Abstract
Objective
To investigate antibiotic resistance and carriage class 1 and 2 integrons in clinical isolates of Acinetobacter baumannii (A. baumannii) from Tehran, Iran.
Methods
Antimicrobial susceptibility testing was performed according to the Clinical and Laboratory Standards Institute. The presence of integrons was investigated by PCR using specific primers.
Results
Among isolated A. baumannii strains, 82% were multidrug resistant, 27 samples (54%) were resistant to three or more than three antibiotics and 16 samples (32%) showed resistance to two antibiotics. Integrons were detected from 44 of 50 isolates (88%), with classes 1 and 2 being observed in 42% (21/50) and 82% (41/50) of isolates, respectively. Integron-positive A. baumannii isolates showed higher antibiotic resistance than integron-negative isolates and all showed a multidrug-resistant phenotype.
Conclusions
Our findings show that classes 1 and 2 integrons, and especially classes 2 integrons are widely disseminated among A. baumannii strains isolated from Tehran and these structures are playing a major role in the acquisition of multidrug resistance in these strains. So monitoring of drug resistance with investigating carriage class 1 and 2 integrons is very important to plan specific infection control measures due to multidrug resistance A. baumannii in Iran hospitals.
Keywords: Acinetobacter baumannii, Integron, Multidrug resistance
1. Introduction
Acinetobacter baumannii (A. baumannii) is an important opportunistic pathogen which is spreading in different groups of people and responsible for nosocomial infections, especially in intensive-care-unit (ICU) and burn wards[1]–[3]. Although A. baumannii strains are usually found in soil and water, the origin of epidemics strains with multidrug- resistant phenotype is from hospital and they are usually genetically very similar[3]–[6]. Ability of this organism in acquiring different mechanisms of resistance and also become resistant to all commonly available antibiotics and lack of new antimicrobial agents and effective drugs are the most risk factors in these bacteria[6],[7]. Various studies show that most of A. baumannii strains become resistant to most antibiotics and these multidrug-resistant strains are expanding rapidly among hospitalized patients in hospitals[8],[9]. Mobile elements include plasmids, transposons and integrons are the most effective genetic elements which play an important role in acquisition and dissemination of resistance factors in different Gram-negative bacteria, especially A. baumannii strains. Also various studies show that multidrug-resistance in these bacteria is significantly in relation with presence of integron and gene cassettes[10].
Integrons are sequences of conserved DNA that contain an integrase gene (IntI) encoding the IntI integrase and cause transmission and incorporation of gene cassettes via site-specific recombination mechanisms[11]. So far, several classes of integrons have been described in gram-negative and gram-positive bacteria. All of these integrons consist of two conserved segments (5′CS) and (3′CS), the integrase gene and the cassette integration site (attI). Integrons of class 1 are the most common and widely distributed among gram-negative bacteria, and its 3′conserved sequence area (3′CS) includes three open reading frames (ORFs), qaEΔ1 gene which confers resistance to quaternary ammonium compounds and sul1 gene which confers resistance to sulfonamides. Integrons of class 2 are found in transposon Tn7 and relatives. Its 3′ conserved sequence contains five tns genes which are responsible for mobility of transposons. Integrons of class 3 also have been reported but the 3′ conserved sequence is still not well described[10]–[12].
In order to identify the presence of integrons of class 1 and 2 in bacteria, researchers are using typically two regions as target. One of such regions is the integrase enzyme gene and based on sequences of this gene divided integrons to different classes. Therefore this gene could be appropriate target for identification of integrons in the sample and also for detection of integron classes. Another region used by most of researchers, is variable region which located among two conserved regions in integron structure. Gene cassettes which located in this region are surrounded by two conserved sequences (3′-CS and 5′-CS). Primers are designed for the variable region so that the junction region located at the end of the two conserved region and therefore researchers could be aware of the length of the variable regions. The length of variable regions depends on the number of gene cassettes, which inserted in that region, so PCR products have different sizes. This could help scientists in identification of integron classes, gene cassettes and the number of them[10]–[13].
Studies in different parts of the world is done to determine the prevalence of different classes of integrons and their relationship with antibiotic resistance in nosocomial isolates of A. baumannii in hospital[13]. As the prevalence of integrons classes 1 and 2 in A. baumannii is not clear in Iran, this study is performed with the main aim of determination of the prevalence of classes 1 and 2 integrons and their relationship with the presence of antibiotic resistance among nosocomial isolates of A. baumannii in Tehran hospitals.
