scales, we subject the signals to varying degrees of Gaussian averaging (‘blurring’) (σ ranging from 0.5 to 10 pixels, corresponding to lattice sites), using the R statistical computing environment [28] with the packages ‘Hmisc’ for correlation calculations [29] and ‘spatstat’ [30] for the blurring algorithm. Correlation coefficients with p-values greater than or equal to 0.05 were removed from the corresponding calculations.
Signals occurring in the first 200 timesteps are not included in the calculations, as this is considered the transient period and the edges of the lattice are not considered to avoid artefacts from the event recognition software. We perform our simulations on a 100×100 lattice with periodic boundary conditions, using the Euler method (step size = 0.01).
Owing to the highly variable and interconnected nature of the dynamics we observe, our efforts to understand pattern predictability in this study thus combine the close examination of spatio-temporal event sequences and quantitative correlation analyses.
3. Results
3.1. Spatio-temporal organization of pattern events
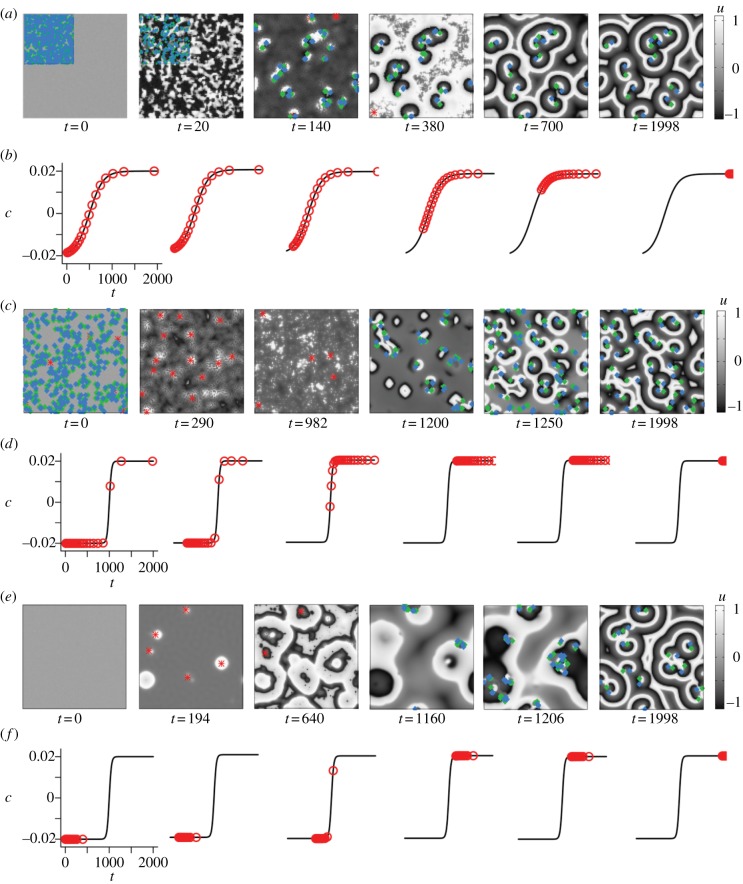
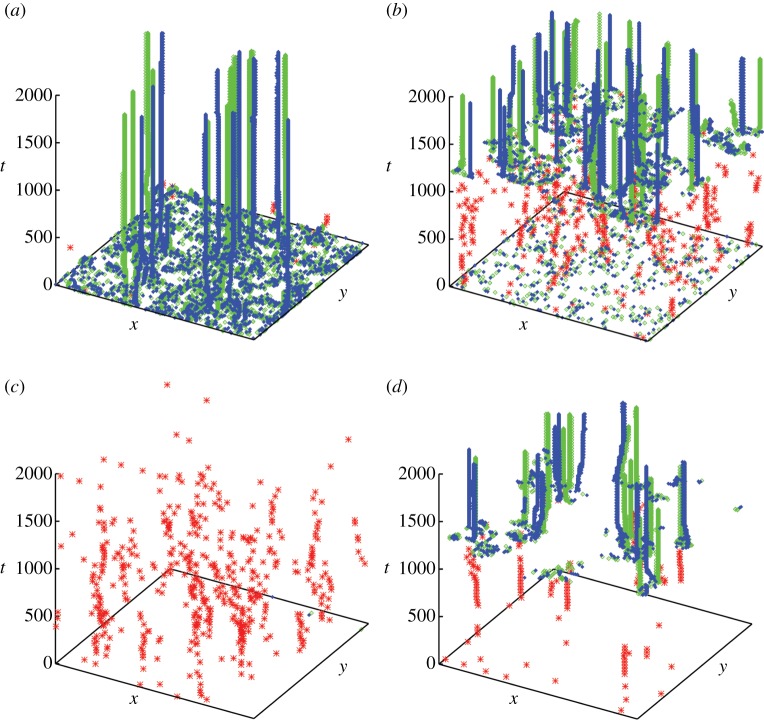
Our first assessment of the developmental paths was based on the dynamics of the spatio-temporal patterns unfolding on the lattice for each of the 100 time offset realizations applied to each path. Examination of the target and spiral wave events reconstructed from the space–time data and represented as space–time event plots allowed us to identify each of the paths as containing distinct and characteristic sequences of events. Typical snapshots of the evolution of the variable u over the lattice for each developmental path are shown in figures 3 and electronic supplementary material, S1. Figure 4 summarizes the spatio-temporal evolution of the characteristic events unfolding in each path. The average lattice occupancy images for target and spiral signals, respectively, at typical fixed time offset distributions and averaged over 100 sets of initial values of u and v, are shown in electronic supplementary material, figures S6 and S7. Electronic supplementary material, figures S8–S11 contain additional examples of the characteristic space–time plots of the events in each of the developmental paths.
