Abstract
Objective
To demonstrate a noninvasive large mammalian genetic sampling method using blood meal obtained from a tabanid fly.
Methods
Blood meal was recovered from the abdomen of an engorged tabanid fly (Haematopota sp.) which was captured immediately after biting a Sumatran rhino in captivity. The blood was applied on to a Whatman FTA® blood card. Subsequent laboratory work was conducted to extract, amplify and sequence the DNA from the sample. Validation was done by sampling the hair follicles and blood samples from the rhinoceros and subjecting it to the same laboratory process.
Results
BLAST search and constructed phylogenetic trees confirmed the blood meal samples were indeed from the rhino.
Conclusions
This method could be used in the field application to noninvasively collect genetic samples. Collection of tabanids and other haematophagous arthropods (e.g. mosquitoes and ticks) and other blood-sucking parasites (e.g. leeches and worms) could also provide information on vector-borne diseases.
Keywords: Noninvasive DNA sampling, Blood meal, Tabanid fly, Sumatran rhino
1. Introduction
Collecting genetic material from free-ranging, elusive and rare animal species that live in remote tropical rainforests is difficult or even impossible. Coupled with the dense nature of the rainforest, even spotting an individual in the wild is almost impossible, making the acquisition of DNA samples almost extremely difficult and in practice prohibitively expensive in terms of manpower, money and time mosquitoes and ticks. Even collection of faecal samples has proven almost impossible, under conditions of frequent rain on remote, extensive forested hills. However, plenty of studies have demonstrated the usefulness of the noninvasive genetic sampling in the research on free-ranging and elusive animals through the ability to use low quantity (and sometimes quality) of shed genetic materials with the advent of polymerase chain reaction (PCR)-based technology[1]–[5]. This advancement had truly escalated the field of conservation biology particularly when dealing with rare and elusive, endangered, or cryptic fauna[1]–[3],[5]–[8].
Noninvasive sampling is described as a method by which scientists gather the genetic materials shed by animals through sources that can be collected without having to catch or disturb the animal[9]. Among the types of samples that may contain genetic materials are dung, shed hairs and feathers, urine, snake skins, sloughed skin, eggshells and even skulls in owl pellets[9],[10]. However, blood meal collected from haematophagous insects have not yet been listed in public domain as a potential practical source for noninvasive samplings.
Analysis of blood meals of haematophagous insects (e.g. mosquiotes, tsetse flies, and sand flies) has been widely used to determine their main hosts, host preferences and feeding patterns, but very little research has been conducted on blood meal from horseflies of the Family Tabanidae[11]. In addition, limited or no studies have been done to look at the effects of tabanids bites on wildlife species.
The genus Haematopota (Insecta: Tabanidae) consists of a growing number of species, females of which suck bloods from mammalian hosts. Although it is a common froup livestock attacking flies, members of this genus still poorly understood. A large number of specimens are available in various Southeast Asian museums but lacking reliable taxonomic work that allows specific identification.
In this study, we demonstrate the use of a noninvasive technique to sample genetic material from the critically endangered Sumatran rhinoceros (Dicerorhinus sumatrensis) (D. sumatrensis) by collecting blood meals from a tabanid horsefly (Haematopota sp.) which was captured immediately after biting and feeding on a captive individual Sumatran rhino.
2. Materials and methods
2.1. Study site and sample collection
This work was conducted at the rhino captive facility managed by the Borneo Rhino Alliance (BORA) in Tabin Wildlife Reserve, Lahad Datu District, Sabah. Two captive Sumatran rhinos were housed at the facility (Table 1). Flies were collected from around the vicinity and identified to the genus level (Figure 1). Blood samples were withdrawn from the abdomen of blood-engorged flies and applied on to a FTA® Blood Card (Whatman) (Figure 2). For the purpose of validation, hair follicles and blood samples were taken from both rhinos and similar laboratory processes were conducted for all sample types. The hair follicles were preserved in 95% ethanol while the blood spots were left to air-dry and later stored at room temperature.
