Abstract
More than 70 years after its initial report, caloric restriction stands strong as the most consistent non-pharmacological intervention increasing lifespan and protecting against metabolic disease. Among the different mechanisms by which caloric restriction may act, Sir2/SIRT1 (Silent information regulator 2/Silent information regulator T1) has gained major attention due to its ability to integrate the sensing of the metabolic status with adaptative transcriptional outputs. This review focuses on gathered evidence suggesting that Sir2/SIRT1 is a key mediator of the beneficial effects of caloric restriction and addresses the main questions that still need to be answered to consolidate this hypothesis.
Aging: eat less, live longer
“Age is not a particularly interesting subject. Anyone can get old. All you have to do is live long enough” (Groucho Marx, 1890-1977). Ironically, very little did Groucho know then just how difficult it still is today for the scientific community to precisely understand how to live a long life. This is probably due to the fact that aging may be a reflection of the progressive functional decay of not just one, but a complex ensemble of physiological functions. However, a major cornerstone in the aging field remains true more than 70 years after its initial report [1], i.e. that caloric restriction (CR) is the most consistent non-pharmacological intervention increasing lifespan and protecting against the deterioration of biological functions.
CR is usually defined as a moderate (normally, 20-40%) reduction in caloric intake as compared with an ad libitum diet, without compromising the maintenance of all essential nutrients [2]. Since the initial finding that CR increases maximal longevity in rats [1], multiple lines of evidence indicate that the effects of CR on lifespan extension stretch all along the evolutionary scale. Up to a 50% increase in maximum lifespan has been reported in caloric restricted yeast, rotifers, spiders, worms, flies, fish, mice and rats [3]. There are two ongoing studies on the effects of CR in Rhesus monkeys. While the final outcome and conclusions of these experiments may still take a few decades to be realized, current data already indicate that caloric restricted monkeys are protected from many age-associated pathophysiological changes, such as the development of insulin resistance and type 2 diabetes [4, 5], atherosclerosis [6], and reductions in their basal metabolic rate [7], body temperature [8], oxidative damage [9] and senescence of the immune system [10]. Despite that we only have very preliminary evidence based on surrogate measures on how CR may impact on human longevity, most available data sets indicate that CR exerts similar adaptive responses in humans as in laboratory animals, reducing the risk of developing age-associated pathological complications (see [11] for review).
Why and how can CR modulate lifespan?
According to evolutionary biology, two main factors may contribute to the progressive decline of physiological functions during aging [12]. First, natural selection loses its pressure in the post reproductive phase of life, when the influence on next generations gradually becomes more indirect. In addition to that, it must be considered that the average lifespan of a species in the wild is much shorter than that reached in captivity [13]. Hence, the decline in vital processes should be an explainable event, since it does not seem likely that natural selection influences age-related phenotypes. This speculation is in line with the fact that aging correlates with the failure of many virtually independent systems, leading to complex pathologies such as cancer or diabetes. Evolutionarily, the effects observed during CR experiments have been speculated to represent adaptation to scarcity [14, 15]. By slowing down aging and reproduction in times of scarcity, organisms save energy to favor proper survival and maximal possibility to reproduce when food becomes available. One example of this is in lower organisms, where formation of spores or larvae acts as a specialized survival strategy during times of scarcity (see [16] for example).
What triggers this decay in physiological fitness during aging? Several theories of aging have been postulated, involving accumulative oxidative damage, inflammation, mitochondrial dysfunction, lack of protein turnover or increased covalent modification of proteins by glucose derivates [17]. These are all parameters that normally correlate with aging, but none of them has clearly been demonstrated to cause aging by itself [3, 18]. Given the multifactorial nature of aging, it was surprising to find that single-gene mutations markedly contribute to extend lifespan in diverse models including yeast, worms and flies. These findings in many species led to the idea that CR could initiate a global effect on lifespan by affecting just one or a few genes and suggested that the effect of CR on lifespan is evolutionary conserved and involves the direct or indirect sensing of lower caloric availability and orchestration of adaptive responses.
Yeast and Sir2: the rise of the magic medicine
Interestingly, one cause of aging in yeast is believed to be the progressive accumulation of extrachromosomal ribosomal DNA (rDNA) circles due to recombination within homologous repeats [19]. The recombination at this locus is repressed by the yeast SIR2 gene [20]. In fact, increasing the dosage of SIR2 prevents recombination at the rDNA locus and increases lifespan, while ablation has the opposite effects, reducing lifespan by 50% [20-22]. The question remained - what exactly was SIR2 and what was it doing?
SIR2 was initially identified as a gene that silenced the extra copies of the mating-type information in yeast [23]. Simultaneous work by three independent groups established afterwards that the SIR2 gene product was a protein, Sir2, containing an unusual NAD+-dependent enzymatic histone deacetylase activity [24]. The deacetylation reaction catalyzed by Sir2 is coordinated with the cleavage of NAD+ into nicotinamide and 1-O-acetyl-ADP-ribose [25]. Since NAD+, or its reduced form NADH, acts as a cofactor in various key metabolic reactions, it was proposed that Sir2 could act as a metabolic sensor, capable of modulating gene expression accordingly to the metabolic state of the cell [26]. Supporting this hypothesis, several studies indicated that Sir2 could be a critical mediator of the effects of CR on yeast lifespan [27, 28]. In yeast, mimicking CR by reducing glucose concentration of media from 2 to 0.5% is enough to increase replicative lifespan [28]. Not only was this CR protocol able to extend lifespan to a similar extent as overexpressing Sir2, but, the reduction in glucose was also unable to increase lifespan in yeast where the gene coding for Sir2 was deleted [28]. Sir2 mutants were unresponsive to CR also in models where the formation of rDNA circles was prevented by mutation of another gene, FOB1 (fork blocking 1), indicating that the mechanisms by which CR and Sir2 modulate lifespan are not restricted to the accumulation of rDNA circles [19]. Further evidence on the involvement of Sir2 in yeast aging was gathered by the utilization of resveratrol, an activator of Sir2 [27]. Treatment with resveratrol was enough to increase yeast lifespan, but could not further increase CR-induced lifespan extension, indicating that resveratrol may be using similar pathways as CR to increase lifespan [27]. Altogether, these findings provided reasonable evidence that CR-induced lifespan extension could be mediated through Sir2 activation, since Sir2 is an evolutionary conserved enzyme; these findings also provided a potential link between metabolic sensing and transcriptional adaptations.