Figure 3.
Evolution of (a,c,e) the variable u and (b,d,f) the corresponding distributions of the element property c with global time t in each of the spiral-generating developmental paths, cases (−+), (++) and (+−) (top to bottom). (a) In the case (−+), stable spiral waves develop from the phase singularities that form owing to inter-element variability at an early stage. The insets (first two panels on the left) or overlays indicate left-handed spiral tips (blue diamonds), right-handed spiral tips (green diamonds) or target waves (red asterisks). (b,d,f) Motion of elements on each of the developmental paths in c evolving as a function of the effective time (t+Δtij) of each element. One red circle represents a group of 5% of the elements. (c) In the case (++), many short-lived right- or left-handed spirals emerge initially and are succeeded by a high number of target waves (red asterisks), which then develop into spiral waves. These do not remain closely clustered but drift to form an even spread over the lattice. (e) In the case (+−), a relatively low number of target waves evolve, which give rise to spiral waves. These emerge within previously established zones of phase synchronization particular to each of the well-developed target wave sites and remain closely packed.
Figure 4.
Space–time plots of the sequences of events for each variant of the developmental path in c. Target wave origins are represented by red asterisks, left-handed spiral tips by blue filled diamonds and right-handed spiral tips by green-edged diamonds. (a) Case (−+): phase singularities develop almost instantaneously owing to high desynchronization (Δ = 500); some persist owing to the gentle slope of the path and become stable spirals. (b) Case (++): phase singularities give rise to target waves early on; at about 1200 timesteps many phase singularities are generated of which some persist. (c) Case (−−): target waves develop but the increase in c is not sharp enough to generate breakup; no phase singularities develop initially owing to the lower value of Δ(Δ = 100). (d) Case (+−): target waves lead to spiral production in a way similar to case (++), although fewer target waves and hence fewer spirals are produced.
A higher value of ‘spread’ on a developmental path (higher Δ) appears to give rise to more initial phase singularities. These are not stable for sharply sloping developmental path variants, but are preserved for more gradual slopes. The ‘speed’ of desynchronization on the path determines how abruptly event patterns change along the path.
Case (−+) features a gradually sloping path with high desynchronization on the path (tc = 500, Tc = 300, Δ = 500). Many phase singularities are generated at a very early stage of the system's evolution (before t = 100), without a preceding target wave phase (figure 3a,b). Some of these phase singularities die out as the system evolves, with the remainder persisting and entraining the system. In certain cases, a few spirals develop into short-lived target waves (figure 4); these occur at positions distinct from the persisting spirals. On the overall temporal scale, we thus observe both a positive and a negative relationship of spiral wave tips and target wave origins.
Case (−−) is based on a gradually sloping path with a low value of desynchronization of time offsets on the path (tc = 500, Tc = 300, Δ = 100). This combination gives rise to target waves, which, however, do not propagate over long distances (see the electronic supplementary material, figure S1). This phase then dies out, and the final state is unsynchronized oscillations.
In the case (++), the developmental path is sharply sloping with high desynchronization of time offsets (tc = 1000, Tc = 50, Δ = 500). The high desynchronization gives rise to numerous phase singularities in the first 100 timesteps (figure 3c,d). At approximately 1200 timesteps, target waves then develop from these phase singularities, before eventually giving rise to a second burst of phase singularities. Most of these are short-lived, but some remain stable and persist until the lattice value of c becomes asymptotic (0.02). In contrast to case (+−), visual identification of target wave locations is difficult without the aid of the event recognition software, and clearly defined target waves do not entrain the lattice to the same extent.