2. Material and methods
2.1. Bacterial isolates
In total, 200 bacterial isolates were collected from different patients hospitalized in several hospitals (Imam Khomeini, Milad and Baqiyatallah) in Tehran, Iran during 2009-2010. In laboratory 50 isolates of A. baumannii were isolated from 19 blood cultures, 15 samples of the trachea, six wound swab samples, four samples of urine and five samples with unknown origin. All of this isolates were identified by conventional biochemical and microscopic methods. The isolates were preserved in -80 °C in nutrient broth containing glycerol 50% v/v until the molecular works was done.
2.2. Antibiotic profiles
Antibiotic susceptibility was determined using the disk diffusion method on Mueller Hinton agar according to the Clinical and Laboratory Standards Institute (CLSI guidelines) recommendations using antibiotic disks amikacin (30 µg), ampicillin/sulbactam (10/10 µg), aztreonam (30 µg), cefepime (30 µg), ceftazidime (30 µg), ciprofloxacin(5 µg), gentamycin(10 µg), imipenem (10 µg), meropenem (10 µg), norfloxacin (10 µg), ofloxacin(1 µg), piperacillin/tazobactam (100/10 µg), tobramycin (10 µg) which were obtained from Oxoid Ltd. (Basingstoke, UK). The standard strains of Escherichia coli ATCC 25922 and A. baumannii ATCC 19606 were used as negative and positive controls.
It is mentioned that according to studies performed, isolates of A. baumannii that show resistance to three or more than three categories, including quinolone antibiotics (ciprofloxacin), broad spectrum cephalosporins (ceftazidime and cefepime), combined lactam/lactamase inhibitor (ampicillin/sulbactam), aminoglycosides (amikacin and tobramycin) and carbapenems (imipenem and meropenem) considered as multidrug- resistant (MDR) strains.
2.3. Integron analysis
Genome DNA of all A. baumannii isolates were extracted by using of high pure PCR template preparation Kit (Roche, Germany Construction Co.). Each PCR reaction mixture contained 15 µL master mix 2× (Ampliqon Co, Denmark) including 1× PCR buffer, 1.5 mmol/L MgCl2, 1 µL template DNA (0.5 µg), 0.15 mmol/L dNTP, 1.25 IU Taq DNA polymerase, 20 pmol of each forward and reverse primers and sterile distilled water up to 50 µL.
As described previously by Koeleman et al., for detection of class 1 integron (integron PCR) was used primers 5′CS and 3′CS[14]. Also, for PCR detection of the IntI1 and IntI2 integrase genes (integrase gene PCR), oligonucleotide primers based on the intI1 and intI2 genes were used (Table 1). Primers Int1F/R were used to amplify 160 bp fragments and primers Int2F/R were used to amplify 288 bp fragments.
Table 1. Primers for amplification of genes from Acinetobacter sp. isolates.
| Primer | Nucleotide sequence (5′ to 3′) |
| 5′-CSa | GGC ATC CAA GCA GCA AG |
| 3′-CSa | AAG CAG ACT TGA CCT GA |
| Int1F | CAG TGG ACA TAA GCC TGT TC |
| Int1R | CCC GAG GCA TAG ACT GTA |
| Int2F | TTG CGA GTA TCC ATA ACC TG |
| Int2R | TTA CCT GCA CTG GAT TAA GC |
PCR amplification was performed in a GenAmp PCR system (Ependorf Co., Germany) according to the following program:initial denaturation at 95 °C for 5 min, followed by 30 cycles at 94 °C for 30 seconds, 55 °C for 30 seconds 72 °C for 30 seconds and a final extension at 72 °C for 5 min. All PCR amplification was performed in duplicate.
The PCR products were analyzed using electrophoresis technique on 15 g/L agarose gel for 1 h at 85 Volt and 25 mA, stained by SYBERgreen and visualized under UV transilluminator. Amplification products were further evaluated by sequencing and restriction digestion procedure.
In cases that were needed to evaluate the relationship between antibiotic resistance pattern and integron positive genotype from statistical tests for measuring P value (like X2) was used that P<0.05 was considered as statistically significant data.