Growing up: CR and Sir2 homologs in higher non-mammal eukaryotes
Further glimpses into how Sir2 orthologs affect lifespan came from studies in flies and worms. In Drosophila, CR efficiently extends lifespan and increases Sir2 mRNA [29, 30]. Overexpression of Sir2 was enough to increase lifespan, an effect that was not further increased by CR [31]. Similarly, Sir2 activation by resveratrol was enough to increase fruit fly lifespan [32], but not when they were already caloric restricted. Finally, the most compelling evidence for a role of Sir2 in the mechanisms by which CR extents lifespan in flies came from experiments showing that neither CR nor resveratrol were able to extend lifespan in fruit flies where Sir2 had been genetically ablated [31]. These studies clearly followed the path already trod in yeast models and pointed towards a conserved mechanism in CR-induced extension of lifespan involving Sir2.
Worms have also been proven to be an important model for the study of the effects of CR on aging. Lowering food intake lengthens C. elegans lifespan, indicating that the common mechanism(s) for CR-induced lifespan extension are conserved [33-35]. The C. elegans genome has four genes with similarity to yeast SIR2, among which the most related is Sir-2.1 [36]. Increased dosage of the Sir-2.1 gene is sufficient to increase lifespan in worms [37]. As in yeast and flies, treatment with resveratrol extends the mean adult lifespan of wild-type worms [32, 38], but not of mutant worms lacking Sir-2.1 [32, 38]. An involvement of Sir-2.1 in mediating the effects of CR on lifespan extension in this model organism has, however, not been clearly demonstrated yet.
Caloric restriction, SIRT1 and longevity in mammals
Mammals express up to seven Sir2 homologs (SIRT1-7; also referred as sirtuins) [36]. All of them contain a conserved catalytic core domain, but differ in their subcellular localization and in the protein sequences flanking their catalytic core domain. Four sirtuins, SIRT1, SIRT3, SIRT6 and SIRT7 are mostly, but not exclusively, nuclear proteins that can be found in different subnuclear localizations. SIRT1, for example is detected in non-nucleolar nuclear regions as well as in the cytosol [39, 40], while SIRT6 and SIRT7 are mostly localized in heterochromatic regions and nucleoli, respectively [40, 41]. SIRT2 is generally found in the cytoplasm, but it binds to chromatin during mitosis [41]. SIRT3, -4, and -5 can be found in the mitochondria [40], and SIRT3 can eventually shuttle to the nucleus [42].
Among the sirtuins, SIRT1 has been the most extensively studied. More than a dozen substrates have already been described for SIRT1 [43], among which are other regulators of aging, such as the FOXO family of transcription factors [44]. SIRT1 also plays a major role in energy homeostasis in key metabolic tissues [45], which is potentially a manifestation of its ability to link metabolic status to transcriptional outputs. In line with this, SIRT1 protein levels are increased in response to CR in many key metabolic tissues [46-50]. A growing body of evidence derived from in vivo studies suggests that SIRT1 may mediate the effects of CR in mice. The data obtained from SIRT1 knock-out mice are complicated to interpret, since these mice display developmental and growth defects as well as altered birth rate [51, 52]. SIRT1 knock-out mice that survive until adulthood, however, do not display some of the metabolic responses normally triggered by CR [53]. Work by the McBurney lab showed that mice lacking SIRT1 are metabolically inefficient and, importantly, the longevity response to CR is blunted in these mice [54]. In contrast, transgenic mice constitutively expressing SIRT1 display several phenotypes that resemble CR mice: they are leaner, metabolically more active and show reduced insulin and fasted blood glucose paired with increased glucose tolerance [55]. Mice mildly overexpressing SIRT1 seem to be protected from the development of metabolic disease when challenged with high-fat diets [56, 57]. Similarly, wild-type mice fed resveratrol displayed prolonged lifespan and were protected against the development of metabolic disease [58, 59]. Treatment of wild-type mice with SRT1720, a newly developed SIRT1 agonist [60], also mimicked the metabolic adaptations triggered by CR and conferred protection against metabolic disease [60, 61].
In light of these observations, it will be interesting to determine how SIRT1 impacts human physiology. It has already been reported that SIRT1 protein levels are increased in muscle of caloric restricted individuals [62], and that SIRT1 gene variants are associated with energy expenditure [58] and the development of metabolic responses to lifestyle interventions [63]. This evidence implies a critical role for SIRT1 in mediating metabolic responses to CR that contribute to enhanced longevity not only in lower eukaryotes, but also in mammals.
SIRT1 and longevity: turbulence in the hypothesis
Evidence linking SIRT1 with longevity is not exempt from doubt. This is not only due to a number of gaps in our understanding of Sir2/SIRT1 biology, but also to controversial and even contradictory data arising in recent years.