Table 1. Information of the Sumatran rhinoceros housed at the rhino captive centre and details of the other rhinoceros species and representatives of the Order Perissodactyla obtained from the GenBank.
| No | Species | Sample abbr. | Common name | House name | Population | Studbook No./NCBI Acc. No. |
| 1 | D. sumatrensis | SR04 | Sumatran rhino | Kretam | Sabah | N/A/JF290494, JQ281903 |
| 2 | D. sumatrensis | SR05 | Sumatran rhino | Gologob | Sabah | #40/JF290495, JQ281904 |
| 3 | D. sumatrensis | NC012684 | Sumatran rhino | Suci | Sumatra | #43/NC012684 |
| 4 | D. sumatrensis | AJ245723 | Sumatran rhino | N/A | Sumatra | AJ245723 |
| 5 | D. sumatrensis | JF718875 | Sumatran rhino | N/A | Sumatra | N/A/JF718875 |
| 6 | Coelodonta antiquitatis | FJ905813 | Woolly rhino | FJ905813 | ||
| 7 | Coelodonta antiquitatis | NC012681 | Woolly rhino | NC012681 | ||
| 8 | Ceratotherium simum | NC001808 | White rhino | NC001808 | ||
| 9 | Ceratotherium simum | Y07726 | White rhino | Y07726 | ||
| 10 | Diceros bicornis | NC012682 | Black rhino | NC012682 | ||
| 11 | Diceros bicornis | FJ905814 | Black rhino | FJ905814 | ||
| 12 | Rhinoceros sondaicus | NC012683 | Javan rhino | NC012683 | ||
| 13 | Rhinoceros sondaicus | FJ905815 | Javan rhino | FJ905815 | ||
| 14 | Rhinoceros unicornis | X97336 | Indian rhino | X97336 | ||
| 15 | Rhinoceros unicornis | NC001779 | Indian rhino | NC001779 | ||
| 16 | Tapirus indicus | AF145734 | Malayan tapir | AF145734 | ||
| 17 | Tapirus terrestris | AJ428947 | Brazillian tapir | AF056030 | ||
| 18 | Tapirus pinchaque | GQ259955 | Mountain tapir | GQ259957 | ||
| 19 | Equus asinus | NC001788 | Donkey | NC001788 | ||
| 20 | Equus caballus | NC001640 | Horse | DQ297663 | ||
| 21 | Equus przewalskii | HQ439484 | Przewalskii horse | DQ223534 | ||
| 22 | Equus hemionus | NC016061 | Plains zebra | DQ470805 |
Figure 1. The tabanid flies collected (Haematopota sp.) (Photo by Zainal Zahari Zainuddin).
Figure 2. Collecting the blood meal from the Haematopota sp.
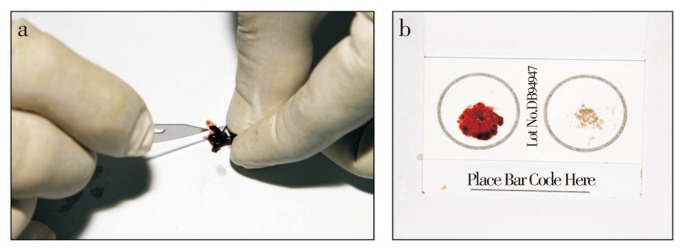
(a) Extracting the blood meal from the fly's abdomen. (b) Spreading of the blood meal onto the blood card (Photos by Zainal Zahari Zainuddin).
2.2. Laboratory analysis
The DNA extraction, PCR amplifications and sequencing reaction were conducted on the blood meals from the fly, the hair follicles, and blood samples from both rhinos. DNA extraction from the blood cards was carried out by incorporating the protocol for dried blood spots (for both blood meals from the flies and blood samples from the rhinos) while the hair follicles were extracted using the forensic case work samples (hair roots) protocol implemented in the QIAamp® DNA Micro Kit (Qiagen).