Case (+−) features a sharply sloping path with low desynchronization (tc = 1000, Tc = 50, Δ = 100). Target waves develop initially and entrain the system. Later emerging pacemakers give rise to noise that destabilizes the target waves. The original entrainment zone becomes synchronized, while the original target wave origins no longer act as pacemakers. As in the case (++), a large number of phase singularities then develops at about 1200 timesteps. At this stage, most of the elements have reached their asymptotic value of c, approaching 0.02, and thus would be expected to oscillate with the same frequency, as well as being locally synchronized. Spirals then develop within these entrainment zones of the initial target waves, possibly from interactions involving the few elements in these zones that have lower time offsets compared with the generally asymptotic mean value. The correspondence between the target waves established by about 400 timesteps and the eventual sites of spiral formation is clearly visible in figure 3e.
In both cases (++) and (+−), the exact mechanism of spiral formation is hard to assess, but resembles wave-in-wave formation of the type described in Pálsson & Cox [31], in which a spiral pair forms owing to the interaction of an emerging pulse of excitation with a preceding circular wave.
The mean spatial densities of spiral and target waves over the 100 distinct time offset distributions simulated for each path agree with the visual impression conveyed by the corresponding images (table 1 and figure 3, electronic supplementary material, figures S1 and S4), with almost no spirals observed in the case (−−), maximum spiral density in the case (++) and lower values for cases (−+) and (+−).
Table 1.
Comparison of the mean and standard deviation (s.d.) values of the time-averaged spatial density (mean events per timestep on the 100×100 lattice) of spiral tips (sw) and target wave origins (tw), and Pearson correlation coefficient values (C) between the element property c (at t = 0) and pattern features for the different developmental paths (σ of Gaussian blur = 5 pixels). Values are based on 100 runs per path with different time offset distributions for each run. As virtually no phase singularities were detected for case (−−), the corresponding quantities were not calculated.
| case | tc /Tc | Δ | mean sw (s.d.) | mean tw (s.d.) | C value | C(sw,tw) | C(tw,c) | C(sw,c) |
|---|---|---|---|---|---|---|---|---|
| (−+) | 1.67 | 500 | 14.25 (4.95) | 0.02 (0.02) | min | −0.22 | −0.24 | −0.02 |
| max | 0.54 | 0.33 | 0.67 | |||||
| mean (s.d.) | 0.04 (0.17) | 0.06 (0.13) | 0.35 (0.12) | |||||
| (−−) | 1.67 | 100 | 0 (0) | 0.27 (0.02) | min | — | 0.24 | — |
| max | — | 0.63 | — | |||||
| mean (s.d.) | — | 0.44 (0.08) | — | |||||
| (++) | 20.00 | 500 | 24.83 (3.53) | 0.22 (0.02) | min | −0.21 | 0.24 | −0.19 |
| max | 0.47 | 0.70 | 0.40 | |||||
| mean (s.d.) | 0.13 (0.12) | 0.52 (0.09) | 0.08 (0.13) | |||||
| (+−) | 20.00 | 100 | 11.08 (2.39) | 0.07 (0.02) | min | −0.22 | − 0.11 | −0.02 |
| max | 0.43 | 0.69 | 0.50 | |||||
| mean (s.d.) | 0.06 (0.15) | 0.18 (0.24) | 0.26 (0.11) |
3.2. Pattern correlation analysis
So far, we have established that the different developmental paths lead to very different event sequences, as the spatio-temporal patterns unfold. Next, we return to the aspect of pattern predictability, which we assess through calculation of the correlation coefficients between each of the spatial distributions: the parameter (c), the target wave origin locations (tw) and the spiral wave tip locations (sw). The variation of the mean correlation coefficients with the degree of local spatial averaging (‘blurring’) of the signals is shown in figure 5. Table 1 contains the computed mean correlation coefficients between the lattice distributions of c and the locations of target wave origins and spiral wave tips (C(sw,tw), C(tw,c) and C(sw,c)) at an intermediate value of signal blurring (σ = 5 pixels). Together, these reveal distinct differences between the four paths, although considerable variation was observed within different runs of each path (differing in the time offset distributions) for the correlation coefficients across all properties measured, for all four developmental paths (corresponding histograms shown in electronic supplementary material, figure S2).
Figure 5.
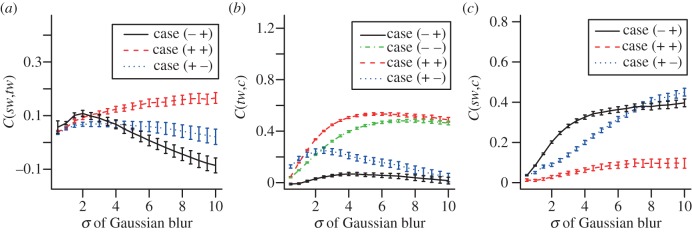
Variation of the mean correlation coefficients between (a) spiral wave tips (sw) and target wave origins (tw), (b) spirals and c (computed from the initial values of t) and (c) targets and c with the value of σ of the Gaussian blur of the respective quantities. Error bars indicate the standard error of the mean.