3. Results
During this study, in total of 70 samples of Acinetobacter were isolated from 500 collected samples. Fifty samples of patients were identified as A. baumannii (71%), 12 samples were Acinetobacter Lwoffii (17.1%) and 8 samples (11.4%) were other Acinetobacter species.
Among of isolated A. baumannii strains, 82% were multidrug- resistant. Results of this study showed that 27 samples of A. baumannii (54%) were resistant to three or more than three antibiotics and 16 samples (32%) showed resistance to two antibiotics. Also, none of resistant strains were showed complete resistance to all antibiotics. It was mentioned that in this study approximately all samples were resistant to ceftazidime and cefepime and tobramycin and meropenem considered as effective drugs.
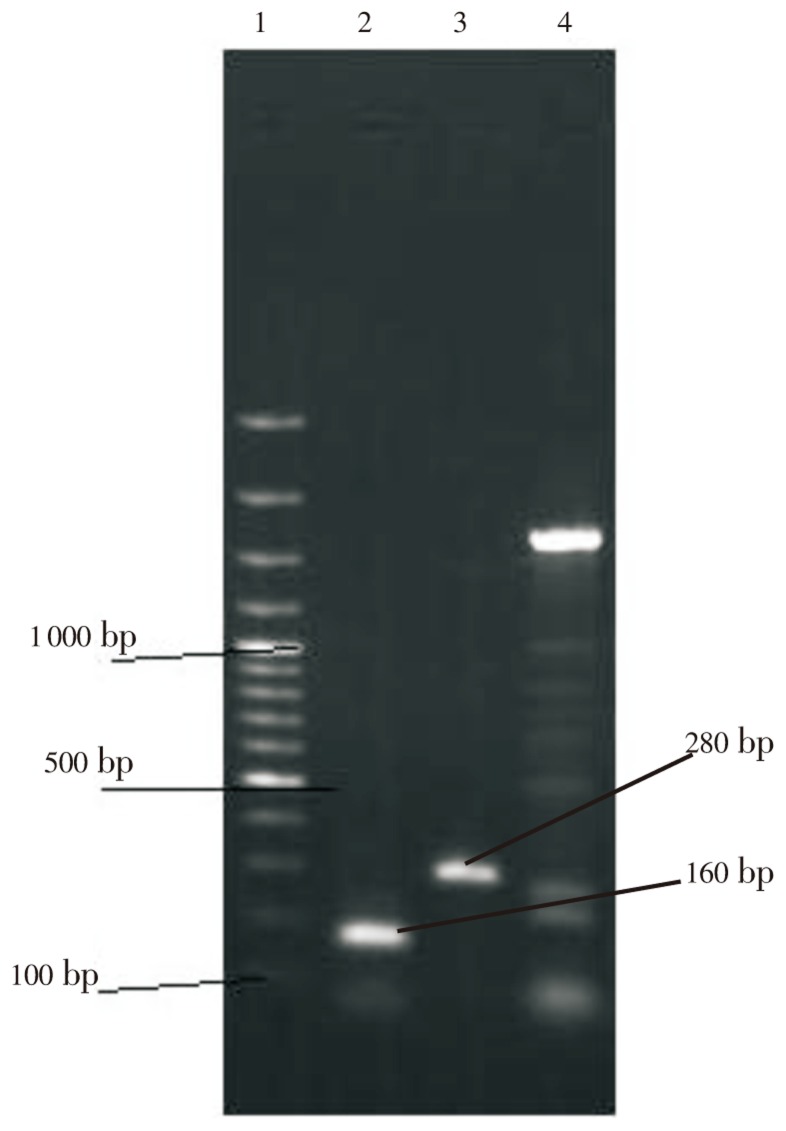
Amplification PCR with primers for the 5′- and 3′-CS primers was performed for detection of complete integron class 1. This amplification PCR also permitted the determination of the size of inserted gene cassette. Results showed that the inserted gene cassettes of class 1 integron ranged with variable sizes (220, 520, 750, 1 031, 1 250, 1 600, 2 200, 3 000 bp) and were found in 44 of 50 isolates (88%) (Figure 1).
Figure 1. Agarose gel electrophoresis of PCR amplified products generated from DNA samples.
Lanes 1 DNA size marker (100 bp DNA ladder, SM#333). Lane 2 and 3 show 160 bp integron class 1 and 280 bp integron class 2 amplification product. Lane 4 shows amplification product of integron by primers cs.