Two primer sets were used to amplify the cytochrome b (cytb) and the control region (CR) of the mitochondrial DNA (mtDNA) gene. The cytb primer set are known as, GluDG-L, 5′-TGA CTT GAA RAA CCA YCG TT-3′ and CB2-H, 5′-CCC TCA GAA TGA TAT TTG TCC TCA-3′[12], while the CR primer set are known as L15 926, 5′-TAC ACT GGT CTT GTA-3′, and H00 651, 5′-AAG GCT AGG ACC AAA CCT-3′[13]. The PCR amplifications (reaction mixtures and PCR profile) and sequencing reaction are described elsewhere[14].
2.3. DNA characterization and phylogenetic analyses
Alignments of the sequences were done by using the program Geneious v5.5[15]. Pair-wise distance analysis using the Kimura two-parameter model was done to estimate the genetic distances among the sequences as performed using MEGA version 5[16],[17]. Sequence characterization (variable sites, conserved sites and parsimony-informative sites) were also done using MEGA.
The sequences were later aligned with the Basic Local Alignment Search Tool (BLAST) program[18] to match the query sequence with the available sequence database in GenBank which is produced and maintained by the National Center for Biotechnology Information (NCBI) (available at http://www.ncbi.nlm.nih.gov/). Depending on the availability of the sequences at the database, BLAST will provide the closest matching sequence to the sequence under query, hence, suggesting the species identification.
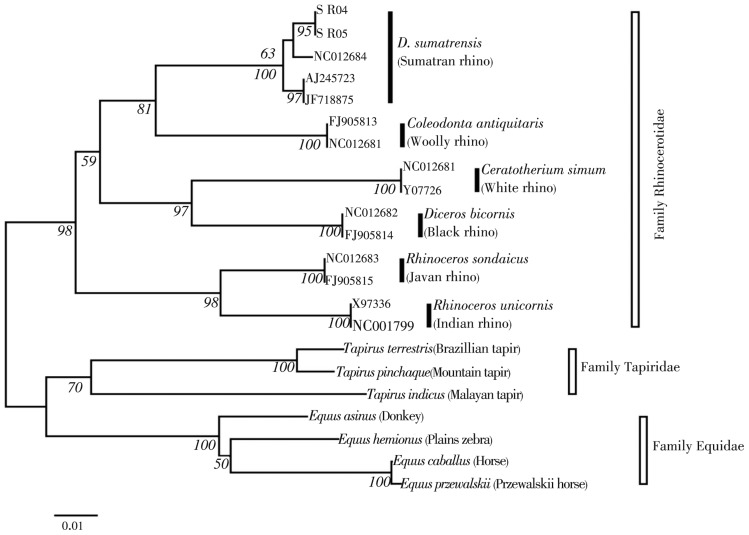
Phylogenetic tree was later constructed for both the cytb and CR segment by using the neighbour-joining (NJ) method implemented in MEGA. The NJ clustering was performed by using the Kimura 2-parameter distance model[16] with pair-wise deletion option. Sequences of the D. sumatrensis and the representatives of the other species from the Order Perissodactyla were also obtained from the GenBank (Table 1). Phylogenetic confidence was estimated by bootstrapping[19] with 1 000 replicate data sets.
3. Results
DNA fragments with the sizes of 446 bp and 500 bp from the cytb and CR respectively were successfully amplified and sequenced from all sample types. All sequences generated from this study were submitted to the GenBank and given the accession number JQ281903-JQ281904, JF290494-JF290495. Sequences from the different types of samples (blood meal, hair follicles, and host blood) from both rhinos produced identical DNA sequences.