For case (−+), spiral wave tip locations were generally positively correlated to the element property c, while spirals and targets and targets and c displayed essentially zero correlation.
The path that yields no spirals, case (−−), evidences a relatively strong positive relationship between targets and c. Case (++)'s strongest observed relationship is the positive correlation between target wave origins and c, whereas C(sw,tw) displays a lower positive value and C(sw,c) is only slightly positive. At intermediate signal blurring in the case (+−), positive correlations exist between spirals and c and targets and c; targets and spirals are only slightly positively correlated.
3.3. Pattern predictability
Based on the distributions of the correlation coefficients obtained for the four developmental paths, our spatio-temporal observations and figure 5, we now broadly assign the overall relationships between cell properties, target wave origins and spiral wave tips to positive correlation, anticorrelation or no correlation, and further qualify the first two possibilities in terms of their relative strength (see also the details provided in §3.4). Thus, case (−+) is considered to display (relatively) strong positive C(sw,c), weak negative C(sw,tw) and no correlation between c and target waves, whereas case (−−) leads to strong positive correlations between c and target waves; no other correlations can be assessed as no spiral waves result. Case (++) and case (+−) both result in positive correlations between all three quantities, but these differ in relative intensities between the paths: C(sw,tw) and C(tw,c) were stronger in the case (++) than in (+−), whereas C(sw,c) is more notable in (+−). These relationships are summarized in the causality networks shown in figure 6. We hence arrive at a preliminary understanding of pattern predictability: the roles of our two parameters, ‘speed’ and ‘spread’, in shaping the relationships between cell properties and pattern features, as measured by our correlation coefficients and observed in the corresponding spatio-temporal event sequences. In particular, the difference in the correlations between target waves and c for cases (−−) and (+−), which have the same value of Δ but differently shaped slopes of c(t), suggests that slope shape determines how the target waves are generated as a function of cell properties.
Figure 6.
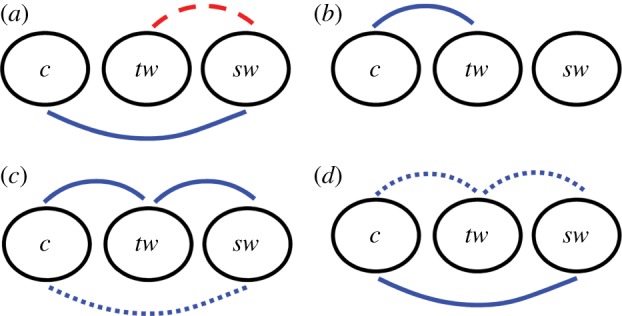
Our interpretation of the relationships between the cell property c, target wave origins (tw) and spiral wave tips (sw) for each of the four case studies: (a) case (−+), (b) case (−−), (c) case (++) and (d) case (+−). Solid lines indicate relatively strong positive correlations, dotted lines indicate relatively weak positive correlations, dashed lines indicate weak negative correlations and the absence of a line between two quantities indicates no distinct relationship. (Online version in colour.)
3.4. Additional observations
Despite the clear spatial correspondence between spiral wave origins and target wave tips visible in the snapshots of the spatio-temporal development of the u field and in the three-dimensional event plots in both cases (++) and (+−) (figures 3c–f and 4), the correlation results did not reflect these observations with the same intensity. In order to clarify these results, we turn once more to the spatio-temporal events observed for these cases. It appears that the lower correlations contributed by some time offset distributions could arise as a consequence of the relationship between the spirals and the target waves that do not develop into spirals but are instead entrained by the spirals. The target waves that develop into spirals appear to make this transition based on a combination of how early in development they become established as pacemakers and their distance from each other: pacemakers that emerge early will not develop into spirals if they are too close to a well-established pacemaker, whereas a later-established pacemaker further away may develop into spirals. By contrast, a time offset distribution giving rise to a maximum correlation does not appear to have a comparable number of small persisting target wave origins (see the electronic supplementary material, figure S3). This may be due to greater spatial co-occurrence of pacemakers, which all then become entrained by the small number of centres which give rise to spiral waves. The resulting mean correlation coefficient between spiral tips and target origins thus echoes the complexity of the possible interactions. The higher positive correlation between spirals and c for case (+−) compared with case (++) may arise owing to the greater meandering of spiral tips in the latter case, which modifies spiral locations from their origins.
Although there is comparable variation between the correlation coefficients of cell properties and target or spiral waves in most cases, further investigation is needed to explore the reasons for these variations (see the electronic supplementary material, figures S4 and S5).