Detection of intI1 and intI2 genes by the integrase gene PCR showed that class I and class II integrons was detected in 42% (21/50) and 82% (41/50) of isolates, respectively. Also 30% (15/50) of isolates were both classes of integrons (integron class I&II).
3.1. Antimicrobial resistance profiles of integron-positive A. baumannii
In this study, relation between presence of integrons and susceptibility to 13 different antibiotics within A. baumannii strains were investigated. Tables 2 and 3 show antimicrobial resistance profiles of integron-positive and integron-negative A. baumannii strains. According to these tables, there was a significant correlation between presence of integrons and resistance to ciprofloxacin, ofloxacin, cefepime, ceftazidime, amikacin, aztreonam and norfloxacin (P<0.005), so that strains integron-positive showed higher resistance to this antibiotics. Also, there was not significant relationship between antibiotics such as gentamycin, piperacillin-tazobactam, imipenem, meropenem, ampicillin-sulbactam and tobramycin and presence of integrons that this subject indicates that there was different resistant mechanism other than integrons about this antibiotics. In this study, integron-positive A. baumannii isolates higher antibiotic resistance than integron-negative isolates.
Table 2. Antibiotic susceptibility of class 1 integron-positive and integron-negative of A. baumannii strains.
| Antimicrobial agents | Total (n=50) R% | Integron negative class I (n=29) |
Integron positive class I (n=21) |
P value | ||||
| R | I | S | R | I | S | |||
| Meropenem | 44 | 33.3 | 33.3 | 33.3 | 45.4 | 22.7 | 31.8 | Ns |
| Tobramycin | 28 | 0.0 | 16.6 | 83.3 | 31.8 | 20.4 | 47.7 | Ns |
| Ampicillin-Sulbactam | 62 | 33.3 | 50.0 | 16.6 | 65.9 | 22.7 | 11.3 | Ns |
| Imipenem | 78 | 66.6 | 16.6 | 16.6 | 79.5 | 11.3 | 9.0 | Ns |
| Aztreonam | 98 | 100.0 | 0.0 | 0.0 | 97.7 | 2.2 | 0.0 | 0.05 |
| Amikacin | 90 | 100.0 | 0.0 | 0.0 | 88.6 | 4.5 | 6.8 | 0.05 |
| Piperacillin/tazobactam | 48 | 33.3 | 33.3 | 33.3 | 50.0 | 34.0 | 16.9 | Ns |
| Ceftazidime | 100 | 100.0 | 0.0 | 0.0 | 100.0 | 0.0 | 0.0 | 0.05 |
| Norfloxacin | 96 | 100.0 | 0.0 | 0.0 | 95.4 | 0.0 | 4.5 | 0.05 |
| Cefepime | 100 | 100.0 | 0.0 | 0.0 | 100.0 | 0.0 | 0.0 | 0.05 |
| Ofloxacin | 92 | 83.3 | 16.6 | 0.0 | 93.1 | 2.27 | 4.5 | 0.05 |
| Gentamycin | 64 | 50.0 | 0.0 | 50.0 | 65.9 | 0.00 | 34.0 | Ns |
| Ciprofloxacin | 92 | 66.6 | 0.0 | 33.3 | 95.4 | 0.00 | 45.4 | 0.05 |
Table 3. Antibiotic susceptibility of class 2 integron-positive and integron-negative of A. baumannii strains.