In total, 19 variable sites with no parsimony-informative sites (Table 2) were observed when the rhino sequences from both gene segments were aligned against the species D. sumatrensis from the NCBI database (Accession No.: NC012684). Genetic distances calculated between the rhinos from Sabah and Sumatra showed the value of 1.0-1.2% for the cytb gene segment while in the CR segment the value ranges from 2.7-3.1%. BLAST search matched the cytb segment with a 99.1% of similarity (both SR04 and SR05) while the sequences of the CR segments matched 97.2% (SR04) and 96.8% (SR05). Phylogenetic tree constructed using the NJ method (Figure 3) further confirmed that the blood meal samples were indeed from the rhino.
Table 2. Variable sites observed in the cytb and CR gene segment of the rhinos in the present study as compared to the D. sumatrensis from the GenBank. Bold nucleotides show the variable sites between the rhinos in the present study.
| Gene |
CYTB |
CR |
||||||||||||||||||
| Position | 14220 | 14350 | 14530 | 14584 | 15475 | 15577 | 15582 | 15614 | 15617 | 15632 | 15658 | 15672 | 15685 | 15695 | 15718 | 15733 | 15774 | 15775 | 15841 | 15897 |
| NC012684 | G | C | C | T | - | A | A | A | A | A | G | A | T | T | T | A | A | T | C | C |
| SR04 | A | T | T | A | A | G | G | G | G | G | A | G | C | C | C | G | A | C | T | C |
| SR05 | A | T | T | A | A | G | G | G | G | G | A | G | C | C | C | G | C | C | T | T |
Figure 3. Phylogenetic tree (NJ method) constructed using the cytb gene segment.
4. Discussion
Our original interest in sampling Sumatran rhino blood was linked to our concern over the implications of a rigid belief in a perceived need to keep separate the different Sumatran rhino populations (on Borneo and Sumatra islands) based on classical divisions into sub-species. Phylogenetic tree constructed supports previous work where similarly, three sister-pairings were observed: (i) the black/white, (ii) the woolly/Sumatran, and (iii) the Javan/Indian. Furthermore, the tree showed a weak support (63%) for the structuring between the Sabah and Sumatra rhino populations suggesting the close relationship between them[20]. Three subspecies of Sumatran rhinos are currently recognized based on morphology: (i) D. sumatrensis (Peninsular Malaysia and Sumatran population), D. harrissoni (Sabah population) and, D. s. lasiotis (unconfirmed reports from Burma, extinct in India and Bangladesh). With the current population number both in the wild and captivity showing continuous and alarming decline over the past few decades[21],[22], there is a dire need to boost the number of rhinos through population inter-mixing regardless of their subspecies designation.
Our interest and approach led to the findings reported here, which demonstrate the usefulness of using blood meals collected from tabanid flies in determining the identity of the host species, using PCR. A recent study conducted demonstrates the use of blood meals to identify host species through PCR[23]. This approach is potentially important when working with rare, elusive and endangered species like the Sumatran rhino. A recent work describing a new screening tool using haematophagous leeches (Haemadipsa spp.) also demonstrated the effectiveness of using micropredators to assess mammalian biodiversity by DNA profiling[5]. Due to the non-destructive nature of this method, we suggest that the sampling of blood meals from haematophagous mircopredators should be considered as a targeted noninvasive sampling technique for Sumatran rhinos and other rare rainforest mammals. In our experience, tabanid flies tend to be associated closely with their normal hosts (large wild mammals) in rainforests, but these flies do sometimes locate and bite humans who are seeking the animals in the forest. Thus, researchers and wildlife patrol and monitoring teams do have the opportunity to collect horsefly blood meals, if they are often out in the forest seeking signs of mammals such as Sumatran rhinos.
We conclude that this method could be used in the field application to noninvasively collect genetic samples especially on the rare, elusive and endangered species. Collection of haematophagous mosquitoes, ticks, leeches and worms from the wild could be conducted at sites frequented by animals including salt-licks, wallows, wildlife main trails or “highways” and river banks. Additionally, information on vector-borne diseases could also be obtained through analyses of the blood meals. Further research needs to be conducted, however, to evaluate and measure the longevity of a particular blood meal in order for positive detection to occur.