For the case (−+), the positive value of C(sw,c) supports the assumption that these spirals emerge as relatively direct consequences of local cell properties. There are indications of a lower, locally positive correlation between spirals and targets, which becomes repulsive over greater distances, and the histogram (see the electronic supplementary material, figure S2) suggests a slightly negative overall correlation. This is supported by the observed event sequence in this path: spirals do not evolve from target waves, although the reverse may occur on occasion. There is thus a generally antagonistic, although complex, interaction between the waveforms here: a lattice site giving rise to target waves will diminish the likelihood of persistence of local spirals, whereas spiral sites appear to make a relatively early (by t = 1000) ‘choice’ between dying out, developing into target waves, or persisting. The positive correlation between spirals and c, and the lack of correlation between targets and c, also suggests that cell properties influence target wave location only indirectly, through determining spiral locations, in this path. It is also possible that the initial phase singularities do emerge from pacemaker candidates, but that spiral development occurs so rapidly that no preceding target wave event is recognized.
In general, the signals show little correlation at very low spatial averaging, while the correlations diverged substantially over the paths as the amount of averaging increased. This indicates that an interaction of cell properties or events over a certain spatial scale, rather than the effect of isolated units, is useful in evaluating relationships in the emergence of the spatio-temporal patterns.
4. Discussion
With the minimal model of spiral wave formation guided by a developmental path presented here, we have shown that strong correlations between cell properties and pattern features emerge, allowing for a certain degree of predictability of patterns from cell properties. Moreover, we have explored in detail how the predictability of self-organized patterns from cell properties depends on the features of the dynamical system. In this way, it may be possible to collect information on, e.g. regulatory properties from the observed relationships between the distribution of cell properties and the emerging spatio-temporal patterns.
The cases (−−) and (−+) as well as the cases (+−) and (++) lie respectively on two extremes of values of Δ. The appearance of a high number of phase singularities at an early stage owing to the higher desynchronization of cell properties in the case (++) generates a relatively high number of target waves, which in turn results in a higher density of persisting spiral waves. In the case (−+), the generation of phase singularities arises from the high value of Δ (Δ = 500). Favourable local cell-to-cell differences lead to the development of spirals, of which some persist. The lower value of Δ used in the implementation of case (−−) (Δ = 100) results in differences in Δtij that are too small to lead to the generation of phase singularities at an early stage. Although target waves arise, the developmental path-derived changes in c are too gradual to result in the formation of phase singularities from the target waves. Cases (++) and (+−), incorporating a sudden change of element properties, appear to result in a sequence of events most similar to that observed in real D. discoideum populations, and modelled in references [19,26].
Our observations thus suggest that the spiral waves observed here develop according to one of two basic mechanisms. The first involves a relatively high initial desynchronization of element properties, accompanied by a gradual change of these properties, modelled here as high ‘spread’. The second results from the generation of target waves that subsequently break and give rise to spirals owing to a comparatively abrupt change in element properties, considered high ‘speed’. In the former mechanism, represented by case (−+), there appears to be a more direct influence of cell properties on the location of spiral wave tips, whereas in the second (cases (++) and (+−)), the details of the influence of cell properties on the location of pattern features remain unclear, and are likely to be complex. The lack of spiral evolution in the case (−−) demonstrates that a threshold value of ‘speed’ is necessary for target waves to fracture and develop into spirals. The interaction of ‘spread’ and ‘speed’ determines the predictability of spiral wave tip and target wave origin locations from cell properties, and the relationship between the two wave patterns. It appears that high ‘speed’ of desynchronization on a developmental path results in spiral wave development from target waves, with positive correlation between these two patterns and between cell property values and spiral tips. Higher accompanying ‘spread’ leads to a stronger positive correlation of these quantities, but a lower correlation between spirals and cell properties. Our path with lower ‘speed’ results in a positive correlation between cell properties and spiral tips, and a likely negative relationship of spirals and target waves.
The variation in values of Δ that all lead to a set of stable and sustained signalling events suggests that a sudden slope offers a more robust evolution of behaviour, and would thus be expected to be more widely found in real biological systems using spiral wave signalling. Our results also broadly suggest that a higher value of desynchronization on a developmental path (at least within certain ranges), and therefore higher intrinsic heterogeneity, shifts the asymptotic state towards spirals. The finding in Lauzeral et al. [19] that lowering the value of Δ reduces or eliminates spiral wave formation matches our observation in the simpler model explored here, namely the differences in spiral numbers seen between the cases (−−) and (−+) and between the cases (+−) and (++).
The developmental paths presented here are only a few of a likely vast number of different possible paths. The establishment of the four distinct case studies was based on an examination of 50 different time offsets in each case to ascertain that each fell into one of the four distinct patterns of events. Outside the parameter ranges discussed in detail in these four case studies, developmental paths exist that result in behaviour that could be described as consisting of combinations of these dynamics. For a variant of case (++), with a value of Δ of 1000 (not shown), the initial variability derived from desynchronization is sufficiently high to result in a varying number of stable phase singularities, which entrain the field. Depending on the number which persist, the remaining area of the lattice may give rise to a target phase followed by spirals in the same way as observed in the case (++).