| Antimicrobial agents | Total (n=50) R% | Integron negative Class II (n=9) |
Integron positive Class II (n=41) |
P value | ||||
| R | I | S | R | I | S | |||
| Meropenem | 44 | 41.3 | 27.5 | 31.2 | 47.6 | 19.0 | 33.3 | Ns |
| Tobramycin | 28 | 20.6 | 20.6 | 58.6 | 38.0 | 14.2 | 42.8 | Ns |
| Ampicillin-sulbactam | 62 | 51.7 | 31.0 | 17.2 | 76.1 | 19.0 | 4.0 | Ns |
| Imipenem | 78 | 72.4 | 17.2 | 10.3 | 85.7 | 4.0 | 9.0 | Ns |
| Aztreonam | 98 | 96.5 | 3.4 | 6.0 | 100.0 | 0.0 | 0.0 | 0.05 |
| Amikacin | 90 | 89.6 | 3.4 | 6.0 | 90.4 | 4.0 | 4.0 | 0.05 |
| Piperacillin/tazobactam | 48 | 41.3 | 34.4 | 24.1 | 57.1 | 33.3 | 9.0 | Ns |
| Ceftazidime | 100 | 100.0 | 0.0 | 0.0 | 100.0 | 0.0 | 0.0 | 0.05 |
| Norfloxacin | 96 | 93.1 | 0.0 | 6.8 | 100.0 | 0.0 | 0.0 | 0.05 |
| Cefepime | 100 | 100.0 | 0.0 | 0.0 | 100.0 | 0.0 | 0.0 | 0.05 |
| Ofloxacin | 92 | 89.6 | 3.4 | 6.8 | 95.2 | 4.7 | 0.0 | 0.05 |
| Gentamycin | 64 | 62.0 | 0.0 | 37.9 | 66.6 | 0.0 | 33.3 | Ns |
| Ciprofloxacin | 92 | 86.2 | 0.0 | 13.7 | 100.0 | 0.0 | 0.0 | 0.05 |
4. Discussion
In recent years, dissemination of antibiotic resistance genes through integrons in A. baumannii strains is a major problem in treatment of infections caused by these bacteria[9],[10]. This study is designed to determine the prevalence of integron classes 1 and 2 and their relationship with the presence of antibiotic resistance in nosocomial isolates of A. baumannii.
The findings of this study like to Bayugo and Joshìs studies showed that antibiotic resistance in A. baumannii isolates is increasing due to uncontrolled usage of drugs, so that 82% of investigated A. baumannii isolates Indicated multidrug- resistance (MDR) phenotype. Bayugo et al and Joshi et al in their studies reported that 45% to 75% (respectively) of A. baumannii strains as multidrug-resistant[15],[16].
In the present study, most resistant pattern observed in cefepime, ceftazidime, aztreonam, norfloxacin, ofloxacin, ciprofloxacin and amikacin and antibiotics such as piperacillin-tazobactam, meropenem, and imipenem were considered as the most effective drugs against A. baumannii strains that this findings largely agrees to results Ayan et al. about cefepime, ceftazidime, aztreonam, ampicillin-sulbactam and the results of Wang et al. about aztreonam, ceftazidime, ciprofloxacin and cefepime and also with studies of Rahbar et al. about ceftazidime, amikacin and ciprofloxacin in Iran[17]–[19].
In the present study, similar to Koeleman et al., Gonzale et al. and Ploy et al. studies, two different PCR assays were used to detect either class 1 integrons by amplification of any inserted gene cassette (by integron PCR) or class 1 and class 2 integrons by identification of the specific intI1 and intI2 genes (by integrase gene PCR)[14],[20],[21]. Overall, these studies showed that integrase gene PCR was more powerful and sensitive in detection of different classes of integrons than integron PCR.
Various studies around the world indicate that prevalence of integrons in A. baumannii strains, by using primers CS from 5% to 80% is variable. In the present study using of these primers, 88% of samples containing integron with size between 280 to 3 000 bp which this is in contrast with results of Ruiz et al, Ribera et al. and Koeleman et al. studies[14],[22],[23]. They expressed that rate of integrons with different sizes (less than 3 000 bp) was between 27.5% to 44%. Primers which were used in this study and above researches are same so this difference can be due to high prevalence of this type of integrons with different gene cassettes in A. baumannii strains which were isolated from clinical samples.
Different studies indicate that the rate of prevalence of integron class 1 in A. baumannii strains around the world is different and is variable between 5% to 84%. In this study, like koeleman et al. study in Netherland the rate of prevalence of integron class 1 in A. baumannii strains were determined by using integrase 1 gene (intI1) 44%[14]. In general, these differences could be due to prevalent epidemic strains and their discrimination in different parts of the world.
In the present study like Gonzalez et al. study and unlike Koeleman et al. study, the rate of detection of integron class 2 was more than integron class 1[14],[20]. In this study, 84% of A. baumannii strains were containing integron class 2 which were more than reports of other researchers around the world, that the rate of integron class 2 has been reported between 0 to 52.6%. These differences may due to study method[24]–[28].