Acknowledgments
The authors are grateful to the Department of Wildlife and National Parks (DWNP) of Peninsular Malaysia and the Sabah Wildlife Department for their support and for providing the research facilities and permits for this study. This work was financially supported by the Sime Darby Foundation (Grant code: P23 071000490001); years 2009-2015 grant to Borneo Rhino Sanctuary programme. We also thank MT Abdullah from the Universiti Malaysia Sarawak (UNIMAS) for critical comments, the Universiti Malaysia Sabah (UMS) and the Borneo Rhino Alliance (BORA) for providing travel assistance and accommodation for Rovie-Ryan JJ during the sampling work. Also many thanks to the staff of BORA for their assistance in sample acquisition.
Comments
Background
Noninvasive Genetic Sampling on the Rare Sumatran Rhinoceros (Dicerorhinus sumatrensis): Identification of the Host Species from the Blood Meal Collected from the tabanid fly (Tabanidae: Haematopota sp.).
Sampling of the rare and endangered Sumatran rhinoceros is a very costly and difficult task in the thick tropical rain forest in Borneo, and the normal method of indirect observation methods can only provide rough estimate of the population size.
Research frontiers
This noninvasive genetic sampling using specific molecular marker method is novel and innovative and could solve the problem of sampling of the illusive rhinos and other wildlife in the thick Bornean rain forest.
Related reports
Molecular methods had been used to identify species and species boundary. But the use of tabanids as a source of DNA for wildlife species has never been used in Malaysia. There was a similar attempt to use leeches to sample mammals in Vietnam.
Innovations and breakthroughs
The noninvasive genetic sampling using specific molecular marker method is novel and innovative.
Applications
Having high potentials for noninvasive genetic sampling of other animals such as tigers, elephants, wild gaur, wild pigs, serow and tapirs.
Peer review
I strongly support this paper to be published as it has improved the field method by cleverly using molecular genetics via tabanids feeding on host species of rhinos. It has greater use to other species of wildlife in the thick rain forest.
Footnotes
Foundation Project: This work was financially supported by the Sime Darby Foundation (Grant code: P23 071000490001).
Conflict of interest statement: We declare that we have no conflict of interest.
References
- 1.Fernando P, Vidya TNC, Rajapakse C, Dangolla A, Melnick DJ. Reliable noninvasive genotyping: Fantasy or reality? J Hered. 2003;94(2):115–123. doi: 10.1093/jhered/esg022. [DOI] [PubMed] [Google Scholar]
- 2.Stenglein JL, De Barba M, Ausband DE, Waits LP. Impacts of sampling location within a faeces on DNA quality in two carnivore species. Mol Ecol Res. 2010;10:109–114. doi: 10.1111/j.1755-0998.2009.02670.x. [DOI] [PubMed] [Google Scholar]
- 3.Harris RB, Winnie J, Jr, Amish SJ, Beja Pereira A, Godinho R, Costa V, Luikart G. Argali abundance in the Afghan Pamir using capture-recapture modeling from fecal DNA. J Wildl Manag. 2010;74(4):668–677. [Google Scholar]
- 4.Brøseth H, Flagstad Ø, Wärdig C, Johansson M, Ellegren H. Large-scale noninvasive genetic monitoring of wolverines using scats reveals density dependent adult survival. Biol Conserv. 2010;143:113–120. [Google Scholar]
- 5.Schnell IB, Thomsen PF, Wilkinson N, Rasmussen M, Jensen LRD, Willerslev E, et al. Screening mammal biodiversity using DNA from leeches. Curr Biol. 2012;22(8):R262–R263. doi: 10.1016/j.cub.2012.02.058. [DOI] [PubMed] [Google Scholar]