An open question that deserves far more attention is how the nonlinearities of the model affect pattern predictability and, more generally, the translation of cell properties into pattern events. We hope to address some of these aspects in our future work.
In Martiel & Goldbeter [22], a two-dimensional version of the model used in Lauzeral et al. [19] has also been discussed (however, not in the context of spatio-temporal patterns or a developmental path), showing an excitable behaviour similar to the FHN model analysed here. The more stylized structure of the FHN model (in particular, the combination of a cubic and a linear nullcline) allows us to study the different effects of desynchronization and drift speed of cell properties along the developmental path with a higher level of genericity. When exploring the effect of the model's nonlinearities on the pattern events triggered by variability, a comparison of the two models may be a good starting point, owing to the detailed biological motivation of the model from Martiel & Goldbeter [22] and the availability of higher dimensional models it has been derived from. Our results obtained so far emphasize the generic nature of the concept put forth in Lauzeral et al. [19] of desynchronization of element properties on a developmental path as a convincing model-independent explanation of the origins of pattern formation in a variety of systems.
The details of the dynamics of wave initiation in D. discoideum colonies continue to receive intense research scrutiny. Although the picture is becoming clearer, with indications that the transition to synchronized, oscillatory cAMP secretion is an outcome of collective responses [32], rather than owing to signalling initiated by specialized pacemaker cells as studied in references [6,15], the role of the variability of individual cell properties in this process is not yet definite. Our work supports the hypothesis that a continuum of desynchronized cell properties leads to spiral wave formation. While in the cases (++) and (+−) we found that higher values of our varying cell property, c, at a location are likely to predispose the location to seeding target and spiral waves, the greater visibility of this effect over a degree of spatial averaging of the respective signals suggests that the interaction of properties over a local spatial range is important in determining the evolution of the patterns. There is also considerable variation in the value of the correlations between c and wave locations, which must be further explored to achieve a fuller understanding of the underlying processes.
In general, the interdependencies summarized in figure 6 indicate a level of pattern predictability in the system, with characteristic differences in the emerging correlations between cell properties and target or spiral wave patterns, depending on the levels of spread and drift speed associated with the corresponding developmental path. The findings described here could be exploited to design patterns by appropriate construction of the forms of parameter variation guiding the pattern evolution. Also, one can ‘reverse-engineer’ properties of the system and/or the underlying developmental path from the observation of such correlations: in principle, one can now determine from the observed correlations, whether the variability contained in the system can be expected to be high-drift or high-spread, etc.
Returning to our original inspiration for this investigation, the development of spiral waves in real colonies of D. discoideum, we now discuss whether these dynamics can be broadly classed into one of the developmental path case studies presented here. The observed sequence of events in real D. discoideum colonies is the transition from non-relay to relay to oscillations to relay [33]. This would appear to most closely resemble case (++) or (+−), although as both of these developmental paths appear to have relatively long target-wave phases, a more realistic approximation in the scheme of our case studies might involve a shortening of the initial asymptotic part of the path (i.e. increase in the value of tc/Tc ).
A current restriction of our work is that the design of the paths was based on the dynamics of the single FHN oscillator. The behaviour of the diffusively coupled elements is expected to differ, understandable as effective shifts in the nullclines of the component oscillators [7]. Finally, the application of point process statistics ([34], see also [17]) to the analysis of the developmental path events is seen as an extension that would potentially yield additional insights into the relationships observed between different wave events over the courses of the various paths.
This study shows that even within a single, relatively simple mathematical model, there is a startlingly rich diversity of possible routes to spiral formation, with different resulting relationships between the relevant (varying) element properties and the positions of spiral tips. The way in which variability enters and unfolds over the system's trajectory in time thus determines the fate of the system.
5. Conclusions
We demonstrate the existence of various possible routes of spiral formation in a lattice of FHN oscillators, which are selected by the strength and form of variability entering the system. The interaction of drift speed and desynchronization leads to systematically different and predictable sequences of events in each of these developmental paths, and different relationships between cell properties and pattern features. Our findings thus offer an event-oriented overview of pattern formation, arising from a symbolic encoding of two basic underlying parameters that direct this process. The observed networks of events could be further investigated to extend our insights into pattern predictability. We conclude that there are fundamental parameters that govern spatio-temporal pattern formation and determine distinct possible fates. These ‘stylized facts’ about the role of variability can be extrapolated to more realistic scenarios of pattern development in general and D. discoideum signalling in particular. As the state of knowledge about the complex biochemical details underlying Dictyostelium dynamics continues to develop, our results can be engineered and re-implemented in more complex mathematical models that capture new findings. Understanding the relationship between the properties of individual elements in spatio-temporal pattern formation, and macroscopic features of the pattern, has implications for a diverse range of fields, including applications in ecological and social systems, and the medical realm, where early-stage detection of the locations of spiral wave tips in cardiac tissue is of great interest.
