Abstract
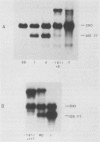
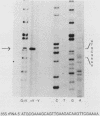
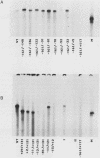
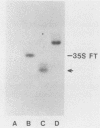
Sequences within the spacer region of yeast rRNA cistrons stimulate synthesis of the major 35S rRNA precursor in vivo 10- to 30-fold (E. A. Elion and J. R. Warner, Cell 39:663-673, 1984). Spacer sequences that mediate this stimulatory activity are located approximately 2.2 kilobases upstream from sequences that encode the 5' terminus of the 35S rRNA precursor. By utilizing a centromere-containing plasmid carrying a 35S rRNA minigene, a 160-base-pair region of spacer rDNA was identified by deletion mapping that is required for efficient stimulation of 35S rRNA synthesis in vivo. A 22-base-pair sequence, previously shown to support RNA polymerase I-dependent selective initiation of transcription in vitro, was located 15 base pairs upstream from the 3' boundary of the stimulatory region. A 77-base pair region of spacer DNA that mediates transcriptional terminator activity in vivo was identified immediately downstream from the 5' boundary of the stimulatory region. Deletion mutations extending downstream from the 5' boundary of the 160-base-pair stimulatory region simultaneously interfere with terminator activity and stimulation of 35S rRNA synthesis from the minigene. The terminator region supported termination of transcripts initiated by RNA polymerase I in vivo. The organization of sequences that support terminator and promoter activities within the 160-base-pair stimulatory region is similar to the organization of rDNA gene promoters in higher organisms. Possible mechanisms for spacer-sequence-dependent stimulation of yeast 35S rRNA synthesis in vivo are discussed.
Full text
PDF
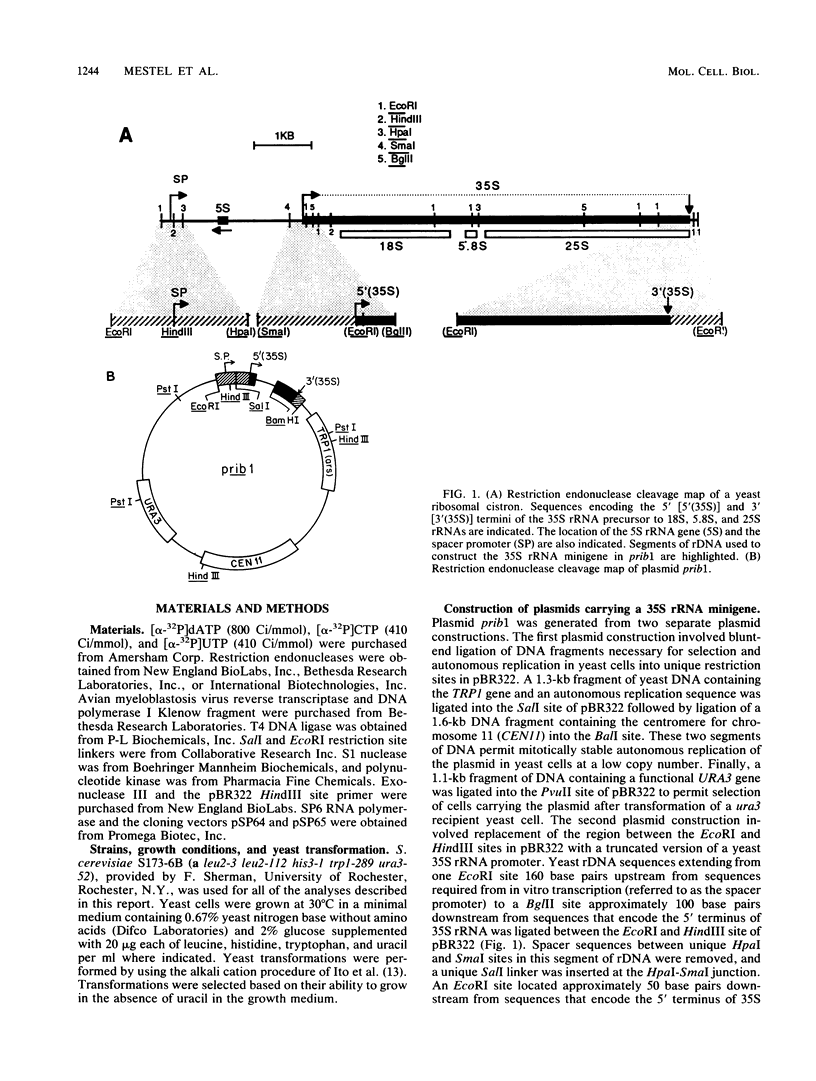







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bayev A. A., Georgiev O. I., Hadjiolov A. A., Kermekchiev M. B., Nikolaev N., Skryabin K. G., Zakharyev V. M. The structure of the yeast ribosomal RNA genes. 2. The nucleotide sequence of the initiation site for ribosomal RNA transcription. Nucleic Acids Res. 1980 Nov 11;8(21):4919–4926. doi: 10.1093/nar/8.21.4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy B. G., Yang-Yen H. F., Rothblum L. I. Additional RNA polymerase I initiation site within the nontranscribed spacer region of the rat rRNA gene. Mol Cell Biol. 1987 Jul;7(7):2388–2396. doi: 10.1128/mcb.7.7.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Winter R. F., Moss T. Spacer promoters are essential for efficient enhancement of X. laevis ribosomal transcription. Cell. 1986 Jan 31;44(2):313–318. doi: 10.1016/0092-8674(86)90765-8. [DOI] [PubMed] [Google Scholar]
- De Winter R. F., Moss T. The ribosomal spacer in Xenopus laevis is transcribed as part of the primary ribosomal RNA. Nucleic Acids Res. 1986 Aug 11;14(15):6041–6051. doi: 10.1093/nar/14.15.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion E. A., Warner J. R. An RNA polymerase I enhancer in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jun;6(6):2089–2097. doi: 10.1128/mcb.6.6.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion E. A., Warner J. R. The major promoter element of rRNA transcription in yeast lies 2 kb upstream. Cell. 1984 Dec;39(3 Pt 2):663–673. doi: 10.1016/0092-8674(84)90473-2. [DOI] [PubMed] [Google Scholar]