- 6.Jalil MF, Cable J, Sinyor J, Lackman Ancrenaz I, Ancrenaz M, Bruford MW, et al. Riverine effect on mitochondrial structure of Bornean orang-utans (Pongo pygmaeus) at two spatial scales. Mol Ecol. 2008;17:2898–2909. doi: 10.1111/j.1365-294X.2008.03793.x. [DOI] [PubMed] [Google Scholar]
- 7.Borthakur U, Barman RD, Das C, Basumatary A, Talukdar A, Ahmed MF, et al. Noninvasive genetic monitoring of tiger (Panthera tigris tigris) population of Orang National Park in the Brahmaputra floodplain, Assam, India. Eur J Wildlife Res. 2011;57(3):603–613. [Google Scholar]
- 8.Galaverni M, Palumbo D, Fabbri E, Caniglia R, Greco C, Randi E. Monitoring wolves (Canis lupus) by non-invasive genetics and camera trapping: a small-scale pilot study. Eur J Wildl Res. 2012;58(1):47–58. [Google Scholar]
- 9.Taberlet P, Waits LP, Luikart G. Noninvasive genetic sampling: look before you leap. Trends Ecol Evol. 1999;14:323–327. doi: 10.1016/s0169-5347(99)01637-7. [DOI] [PubMed] [Google Scholar]
- 10.Kohn M, Wayne RK. Facts from feces revisited. Tree. 1997;12:223–227. doi: 10.1016/s0169-5347(97)01050-1. [DOI] [PubMed] [Google Scholar]
- 11.Muzari MO, Burgess GW, Skerratt LF, Jones RE, Duran TL. Host preferences of tabanid flies based on identification of blood meals by ELISA. Vet Parasitol. 2010;174:191–198. doi: 10.1016/j.vetpar.2010.08.040. [DOI] [PubMed] [Google Scholar]
- 12.Palumbi S, Martin A, Romano S, McMillan WO, Stice L, Grabowski G. The simple fool's guide to PCR version 2.0. 1991:1–45. Department of Zoology and Kewalo Marine Laboratory, University of Hawaii, Honolulu. [Google Scholar]
- 13.Morales JC, Andau PM, Supriatna J, Zainuddin Z-Z, Melnick DJ. Mitochondrial DNA variability and conservation genetics of the Sumatran rhinoceros. Conserv Biol. 1997;11(2):539–543. [Google Scholar]
- 14.Rovie-Ryan JJ, Guan AKH, Kumaran JV, Esa YB, Sallehin AA, Abdullah MT. Malaysian fruit bats phylogeny inferred using ribosomal RNA. Pertanika J Trop Agric Sci. 2008;31(1):67–77. [Google Scholar]
- 15.Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, et al. Geneious v5.5. 2010. Available from http://www.geneious.com.
- 16.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotides sequence. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 17.Tamura K, Dudley J, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:1193–1204. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 20.Willerslev E, Gilbert MTP, Binladen J, Ho SYW, Campos PF, Ratan A, et al. Analysis of complete mitochondrial genomes from extinct and extant rhinoceros reveals lack of phylogenetic resolution. BMC Evol Biol. 2009;9:95. doi: 10.1186/1471-2148-9-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmad Zafir AW, Payne J, Mohamed A, Lau CF, Sharma DSK, Alfred R, et al. Now or never: what will it take to save the Sumatran rhinoceros Dicerorhinus sumatrensis from extinction? Oryx. 2011;45(2):225–233. [Google Scholar]
- 22.Clements R, Rayan DM, Ahmad Zafir AW, Venkataraman A, Alfred R, Payne J, Ambu L, Sharma DSK. Trio under threat: can we secure the future of rhinos, elephants and tigers in Malaysia? Biodivers Conserv. 2010;19:1115–1136. [Google Scholar]
- 23.Ernieenor FCL, Ahamad M, Haron MS, Ming HT. Establishment of a molecular tool for blood meal identification in Malaysia. Asian Pac J Trop Biomed. 2012;2(3):223–227. doi: 10.1016/S2221-1691(12)60046-X. [DOI] [PMC free article] [PubMed] [Google Scholar]