Like Koeleman et al. study, this survey indicated that strains containing integrons were significantly associated with resistance to multiple antibiotics that this may be resulted by antibiotics resistant gene cassettes which can be code resistance to several antibiotics[14].
In the present study, like Lin et al. study were observed a significant correlation between presence of integrons and resistance to ciprofloxacin, ofloxacin, cefepime, ceftazidime, aztreonam, amikacin and norfloxacin so that integron-positive showed more resistance to these antibiotics[13]. Also in study which performed by Koeleman et al. indicated significant relationship between the presence of integrons and resistance to amikacin, ciprofloxacin and ceftazidime and such correlation between presence of integrons and resistance to imipenem and meropenem was not statistically justified that is completely match with the present study[14]. The results of Guar et al. study demonstrated same results with the present study that show correlation between presence of integrons and resistance to amikacin, cefepime and ciprofloxacin[12]. In cases that significant relationship between the presence of integrons and antibiotic resistance were not observed, resistance could be achieved by different ways such as deficiency in cell wall enzymes or resistance under plasmid or chromosome control[3],[14].
In conclusion, we have shown that different classes of integrons are widely disseminated among A. baumannii strains isolated from Tehran hospitals and these structures are playing a major role in the acquisition of multidrug-resistance in these strains. The most resistance in integron-positive strains is related to aminoglycosides, cephalosporins, quinolones and monobactams and about other antibiotics may involved other mechanisms of resistance. In this survey, regardless of whether resistance genes are present or not, strong relation between presence of integrons and reducing sensitivity to many groups of antibiotics were observed and this could be challenging because these structures can displacement of the genes involved in resistance among strains and therefore they become resistant to new antibiotics. So monitoring of drug resistance with use of gene integrase PCR is very important to plan specific infection control measures due to multidrug- resistance A. baumannii in Iran hospitals. Although, further studies on the prevalence integrons should be done in other parts from Iran.
Acknowledgments
This study was financially supported by Cell and Molecular Biology Research Center and Microbiology Group of Tehran Medicine University, with grant number TUMS/CMBRC- 89-007.
Comments
Background
A. baumannii is an important opportunistic pathogen, which is responsible for nosocomial infections. Currently, the strains with multidrug-resistant are a complicate problem in the hospitals. Therefore, epidemiologic study about A. baumanni is important. The manuscript report an antibiotic resistance and carriage class 1 and 2 integrons in clinical isolates from hospitalised patients.
Research frontiers
Acinetobacter is one of nosocomial infection agent in Iran. Multidrug resistance strains is one of major problems in hospitalized patients. Therefore, this is important that epidemiologic profile of this bacterium was defined in Iran.
Related reports
Study of prevalence integrons class 1 and 2 in A. baumannii is very low in Tehran. but some reports were existed about acinetobacter colonization and drug resistance from iran.
Innovations and breakthroughs
The prevalence of different classes of integrons an relationship with antibiotic resistance in nosocomial isolates of A. baumannii in hospital is done the prevalence integrons classes 1 and 2 in A. baumannii is not clear in Iran. This study is performed with the main aim of determinat of the prevalence of classes 1 and 2 integrons and the relationship with the presence of antibiotic resistance amonosocomial isolates of A. baumannii in Tehran hospitals.
Applications
It may be significant to know the distribution of A. baumannii hospitalized patients. The results of the present study suggest that most resistant pattern observed in cefepime, ceftazidime, aztreonam, norfloxacin, ofloxacin, ciprofloxacin and amikacin and antibiotics such as piperacillin-tazobactam, meropenem, and imipenem were considered as the most effective drugs against A. baumannii strains. Therefore, they become resistant to new antibiotics. On the other hand, monitoring of drug resistance with use of gene integrase PCR is very important to plan specific infection control measures due.
Peer review
This is a good study in which the authors evaluated the antibiotic resistance and carriage class 1 and 2 integrons in clinical. Isolates of A. baumannii from Tehran, Iran. The results are interesting and suggested that class 1 and 2 integrons are present especially in A. baumannii isolated from clinical specimen.
Footnotes
Foundation Project: This study was financially supported by Cell and Molecular Biology Research Center and Microbiology group of Tehran Medicine University, with grant number TUMS/CMBRC-89-007.
Conflict of interest statement: We declare that we have no conflict of interest.