Acknowledgements
The authors thank Daniel Geberth for valuable discussions about the software suite from Geberth & Hütt M-Th [26]. This project was funded by the Deutsche Forschungsgemeinschaft (DFG; grant no. HU 937/4) and the centre for Mathematics, Modeling and Computing (MAMOC) at Jacobs University Bremen.
References
- 1.Epstein IR, Showalter K. 1996. Nonlinear chemical dynamics: oscillations, patterns, and chaos. J. Phys. Chem. 100, 13 132–13 147 10.1021/jp953547m (doi:10.1021/jp953547m) [DOI] [Google Scholar]
- 2.Cross M, Greenside H. 2009. Pattern formation and dynamics in nonequilibrium systems. Cambridge, UK: Cambridge University Press [Google Scholar]
- 3.Karsenti E. 2008. Self-organization in cell biology: a brief history. Nat. Rev. Mol. Cell Biol. 9, 255–262 10.1038/nrm2357 (doi:10.1038/nrm2357) [DOI] [PubMed] [Google Scholar]
- 4.Hess B. 2000. Periodic patterns in biology. Naturwissenschaften 87, 199–211 10.1007/s001140050704 (doi:10.1007/s001140050704) [DOI] [PubMed] [Google Scholar]
- 5.Samadani A, Mettetal J, van Oudenaarden A. 2006. Cellular asymmetry and individuality in directional sensing. Proc. Natl Acad. Sci. USA 103, 11 549–11 554 10.1073/pnas.0601909103 (doi:10.1073/pnas.0601909103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sawai S, Thomason PA, Cox EC. 2005. An autoregulatory circuit for long-range self-organization in Dictyostelium cell populations. Nature 433, 323–326 10.1038/nature03228 (doi:10.1038/nature03228) [DOI] [PubMed] [Google Scholar]
- 7.Glatt E, Gassel M, Kaiser F. 2006. Variability-induced transition in a net of neural elements: from oscillatory to excitable behaviour. Phys. Rev. E. Stat. Nonlinear Soft Matter Phys. 73, 066230. 10.1103/PhysRevE.73.066230 (doi:10.1103/PhysRevE.73.066230) [DOI] [PubMed] [Google Scholar]
- 8.Hütt M-Th, Busch H, Kaiser F. 2003. The effect of biological variability on spatiotemporal patterns: model simulations for a network of biochemical oscillators. Nova Acta Leopold 332, 381–404 [Google Scholar]
- 9.Tessone CJ, Mirasso C, Toral R, Gunton J. 2006. Diversity-induced resonance. Phys. Rev. Lett. 97, 194101. 10.1103/PhysRevLett.97.194101 (doi:10.1103/PhysRevLett.97.194101) [DOI] [PubMed] [Google Scholar]
- 10.Gassel M, Glatt E, Kaiser F. 2007. Doubly diversity-induced resonance. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 76, 016203. 10.1103/PhysRevE.76.016203 (doi:10.1103/PhysRevE.76.016203). [DOI] [PubMed] [Google Scholar]
- 11.Tessone CJ, Toral R. 2009. Diversity-induced resonance in a model for opinion formation. Eur. Phys. J. B 71, 549–555 10.1140/epjb/e2009-00343-8 (doi:10.1140/epjb/e2009-00343-8) [DOI] [Google Scholar]
- 12.Marko G. 2009. Cellular diversity promotes intercellular Ca2+ wave propagation. Biophys. Chem. 139, 53–56 10.1016/j.bpc.2008.10.001 (doi:10.1016/j.bpc.2008.10.001) [DOI] [PubMed] [Google Scholar]
- 13.Glatt E, Gassel M, Kaiser F. 2007. Variability-sustained pattern formation in subexcitable media. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 75, 026206. 10.1103/PhysRevE.75.026206 (doi:10.1103/PhysRevE.75.026206) [DOI] [PubMed] [Google Scholar]
- 14.Hilgardt C, Müller SC, Hütt M-Th. 2007. Reconstruction of cellular variability from spatiotemporal patterns of Dictyostelium discoideum. Nonlinear Biomed. Phys. 1, 10. 10.1186/1753-4631-1-10 (doi:10.1186/1753-4631-1-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geberth D, Hütt M-Th. 2008. Predicting spiral wave patterns from cell properties in a model of biological self-organization. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 78, e031917. 10.1103/PhysRevE.78.031917 (doi:10.1103/PhysRevE.78.031917). [DOI] [PubMed] [Google Scholar]
- 16.Geberth D, Hilgardt C, Hütt M-Th. 2009. Systematics of spatial heterogeneity regulation of large-scale patterns by biological variability. Nova Acta Leopold 96, 145–159 [Google Scholar]