1. How does CR activate Sir2/SIRT1?
The mechanism by which CR impacts Sir2/SIRT1 activity is still a matter of debate. The initial hypothesis by the Guarente lab proposed that the metabolic shift from fermentation to respiration occurring in response to CR results in increased NAD+ intracellular levels [64], which would in turn increase Sir2 activity. The respiratory increase would decrease NADH, which acts as an competitive inhibitor of Sir2 deacetylase activity [65], even though whether physiological levels of NADH could inhibit Sir2 is still a matter of debate [66]. This hypothesis however implies that CR should not increase lifespan in cells that are incapable of undergoing a metabolic shift from fermentation to respiration. Challenging this prediction, CR efficiently increases lifespan in several respiratory deficient yeast strains (see [67] for review). Similarly, some studies in worms also support that reduced mitochondrial function actually may increase lifespan [68-70]. In contrast, there is also compelling evidence supporting that mitochondrial activity is positively correlated to lifespan (see [71] for review). A conciliatory view would be that mitochondrial respiration might be fine-tuned during diverse life stages for maximal lifespan extension and that, while the shift from fermentation to respiration might not be mandatory for lifespan extension during CR, it does not mean that mitochondrial respiration is not important for proper longevity. How these requirements for mitochondrial respiration might affect NAD+/NADH levels is still largely unknown. After all, NAD+/NADH levels are determined by complex networks of reactions [72] and to know when, how and where NAD+/NADH levels are modulated is an extremely difficult goal to achieve, but, nonetheless, necessary to evaluate whether Sir2/SIRT1 acts as a true metabolic sensor of this redox balance. Interestingly, recent evidence suggests that NADH shuttles may participate in the mechanisms by which CR regulates lifespan, highlighting the relevance of compartmentalization of this redox balance in affecting lifespan [73].
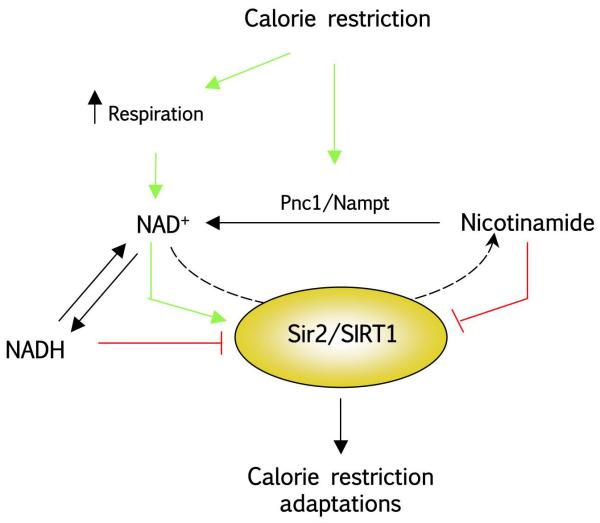
In light of the caveats surrounding the NAD+/NADH hypothesis, research pioneered by Sinclair and co-workers proposed an alternative hypothesis in which CR increases Sir2 activity by decreasing intracellular concentrations of nicotinamide (NAM), a product of the deacetylation reaction catalyzed by Sir2, which inhibits Sir2 deacetylatse activity [74]. The reduction in nicotinamide content would happen due to an increase in PNC1 (Pyrazinamidase and NiCotinamidase 1) levels, a nicotinamidase enzyme, which catalyzes the initial reaction leading the regeneration of NAD+ from NAM, also known as NAM salvage pathway [75]. Consistent with this hypothesis, the effects of CR on lifespan are blunted in yeast devoid of PNC1, while increased PNC1 dosage is sufficient to increase yeast lifespan [74, 76]. Similarly, PNC1 homologs in flies [77] and worms [78] modulate lifespan. In higher eukaryotes, the NAM salvage pathway is slightly differerent and the initial rate-limiting enzyme initiating NAD+ resynthesis from NAM is Nampt (for nicotinamide phosphoribosyltransferase), which has also been shown to increase intracellular NAD+ levels [79], SIRT1 activity [79] and replicative senescence in mammalian cells [80]. Recent evidence clearly supports the physiological relevance of Nampt and other intermediaries of the NAM salvage pathway in regulating NAD+ levels and SIRT1 function in mammals [81-83]. Altogether, these results suggest that, rather than increased respiration, lifespan extension from CR could be due to Sir2/SIRT1 activation in response to increased NAM clearance. Most of the experiments on PNC1 carried out in yeast, however, have been performed in a strain were Sir2 overexpression fails to increase lifespan [84], thus raising the possibility that PNC1/Nampt action and NAM clearance may increase lifespan through a Sir2/SIRT1-independent mechanism. In Figure 1, a scheme of the different ways allowing Sir2/SIRT1 activation in response to CR are depicted, even though, as explained above, further research will be required to fully understand the activation of SIRT1 during CR.
Figure 1. How does caloric restriction (CR) impact on SIRT1 activity?