- Grummt I., Kuhn A., Bartsch I., Rosenbauer H. A transcription terminator located upstream of the mouse rDNA initiation site affects rRNA synthesis. Cell. 1986 Dec 26;47(6):901–911. doi: 10.1016/0092-8674(86)90805-6. [DOI] [PubMed] [Google Scholar]
- Grummt I., Maier U., Ohrlein A., Hassouna N., Bachellerie J. P. Transcription of mouse rDNA terminates downstream of the 3' end of 28S RNA and involves interaction of factors with repeated sequences in the 3' spacer. Cell. 1985 Dec;43(3 Pt 2):801–810. doi: 10.1016/0092-8674(85)90253-3. [DOI] [PubMed] [Google Scholar]
- Hammond C. I., Holland M. J. Purification of yeast RNA polymerases using heparin agarose affinity chromatography. Transcriptional properties of the purified enzymes on defined templates. J Biol Chem. 1983 Mar 10;258(5):3230–3241. [PubMed] [Google Scholar]
- Henderson S., Sollner-Webb B. A transcriptional terminator is a novel element of the promoter of the mouse ribosomal RNA gene. Cell. 1986 Dec 26;47(6):891–900. doi: 10.1016/0092-8674(86)90804-4. [DOI] [PubMed] [Google Scholar]
- Holland M. J., Hager G. L., Rutter W. J. Transcription of yeast DNA by homologous RNA polymerases I and II: selective transcription of ribosomal genes by RNA polymerase I. Biochemistry. 1977 Jan 11;16(1):16–24. doi: 10.1021/bi00620a003. [DOI] [PubMed] [Google Scholar]
- Holland M. J., Holland J. P., Thill G. P., Jackson K. A. The primary structures of two yeast enolase genes. Homology between the 5' noncoding flanking regions of yeast enolase and glyceraldehyde-3-phosphate dehydrogenase genes. J Biol Chem. 1981 Feb 10;256(3):1385–1395. [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempers-Veenstra A. E., Oliemans J., Offenberg H., Dekker A. F., Piper P. W., Planta R. J., Klootwijk J. 3'-End formation of transcripts from the yeast rRNA operon. EMBO J. 1986 Oct;5(10):2703–2710. doi: 10.1002/j.1460-2075.1986.tb04554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempers-Veenstra A. E., van Heerikhuizen H., Musters W., Klootwijk J., Planta R. J. Transcription of an artificial ribosomal RNA gene in yeast. EMBO J. 1984 Jun;3(6):1377–1382. doi: 10.1002/j.1460-2075.1984.tb01980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermekchiev M. B., Grummt I. Natural point mutations within rat rDNA transcription terminator elements reveal the functional importance of single bases for factor binding and termination. Nucleic Acids Res. 1987 May 26;15(10):4131–4143. doi: 10.1093/nar/15.10.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemenz R., Geiduschek E. P. The 5' terminus of the precursor ribosomal RNA of Saccharomyces cerevisiae. Nucleic Acids Res. 1980 Jun 25;8(12):2679–2689. doi: 10.1093/nar/8.12.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klootwijk J., Verbeet M. P., Veldman G. M., de Regt V. C., van Heerikhuizen H., Bogerd J., Planta R. J. The in vivo and in vitro initiation site for transcription of the rRNA operon of Saccharomyces carlsbergensis. Nucleic Acids Res. 1984 Feb 10;12(3):1377–1390. doi: 10.1093/nar/12.3.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klootwijk J., de Jonge P., Planta R. J. The primary transcript of the ribosomal repeating unit in yeast. Nucleic Acids Res. 1979 Jan;6(1):27–39. doi: 10.1093/nar/6.