- 17.Geberth D, Hütt M-Th. 2009. Predicting the distribution of spiral waves from cell properties in a developmental-path model of Dictyostelium pattern formation. PLoS Comput. Biol. 5, e1000422. 10.1371/journal.pcbi.1000422 (doi:10.1371/journal.pcbi.1000422) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levine H, Aranson I, Tsimring L, Truong TV. 1996. Positive genetic feedback governs cAMP spiral wave formation in Dictyostelium. Proc. Natl Acad. Sci. USA 93, 6382–6386 10.1073/pnas.93.13.6382 (doi:10.1073/pnas.93.13.6382) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lauzeral J, Halloy J, Goldbeter A. 1997. Desynchronization of cells on the developmental path triggers the formation of spiral waves of cAMP during Dictyostelium aggregation. Proc. Natl Acad. Sci. USA 94, 9153–9158 10.1073/pnas.94.17.9153 (doi:10.1073/pnas.94.17.9153) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bub G, Shrier A, Glass L. 2002. Spiral wave generation in heterogeneous excitable media. Phys. Rev. Lett. 88, 1–4 10.1103/PhysRevLett.88.058101 (doi:10.1103/PhysRevLett.88.058101) [DOI] [PubMed] [Google Scholar]
- 21.Pálsson E, Lee KJ, Goldstein RE, Franke J, Kessin RH, Cox EC. 1997. Selection for spiral waves in the social amoebae Dictyostelium. Proc. Natl Acad. Sci. USA 94, 13 719–13 723 10.1073/pnas.94.25.13719 (doi:10.1073/pnas.94.25.13719) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martiel JL, Goldbeter A. 1987. A model based on receptor desensitization for cyclic AMP signaling in Dictyostelium cells. Biophys. J. 52, 807–828 10.1016/S0006-3495(87)83275-7 (doi:10.1016/S0006-3495(87)83275-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halloy J, Lauzeral J, Goldbeter A. 1998. Modeling oscillations and waves of cAMP in Dictyostelium discoideum cells. Biophys. Chem. 72, 9–19 10.1016/S0301-4622(98)00119-7 (doi:10.1016/S0301-4622(98)00119-7) [DOI] [PubMed] [Google Scholar]
- 24.FitzHugh R. 1961. Impulses and physiological states in theoretical models of nerve membrane. Biophys. J. 1, 445–466 10.1016/S0006-3495(61)86902-6 (doi:10.1016/S0006-3495(61)86902-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagumo J, Arimoto S, Yoshizawa S. 1962. An active pulse transmission line simulating nerve axon. Proc. IRE 50, 2061–2070 [Google Scholar]
- 26.Geberth D, Hütt M-Th. 2010. Combining spiral and target wave detection to analyze excitable media dynamics. Physica A 389, 249–258 10.1016/j.physa.2009.09.034 (doi:10.1016/j.physa.2009.09.034) [DOI] [Google Scholar]
- 27.Bray MA, Lin SF, Aliev RR, Roth BJ, Wikswo JP. 2001. Experimental and theoretical analysis of phase singularity dynamics in cardiac tissue. J. Cardiovasc. Electrophysiol. 12, 716–722 10.1046/j.1540-8167.2001.00716.x (doi:10.1046/j.1540-8167.2001.00716.x) [DOI] [PubMed] [Google Scholar]
- 28.R Core Team 2012. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: See http://www.R-project.org/. [Google Scholar]
- 29.Harrell FE. 2004. Hmisc S function library. Programs available from http://biostat.mc.vanderbilt.edu/s/Hmisc. [Google Scholar]
- 30.Baddeley A, Turner R. 2005. Spatstat: an R package for analyzing spatial point patterns. J. Stat. Softw. 12, 1–42 [Google Scholar]
- 31.Pálsson E, Cox EC. 1996. Origin and evolution of circular waves and spirals in Dictyostelium discoideum territories. Proc. Natl Acad. Sci. USA 93, 1151–1155 10.1073/pnas.93.3.1151 (doi:10.1073/pnas.93.3.1151) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gregor T, Fujimoto K, Masaki N, Sawai S. 2010. The onset of collective behavior in social amoebae. Science 328, 1021–1025 10.1126/science.1183415 (doi:10.1126/science.1183415) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gerisch G, Malchow D, Roos W, Wick U. 1979. Oscillations of cyclic nucleotide concentrations in relation to the excitability of Dictyostelium cells. J. Exp. Biol. 81, 33–47 [DOI] [PubMed] [Google Scholar]
- 34.Illian J, Penttinen A, Stoyan H, Stoyan D. 2008. Statistical analysis and modelling of spatial point patterns. Hoboken, UK: John Wiley & Sons, Ltd [Google Scholar]