SIRT1 deacetylates proteic substrates in a reaction that cleaves NAD+ to render NAM. NAD+ acts as a positive modulator (green arrows) of SIRT1 activity, while the accumulation of NAM and NADH (the reduced form of NAD+) lead to inhibition (red arrows) of SIRT1. During CR there is a decline in glycolytic rates in favour of respiratory metabolism as the main energy source. This changes displaces the equilibrium of the reduced/oxidized forms of NAD towards NAD+, thus increasing SIRT1 activity. Similarly, CR leads to an increase in PNC1/Nampt expression, which favors the resynthesis of NAD+ from NAM, potentially acting as a major mechanism to drop the leash of SIRT1 inhibition. (SIRT1: Silent Information Regulator T1; NAD: Nicotinamide dinucleotide; NAM: Nicotinamide; CR: Caloric restriction; PNC1: Pyrazinamidase and nicotinamidase 1; Nampt: nicotinamide phosphorybosyl transferase)
2. CR-induced longevity can be Sir2/SIRT1 independent
While we have discussed relevant data showing that Sir2/SIRT1 is necessary for CR-induced increased lifespan, not all studies point in that direction. For example, there are reported cases were CR increases lifespan independent of Sir2 [84-86] as well as in Sir2 overexpressing yeast [87]. This raised the question of whether CR does in fact increase Sir2/SIRT1 activity. Experiments measuring the silencing of Sir2 target genes in yeast have shown that, in some strains, CR does not significantly increase Sir2 activity in vivo [88, 89], and Sir2 overexpressing yeast do not completely phenocopy CR responses [88, 89]. Further investigations on whether overexpression of Sir2 mimics Sir2 activation will be necessary to correctly interpret these data.
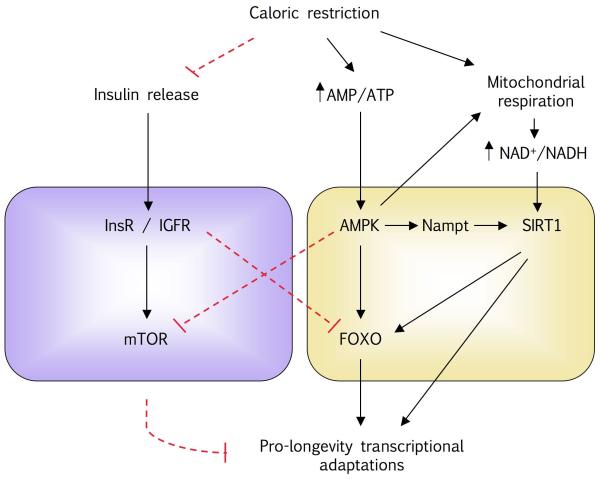
In mammals, the response to CR does not imply an increase in SIRT1 levels in all tissues [48], and SIRT1 has been shown to be dispensable for proper adaptation to CR in liver [48]. Additional conserved mechanisms have been postulated by which CR could act, such as the insulin signalling pathway, the FOXO family of transcription factors or AMP-activated protein kinase (see [90] for review). Future studies will need to establish whether SIRT1 is dispensable or whether these other mechanisms can compensate for the possible limitations of SIRT1 activity/expression by acting either in an independent manner or in a balanced interconnected network, as suggested in Figure 2.
Figure 2. Integrative view of mammalian signalling pathways involved in regulating the effects of caloric restriction (CR).
Despite the evidence linking SIRT1 to the effects of CR on mammals, these effects might be accomplished by acting in consensus with other factors. For example, CR might be not only sensed by SIRT1 as a change in the NAD+/NADH ratio, but also by AMPK as a change in the AMP/ATP ratio. AMPK can regulate mitochondrial respiration, which in turn, can also positively regulate SIRT1. Both AMPK and SIRT1 can impact the activity of FOXO transcription factors, which also have been extensively linked to the regulation of metabolism and longevity. Additionally, CR promotes the downregulation of insulin-derived signals, which also interacts with FOXO transcription factors. Hence, the metabolic and longevity responses to CR, rather than defined by single elements, may be a consequence of the balance of these signalling networks. (CR: caloric restriction; SIRT1: Silent Information Regulator T1; AMPK: AMP-activated protein kinase; FOXO: Forkhead Box O1)
3. Resveratrol: to which extent is it a direct SIRT1 activator?
Given the major role that Sir2/SIRT1 may play on regulating longevity and energy metabolism, there is intense research to identify potential specific agonists. Resveratrol treatment mimics numerous aspects of calorie restriction in all eukaryotes tested [27, 32, 58, 59, 91-93], and in some of them, the action seems to depend on SIRT1 [27, 32, 58]. Consequently, in most [27, 32, 59, 92], albeit not all [91], models tested resveratrol treatment has proven to increase lifespan. Furthermore, CR could not further augment the increase in yeast lifespan triggered by resveratrol [27]. It is, however, not clear whether the actions of resveratrol are due to direct Sir2/SIRT1 activation. Resveratrol, initially reported as a polyphenol that interfered with the mitochondrial respiratory chain [94], was shown to be able to increase SIRT1 catalytic activity in vitro [27]. The ability of resveratrol to directly activate SIRT1, however, was severely questioned by results demonstrating that the non-physiological fluorescent Fluor de Lys moiety used for SIRT1 activity assays can lead to artefactual results [95]. Blinded by the possible effects on SIRT1, resveratrol actions on other possible molecules/pathways have been largely neglected. It is interesting that many reports have recently shown that resveratrol can also activate AMPK [59, 61, 96-98], which is consistent with its possible effect on the mitochondrial respiratory chain. While some groups have reported that resveratrol action on AMPK is SIRT1 dependent [99, 100], other groups, as well as unpublished data from our lab, suggest that, at least in certain cell types, SIRT1 is dispensable for resveratrol-induced AMPK-activation [97, 101]. Importantly, AMPK has also been implicated in eukaryotic regulation of lifespan [102-105]. A conciliatory view of these events would be illustrated by possible SIRT1 activation downstream of AMPK, as suggested recently in cultured myotubes [83, 106] and skeletal muscle [106], by increasing NAD+ levels [83, 106] and Nampt expression [83]. In this regard, SIRT1 may be essential for resveratrol action but as a downstream consequence of AMPK activation, rather than as a direct target of resveratrol.