References
- 1.Fournier PE, Richet H. The epidemiology and control of Acinetobacter baumannii in health care facilities. Clin Infect Dis. 2006;42:692–699. doi: 10.1086/500202. [DOI] [PubMed] [Google Scholar]
- 2.Maragakis LL, Perl TM. Acinetobacter baumannii: epidemiology, antimicrobial resistance, and treatment options. Clin Infect Dis. 2008;46:1254–1263. doi: 10.1086/529198. [DOI] [PubMed] [Google Scholar]
- 3.Nemec A. Multidrug resistant Acinetobacter baumannii. Klin Mikrobiol Infekc Lek. 2008;14:162–167. [PubMed] [Google Scholar]
- 4.Karageorgopoulos DE, Falagas ME. Current control and treatment of multidrug-resistant Acinetobacter baumannii infections. Lancet Infect Dis. 2008;8:751–762. doi: 10.1016/S1473-3099(08)70279-2. [DOI] [PubMed] [Google Scholar]
- 5.Bassetti M, Righi E, Esposito S, Petrosillo N, Nicolini L. Drug treatment for multidrug-resistant Acinetobacter baumannii infections. Future Microbiol. 2008;3:649–660. doi: 10.2217/17460913.3.6.649. [DOI] [PubMed] [Google Scholar]
- 6.Michalopoulos A, Falagas ME. Treatment of Acinetobacter infections. Expert Opin Pharmacother. 2010;11:779–788. doi: 10.1517/14656561003596350. [DOI] [PubMed] [Google Scholar]
- 7.Dijkshoorn L, Nemec A, Seifert H. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol. 2007;5:939–951. doi: 10.1038/nrmicro1789. [DOI] [PubMed] [Google Scholar]
- 8.Neonakis IK, Spandidos DA, Petinaki E. Confronting multidrug-resistant Acinetobacter baumannii: a review. Int J Antimicrob Agents. 2011;37:102–109. doi: 10.1016/j.ijantimicag.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 9.Garnacho-Montero J, Amaya-Villar R. Multiresistant Acinetobacter baumannii infections: epidemiology and management. Curr Opin Infect Dis. 2010;23:332–339. doi: 10.1097/QCO.0b013e32833ae38b. [DOI] [PubMed] [Google Scholar]
- 10.Cambray G, Guerout AM, Mazel D. Integrons. Annu Rev Genet. 2010;44:141–166. doi: 10.1146/annurev-genet-102209-163504. [DOI] [PubMed] [Google Scholar]
- 11.Labbate M, Case RJ, Stokes HW. The integron/gene cassette system: an active player in bacterial adaptation. Methods Mol Biol. 2009;532:103–125. doi: 10.1007/978-1-60327-853-9_6. [DOI] [PubMed] [Google Scholar]
- 12.Gaur A, Prakash P, Anupurba S, Mohapatra TM. Possible role of integrase gene polymerase chain reaction as an epidemiological marker: study of multidrug-resistant Acinetobacter baumannii isolated from nosocomial infections. Int J Antimicrob Agents. 2007;29:446–450. doi: 10.1016/j.ijantimicag.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 13.Lin MF, Chang KC, Yang CY, Yang CM, Xiao CC, Kuo HY, et al. Role of integrons in antimicrobial susceptibility patterns of Acinetobacter baumannii. Jpn J Infect Dis. 2010;63:440–443. [PubMed] [Google Scholar]
- 14.Koeleman JG, Stoof J, Van Der Bijl MW, Vandenbroucke-Grauls CM, Savelkoul PH. Identification of epidemic strains of Acinetobacter baumannii by integrase gene PCR. J Clin Microbiol. 2001;39:8–13. doi: 10.1128/JCM.39.1.8-13.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bayuga S, Zeana C, Sahni J, Della-latta P, El-Sadr W, Larson E. Prevalence and antimicrobial patterns of Acinetobacter baumanii on hands and nares of hospital personnel and patients: the iceberg phenomena again. Heart Lung. 2002;31:382–390. doi: 10.1067/mhl.2002.126103. [DOI] [PubMed] [Google Scholar]
- 16.Joshi SG, Litake GM, Niphadkar KB, Ghole VS. Multidrug resistant Acinetobacter baumannii isolates from a teaching hospital. J Infect Chemother. 2003;9:187–190. doi: 10.1007/s10156-002-0224-4. [DOI] [PubMed] [Google Scholar]