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohorn B. D., Rae P. M. Nontranscribed spacer sequences promote in vitro transcription of Drosophila ribosomal DNA. Nucleic Acids Res. 1982 Nov 11;10(21):6879–6886. doi: 10.1093/nar/10.21.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. Characterization of three sites of RNA 3' end formation in the Xenopus ribosomal gene spacer. Cell. 1986 May 9;45(3):431–443. doi: 10.1016/0092-8674(86)90329-6. [DOI] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. Enhancer-like properties of the 60/81 bp elements in the ribosomal gene spacer of Xenopus laevis. Cell. 1984 May;37(1):285–289. doi: 10.1016/0092-8674(84)90324-6. [DOI] [PubMed] [Google Scholar]
- Lohr D., Ide G. I. In vitro initiation and termination of ribosomal RNA transcription in isolated yeast nuclei. J Biol Chem. 1983 Apr 25;258(8):4668–4671. [PubMed] [Google Scholar]
- McStay B., Reeder R. H. A termination site for Xenopus RNA polymerase I also acts as an element of an adjacent promoter. Cell. 1986 Dec 26;47(6):913–920. doi: 10.1016/0092-8674(86)90806-8. [DOI] [PubMed] [Google Scholar]
- Moss T. A transcriptional function for the repetitive ribosomal spacer in Xenopus laevis. Nature. 1983 Mar 17;302(5905):223–228. doi: 10.1038/302223a0. [DOI] [PubMed] [Google Scholar]
- Nikolaev N., Georgiev O. I., Venkov P. V., Hadjiolov A. A. The 37 S precursor to ribosomal RNA is the primary transcript of ribosomal RNA genes in Saccharomyces cerevisiae. J Mol Biol. 1979 Jan 25;127(3):297–308. doi: 10.1016/0022-2836(79)90331-0. [DOI] [PubMed] [Google Scholar]
- Petes T. D. Molecular genetics of yeast. Annu Rev Biochem. 1980;49:845–876. doi: 10.1146/annurev.bi.49.070180.004213. [DOI] [PubMed] [Google Scholar]
- Quincey R. V., Godfrey R. E. Upstream activation of ribosomal RNA biosynthesis in Saccharomyces cerevisiae. Biochem J. 1985 Nov 15;232(1):205–209. doi: 10.1042/bj2320205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder R. H., Roan J. G., Dunaway M. Spacer regulation of Xenopus ribosomal gene transcription: competition in oocytes. Cell. 1983 Dec;35(2 Pt 1):449–456. doi: 10.1016/0092-8674(83)90178-2. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Tower J. Transcription of cloned eukaryotic ribosomal RNA genes. Annu Rev Biochem. 1986;55:801–830. doi: 10.1146/annurev.bi.55.070186.004101. [DOI] [PubMed] [Google Scholar]
- Swanson M. E., Holland M. J. RNA polymerase I-dependent selective transcription of yeast ribosomal DNA. Identification of a new cellular ribosomal RNA precursor. J Biol Chem. 1983 Mar 10;258(5):3242–3250. [PubMed] [Google Scholar]
- Swanson M. E., Yip M., Holland M. J. Characterization of an RNA polymerase I-dependent promoter within the spacer region of yeast ribosomal cistrons. J Biol Chem. 1985 Aug 15;260(17):9905–9915. [PubMed] [Google Scholar]
- Van Keulen H., Retèl J. Transcription specificity of yeast RNA polymerase A. Highly specific transcription in vitro of the homologous ribosomal transcription units. Eur J Biochem. 1977 Oct 3;79(2):579–588. doi: 10.1111/j.1432-1033.1977.tb11842.x. [DOI] [PubMed] [Google Scholar]
- Windle J. J., Sollner-Webb B. Two distant and precisely positioned domains promote transcription of Xenopus laevis rRNA genes: analysis with linker-scanning mutants. Mol Cell Biol. 1986 Dec;6(12):4585–4593. doi: 10.1128/mcb.6.12.4585. [DOI] [PMC free article] [PubMed] [Google Scholar]