Conclusions and future directions
There is an undeniable amount of data clearly suggesting that Sir2/SIRT1 modulates longevity and that it might be a mechanism by which CR enhances lifespan. Conclusive and coherent evidence supporting this link between CR and Sir2/SIRT1 is, however, still fragmentary and sketchy between the models studied and confounded by effects of other candidate proteins on lifespan regulation. The above mentioned link between AMPK and SIRT1 holds some promise, not only as an important signalling axis itself, but also due to its connections with other pathways such as the insulin signalling pathway and the FOXO family of transcription factors (Figure 1), providing a way to integrate the pathways affecting longevity. Further studies in genetically engineered mouse models will undoubtedly shed light on these theories. For example, skeletal muscle has been shown to be a key target tissue for resveratrol metabolic actions, so it would be interesting to see whether skeletal muscle-specific deletion of AMPK or SIRT1 activity impacts lifespan and/or resveratrol’s effects. Similarly, how CR affects Sir2/SIRT1 and whether Sir2/SIRT1 acts as a metabolic sensor are still unresolved questions. It will be interesting to investigate how modulating the redox balance and cellular NAD+/NADH and NAM levels influence longevity in mammals. Along this line, promising new data is arising from a relatively unexplored pathway that impacts on NAD+/NADH and longevity: the nicotinamide riboside pathway [107, 108]. Finally, and in addition to these elements, we have to keep in mind that mammals have seven different sirtuin proteins. Describing each of their roles in CR and identifying their physiological substrates will be crucial to determine whether sirtuins are key metabolic sensors participating in adaptations to CR.
Acknowledgements
The work in the laboratory of the authors is sponsored by the Ecole Polytechnique Fédérale de Lausanne and an advanced research award by the European Research Council “Ideas” program (231138-Sirtuins). CC is an EMBO fellow.
References
- [1].McCay CM, Crowell MF, Maynard LA. The effect of retarded growth upon the length of life span and upon the ultimate body size. 1935. Nutrition. 1989;5:155–71. discussion 172. [PubMed] [Google Scholar]
- [2].Piper MD, Bartke A. Diet and aging. Cell Metab. 2008;8:99–104. doi: 10.1016/j.cmet.2008.06.012. [DOI] [PubMed] [Google Scholar]
- [3].Koubova J, Guarente L. How does calorie restriction work? Genes Dev. 2003;17:313–21. doi: 10.1101/gad.1052903. [DOI] [PubMed] [Google Scholar]
- [4].Lane MA, Ingram DK, Roth GS. Calorie restriction in nonhuman primates: effects on diabetes and cardiovascular disease risk. Toxicol Sci. 1999;52:41–8. doi: 10.1093/toxsci/52.2.41. [DOI] [PubMed] [Google Scholar]
- [5].Kemnitz JW, Roecker EB, Weindruch R, et al. Dietary restriction increases insulin sensitivity and lowers blood glucose in rhesus monkeys. Am J Physiol. 1994;266:E540–7. doi: 10.1152/ajpendo.1994.266.4.E540. [DOI] [PubMed] [Google Scholar]
- [6].Verdery RB, Ingram DK, Roth GS, Lane MA. Caloric restriction increases HDL2 levels in rhesus monkeys (Macaca mulatta) Am J Physiol. 1997;273:E714–9. doi: 10.1152/ajpendo.1997.273.4.E714. [DOI] [PubMed] [Google Scholar]
- [7].Blanc S, Schoeller D, Kemnitz J, et al. Energy expenditure of rhesus monkeys subjected to 11 years of dietary restriction. J Clin Endocrinol Metab. 2003;88:16–23. doi: 10.1210/jc.2002-020405. [DOI] [PubMed] [Google Scholar]
- [8].Lane MA, Baer DJ, Rumpler WV, et al. Calorie restriction lowers body temperature in rhesus monkeys, consistent with a postulated anti-aging mechanism in rodents. Proc Natl Acad Sci U S A. 1996;93:4159–64. doi: 10.1073/pnas.93.9.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zainal TA, Oberley TD, Allison DB, et al. Caloric restriction of rhesus monkeys lowers oxidative damage in skeletal muscle. Faseb J. 2000;14:1825–36. doi: 10.1096/fj.99-0881com. [DOI] [PubMed] [Google Scholar]
- [10].Messaoudi I, Warner J, Fischer M, et al. Delay of T cell senescence by caloric restriction in aged long-lived nonhuman primates. Proc Natl Acad Sci U S A. 2006;103:19448–53. doi: 10.1073/pnas.0606661103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Holloszy JO, Fontana L. Caloric restriction in humans. Exp Gerontol. 2007;42:709–12. doi: 10.1016/j.exger.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hekimi S, Guarente L. Genetics and the specificity of the aging process. Science. 2003;299:1351–4. doi: 10.1126/science.1082358. [DOI] [PubMed] [Google Scholar]
- [13].Kirkwood TB, Austad SN. Why do we age? Nature. 2000;408:233–8. doi: 10.1038/35041682. [DOI] [PubMed] [Google Scholar]
- [14].Harrison DE, Archer JR. Natural selection for extended longevity from food restriction. Growth Dev Aging. 1989;53:3. [PubMed] [Google Scholar]
- [15].Holliday R. Food, reproduction and longevity: is the extended lifespan of calorie-restricted animals an evolutionary adaptation? Bioessays. 1989;10:125–7. doi: 10.1002/bies.950100408. [DOI] [PubMed] [Google Scholar]
- [16].Golden JW, Riddle DL. The Caenorhabditis elegans dauer larva: developmental effects of pheromone, food, and temperature. Dev Biol. 1984;102:368–78. doi: 10.1016/0012-1606(84)90201-x. [DOI] [PubMed] [Google Scholar]
- [17].Weinert BT, Timiras PS. Invited review: Theories of aging. J Appl Physiol. 2003;95:1706–16. doi: 10.1152/japplphysiol.00288.2003. [DOI] [PubMed] [Google Scholar]
- [18].Carter CS, Hofer T, Seo AY, Leeuwenburgh C. Molecular mechanisms of life- and health-span extension: role of calorie restriction and exercise intervention. Appl Physiol Nutr Metab. 2007;32:954–66. doi: 10.1139/H07-085. [DOI] [PubMed] [Google Scholar]