- 17.Ayan M, Durmaz R, Aktas E, Durmaz B. Bacteriological, clinical and epidemiological characteristics of hospital-acquired Acinetobacter baumannii infection in a teaching hospital. J Hosp Infect. 2003;54:39–45. doi: 10.1016/s0195-6701(03)00076-8. [DOI] [PubMed] [Google Scholar]
- 18.Wang SH, Sheng WH, Chang YY, Wang LH, Lin HC, Chen ML, et al. Healthcare-associated outbreak due to pan-drug resistant Acinetobacter baumanii in a surgical intensive care unit. J Hosp Infect. 2003;53:97–102. doi: 10.1053/jhin.2002.1348. [DOI] [PubMed] [Google Scholar]
- 19.Rahbar M, Mehrgan H, Aliakbari NH. Prevalence of antibiotic-resistant Acinetobacter baumannii in a 1000-bed tertiary care hospital in Tehran, Iran. Indian. J Pathol Microbiol. 2010;53:290–293. doi: 10.4103/0377-4929.64333. [DOI] [PubMed] [Google Scholar]
- 20.Gonzalez G, Sossa K, Bello H, Dominguez M, Mella S, Zemel-man R. Presence of integrons in isolates of different biotypes of Acinetobacter baumannii from Chilean hospitals. FEMS Microbiol Lett. 1998;161:125–128. doi: 10.1111/j.1574-6968.1998.tb12937.x. [DOI] [PubMed] [Google Scholar]
- 21.Ploy MC, Denis F, Courvalin P, Lambert T. Molecular characterization of integrons in Acinetobacter baumannii, description of a hybrid class 2 integron. Antimicrob Agents Chemother. 2000;44:2684–2688. doi: 10.1128/aac.44.10.2684-2688.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruiz J, Navia MM, Casals C, Sierra JM, Jiménez De Anta MT. Integron-mediated antibiotic multiresistance in Acinetobacter baumannii clinical isolates from Spain. Clin Microbiol Infect. 2003;9:907–911. doi: 10.1046/j.1469-0691.2003.00561.x. [DOI] [PubMed] [Google Scholar]
- 23.Ribera A, Vila J, Fernández-Cuenca F, Martínez-Martínez L, Pascual A, Beceiro A, et al. Type 1 integrons in epidemiologically unrelated Acinetobacter baumannii isolates collected at Spanish hospitals. Antimicrob Agents Chemother. 2004;48:364–365. doi: 10.1128/AAC.48.1.364-365.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramírez MS, Stietz MS, Vilacoba E, Jeric P, Limansky AS, Catalano M, et al. Increasing frequency of class 1 and 2 integrons in multidrug-resistant clones of Acinetobacter baumannii reveals the need for continuous molecular surveillance. Int J Antimicrob Agents. 2011;37:175–177. doi: 10.1016/j.ijantimicag.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 25.D'Arezzo S, Capone A, Petrosillo N, Visca P, Ballardini M, Bartolini S, et al. Epidemic multidrug-resistant Acinetobacter baumannii related to European clonal types I and II in Rome (Italy) Clin Microbiol Infect. 2009;15:347–357. doi: 10.1111/j.1469-0691.2009.02668.x. [DOI] [PubMed] [Google Scholar]
- 26.Koo SH, Kwon KC, Cho HH, Sung JY. Genetic basis of multidrug-resistant Acinetobacter baumannii clinical isolates from three university hospitals in Chungcheong Province, Korea. Korean J Lab Med. 2010;30:498–506. doi: 10.3343/kjlm.2010.30.5.498. [DOI] [PubMed] [Google Scholar]
- 27.Kansakar P, Dorji D, Chongtrakool P, Mingmongkolchai S, Mokmake B, Dubbs P. Local dissemination of multidrug-resistant Acinetobacter baumannii clones in a Thai hospital. Microb Drug Resist. 2011;17:109–119. doi: 10.1089/mdr.2010.0062. [DOI] [PubMed] [Google Scholar]
- 28.Turton JF, Kaufmann ME, Glover J, Coelho JM, Warner M, Pike R, et al. Detection and typing of integrons in epidemic strains of Acinetobacter baumannii found in the United Kingdom. J Clin Microbiol. 2005;43:3074–3082. doi: 10.1128/JCM.43.7.3074-3082.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]