- [19].Sinclair DA, Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell. 1997;91:1033–42. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- [20].Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–80. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gottlieb S, Esposito RE. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56:771–6. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- [22].Kennedy BK, Austriaco NR, Jr., Zhang J, Guarente L. Mutation in the silencing gene SIR4 can delay aging in S. cerevisiae. Cell. 1995;80:485–96. doi: 10.1016/0092-8674(95)90499-9. [DOI] [PubMed] [Google Scholar]
- [23].Shore D, Squire M, Nasmyth KA. Characterization of two genes required for the position-effect control of yeast mating-type genes. Embo J. 1984;3:2817–23. doi: 10.1002/j.1460-2075.1984.tb02214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- [25].Tanner KG, Landry J, Sternglanz R, Denu JM. Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc Natl Acad Sci U S A. 2000;97:14178–82. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Guarente L. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 2000;14:1021–6. [PubMed] [Google Scholar]
- [27].Howitz KT, Bitterman KJ, Cohen HY, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–6. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- [28].Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–8. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- [29].Clancy DJ, Gems D, Harshman LG, et al. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292:104–6. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- [30].Rogina B, Helfand SL, Frankel S. Longevity regulation by Drosophila Rpd3 deacetylase and caloric restriction. Science. 2002;298:1745. doi: 10.1126/science.1078986. [DOI] [PubMed] [Google Scholar]
- [31].Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A. 2004;101:15998–6003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wood JG, Rogina B, Lavu S, et al. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–9. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- [33].Klass MR. Aging in the nematode Caenorhabditis elegans: major biological and environmental factors influencing life span. Mech Ageing Dev. 1977;6:413–29. doi: 10.1016/0047-6374(77)90043-4. [DOI] [PubMed] [Google Scholar]
- [34].Hosono R, Nishimoto S, Kuno S. Alterations of life span in the nematode Caenorhabditis elegans under monoxenic culture conditions. Exp Gerontol. 1989;24:251–64. doi: 10.1016/0531-5565(89)90016-8. [DOI] [PubMed] [Google Scholar]
- [35].Lee GD, Wilson MA, Zhu M, et al. Dietary deprivation extends lifespan in Caenorhabditis elegans. Aging Cell. 2006;5:515–24. doi: 10.1111/j.1474-9726.2006.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–8. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- [37].Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–30. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- [38].Viswanathan M, Kim SK, Berdichevsky A, Guarente L. A role for SIR-2.1 regulation of ER stress response genes in determining C. elegans life span. Dev Cell. 2005;9:605–15. doi: 10.1016/j.devcel.2005.09.017. [DOI] [PubMed] [Google Scholar]
- [39].Tanno M, Sakamoto J, Miura T, et al. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J Biol Chem. 2007;282:6823–32. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- [40].Michishita E, Park JY, Burneskis JM, et al. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16:4623–35. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Mostoslavsky R, Chua KF, Lombard DB, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–29. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- [42].Scher MB, Vaquero A, Reinberg D. SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes Dev. 2007;21:920–8. doi: 10.1101/gad.1527307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Feige JN, Auwerx J. Transcriptional targets of sirtuins in the coordination of mammalian physiology. Curr Opin Cell Biol. 2008;20:303–9. doi: 10.1016/j.ceb.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Salih DA, Brunet A. FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr Opin Cell Biol. 2008;20:126–36. doi: 10.1016/j.ceb.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Rodgers JT, Lerin C, Gerhart-Hines Z, Puigserver P. Metabolic adaptations through the PGC-1 alpha and SIRT1 pathways. FEBS Lett. 2008;582:46–53. doi: 10.1016/j.febslet.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Cohen HY, Miller C, Bitterman KJ, et al. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–2. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- [47].Nisoli E, Tonello C, Cardile A, et al. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005;310:314–7. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- [48].Chen D, Bruno J, Easlon E, et al. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 2008;22:1753–7. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Gerhart-Hines Z, Rodgers JT, Bare O, et al. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. Embo J. 2007;26:1913–23. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Rodgers JT, Lerin C, Haas W, et al. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–8. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- [51].McBurney MW, Yang X, Jardine K, et al. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol Cell Biol. 2003;23:38–54. doi: 10.1128/MCB.23.1.38-54.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Cheng HL, Mostoslavsky R, Saito S, et al. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci U S A. 2003;100:10794–9. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Chen D, Steele AD, Lindquist S, Guarente L. Increase in activity during calorie restriction requires Sirt1. Science. 2005;310:1641. doi: 10.1126/science.1118357. [DOI] [PubMed] [Google Scholar]
- [54].Boily G, Seifert EL, Bevilacqua L, et al. SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS ONE. 2008;3:e1759. doi: 10.1371/journal.pone.0001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Bordone L, Cohen D, Robinson A, et al. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell. 2007;6:759–67. doi: 10.1111/j.1474-9726.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- [56].Banks AS, Kon N, Knight C, et al. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 2008;8:333–41. doi: 10.1016/j.cmet.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Pfluger PT, Herranz D, Velasco-Miguel S, et al. Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci U S A. 2008;105:9793–8. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Lagouge M, Argmann C, Gerhart-Hines Z, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–22. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- [59].Baur JA, Pearson KJ, Price NL, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–42. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Milne JC, Lambert PD, Schenk S, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–6. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Feige JN, Lagouge M, Canto C, et al. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008;8:347–58. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- [62].Civitarese AE, Carling S, Heilbronn LK, et al. Calorie restriction increases muscle mitochondrial biogenesis in healthy humans. PLoS Med. 2007;4:e76. doi: 10.1371/journal.pmed.0040076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Weyrich P, Machicao F, Reinhardt J, et al. SIRT1 genetic variants associate with the metabolic response of Caucasians to a controlled lifestyle intervention - the TULIP Study. BMC Med Genet. 2008;9:100. doi: 10.1186/1471-2350-9-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Lin SJ, Kaeberlein M, Andalis AA, et al. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–8. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- [65].Lin SJ, Ford E, Haigis M, et al. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 2004;18:12–6. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Schmidt MT, Smith BC, Jackson MD, Denu JM. Coenzyme specificity of Sir2 protein deacetylases: implications for physiological regulation. J Biol Chem. 2004;279:40122–9. doi: 10.1074/jbc.M407484200. [DOI] [PubMed] [Google Scholar]
- [67].Kaeberlein M, Powers RW., 3rd Sir2 and calorie restriction in yeast: a skeptical perspective. Ageing Res Rev. 2007;6:128–40. doi: 10.1016/j.arr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- [68].Lee SS, Kennedy S, Tolonen AC, Ruvkun G. DAF-16 target genes that control C. elegans life-span and metabolism. Science. 2003;300:644–7. doi: 10.1126/science.1083614. [DOI] [PubMed] [Google Scholar]
- [69].Hansen M, Hsu AL, Dillin A, Kenyon C. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 2005;1:119–28. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Dillin A, Hsu AL, Arantes-Oliveira N, et al. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- [71].Dilova I, Easlon E, Lin SJ. Calorie restriction and the nutrient sensing signaling pathways. Cell Mol Life Sci. 2007;64:752–67. doi: 10.1007/s00018-007-6381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Rongvaux A, Andris F, Van Gool F, Leo O. Reconstructing eukaryotic NAD metabolism. Bioessays. 2003;25:683–90. doi: 10.1002/bies.10297. [DOI] [PubMed] [Google Scholar]
- [73].Easlon E, Tsang F, Skinner C, et al. The malate-aspartate NADH shuttle components are novel metabolic longevity regulators required for calorie restriction-mediated life span extension in yeast. Genes Dev. 2008;22:931–44. doi: 10.1101/gad.1648308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Anderson RM, Bitterman KJ, Wood JG, et al. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature. 2003;423:181–5. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Ghislain M, Talla E, Francois JM. Identification and functional analysis of the Saccharomyces cerevisiae nicotinamidase gene, PNC1. Yeast. 2002;19:215–24. doi: 10.1002/yea.810. [DOI] [PubMed] [Google Scholar]
- [76].Gallo CM, Smith DL, Jr., Smith JS. Nicotinamide clearance by Pnc1 directly regulates Sir2-mediated silencing and longevity. Mol Cell Biol. 2004;24:1301–12. doi: 10.1128/MCB.24.3.1301-1312.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Balan V, Miller GS, Kaplun L, et al. Life span extension and neuronal cell protection by Drosophila nicotinamidase. J Biol Chem. 2008;283:27810–9. doi: 10.1074/jbc.M804681200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].van der Horst A, Schavemaker JM, Pellis-van Berkel W, Burgering BM. The Caenorhabditis elegans nicotinamidase PNC-1 enhances survival. Mech Ageing Dev. 2007;128:346–9. doi: 10.1016/j.mad.2007.01.004. [DOI] [PubMed] [Google Scholar]
- [79].Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem. 2004;279:50754–63. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- [80].van der Veer E, Ho C, O’Neil C, et al. Extension of human cell lifespan by nicotinamide phosphoribosyltransferase. J Biol Chem. 2007;282:10841–5. doi: 10.1074/jbc.C700018200. [DOI] [PubMed] [Google Scholar]
- [81].Yang H, Yang T, Baur JA, et al. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130:1095–107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Revollo JR, Korner A, Mills KF, et al. Nampt/PBEF/Visfatin regulates insulin secretion in beta cells as a systemic NAD biosynthetic enzyme. Cell Metab. 2007;6:363–75. doi: 10.1016/j.cmet.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Fulco M, Cen Y, Zhao P, et al. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell. 2008;14:661–73. doi: 10.1016/j.devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2004;2:E296. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Jiang JC, Jaruga E, Repnevskaya MV, Jazwinski SM. An intervention resembling caloric restriction prolongs life span and retards aging in yeast. Faseb J. 2000;14:2135–7. doi: 10.1096/fj.00-0242fje. [DOI] [PubMed] [Google Scholar]
- [86].Lamming DW, Latorre-Esteves M, Medvedik O, et al. HST2 mediates SIR2-independent life-span extension by calorie restriction. Science. 2005;309:1861–4. doi: 10.1126/science.1113611. [DOI] [PubMed] [Google Scholar]
- [87].Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Genes determining yeast replicative life span in a long-lived genetic background. Mech Ageing Dev. 2005;126:491–504. doi: 10.1016/j.mad.2004.10.007. [DOI] [PubMed] [Google Scholar]
- [88].Kaeberlein M, Powers RW, 3rd, Steffen KK, et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–6. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- [89].Kaeberlein M, Hu D, Kerr EO, et al. Increased life span due to calorie restriction in respiratory-deficient yeast. PLoS Genet. 2005;1:e69. doi: 10.1371/journal.pgen.0010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Russell SJ, Kahn CR. Endocrine regulation of ageing. Nat Rev Mol Cell Biol. 2007;8:681–91. doi: 10.1038/nrm2234. [DOI] [PubMed] [Google Scholar]
- [91].Pearson KJ, Baur JA, Lewis KN, et al. Resveratrol delays age-related deterioration and mimics transcriptional aspects of dietary restriction without extending life span. Cell Metab. 2008;8:157–68. doi: 10.1016/j.cmet.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Valenzano DR, Terzibasi E, Genade T, et al. Resveratrol prolongs lifespan and retards the onset of age-related markers in a short-lived vertebrate. Curr Biol. 2006;16:296–300. doi: 10.1016/j.cub.2005.12.038. [DOI] [PubMed] [Google Scholar]
- [93].Barger JL, Kayo T, Vann JM, et al. A low dose of dietary resveratrol partially mimics caloric restriction and retards aging parameters in mice. PLoS ONE. 2008;3:e2264. doi: 10.1371/journal.pone.0002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Zini R, Morin C, Bertelli A, et al. Effects of resveratrol on the rat brain respiratory chain. Drugs Exp Clin Res. 1999;25:87–97. [PubMed] [Google Scholar]
- [95].Kaeberlein M, McDonagh T, Heltweg B, et al. Substrate-specific activation of sirtuins by resveratrol. J Biol Chem. 2005;280:17038–45. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- [96].Zang M, Xu S, Maitland-Toolan KA, et al. Polyphenols stimulate AMP-activated protein kinase, lower lipids, and inhibit accelerated atherosclerosis in diabetic LDL receptor-deficient mice. Diabetes. 2006;55:2180–91. doi: 10.2337/db05-1188. [DOI] [PubMed] [Google Scholar]
- [97].Dasgupta B, Milbrandt J. Resveratrol stimulates AMP kinase activity in neurons. Proc Natl Acad Sci U S A. 2007;104:7217–22. doi: 10.1073/pnas.0610068104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Park CE, Kim MJ, Lee JH, et al. Resveratrol stimulates glucose transport in C2C12 myotubes by activating AMP-activated protein kinase. Exp Mol Med. 2007;39:222–9. doi: 10.1038/emm.2007.25. [DOI] [PubMed] [Google Scholar]
- [99].Breen DM, Sanli T, Giacca A, Tsiani E. Stimulation of muscle cell glucose uptake by resveratrol through sirtuins and AMPK. Biochem Biophys Res Commun. 2008;374:117–22. doi: 10.1016/j.bbrc.2008.06.104. [DOI] [PubMed] [Google Scholar]
- [100].Hou X, Xu S, Maitland-Toolan KA, et al. SIRT1 regulates hepatocyte lipid metabolism through activating AMP-activated protein kinase. J Biol Chem. 2008;283:20015–26. doi: 10.1074/jbc.M802187200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Chan AY, Dolinsky VW, Soltys CL, et al. Resveratrol inhibits cardiac hypertrophy via AMP-activated protein kinase and Akt. J Biol Chem. 2008;283:24194–201. doi: 10.1074/jbc.M802869200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Ashrafi K, Lin SS, Manchester JK, Gordon JI. Sip2p and its partner snf1p kinase affect aging in S. cerevisiae. Genes Dev. 2000;14:1872–85. [PMC free article] [PubMed] [Google Scholar]
- [103].Curtis R, O’Connor G, DiStefano PS. Aging networks in Caenorhabditis elegans: AMP-activated protein kinase (aak-2) links multiple aging and metabolism pathways. Aging Cell. 2006;5:119–26. doi: 10.1111/j.1474-9726.2006.00205.x. [DOI] [PubMed] [Google Scholar]
- [104].Apfeld J, O’Connor G, McDonagh T, et al. The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes Dev. 2004;18:3004–9. doi: 10.1101/gad.1255404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Greer EL, Dowlatshahi D, Banko MR, et al. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr Biol. 2007;17:1646–56. doi: 10.1016/j.cub.2007.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Canto C, Gerhart-Hines Z, Feige JN, et al. AMPK regulates energy expenditure by modulating NAD(+) metabolism and SIRT1 activity. Nature. 2009 doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Belenky P, Racette FG, Bogan KL, et al. Nicotinamide riboside promotes Sir2 silencing and extends lifespan via Nrk and Urh1/Pnp1/Meu1 pathways to NAD+ Cell. 2007;129:473–84. doi: 10.1016/j.cell.2007.03.024. [DOI] [PubMed] [Google Scholar]
- [108].Bieganowski P, Brenner C. Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss-Handler independent route to NAD+ in fungi and humans. Cell. 2004;117:495–502. doi: 10.1016/s0092-8674(04)00416-7. [DOI] [PubMed] [Google Scholar]