Abstract
Objective
To investigate the infection of hospital- and community-acquired “erythromycin-induced clindamycin resistant” strains or D-test positives of clinical isolates of Staphylococcus aureus (S. aureus) (with and without methicillin resistance) in a hospital.
Methods
Strains of S. aureus isolated from clinical specimens were subjected to D-test and antibiotic profiling.
Results
Of the total 278 isolates, 140 (50.35%) were D-test positives and the rest were D-test negatives. Further, of 140 (100%) positives, 87 (62.14%) and 53 (37.85%) strains were from males and females, respectively. Of 140 (100%) positives, 117 (83.57%) were methicillin resistant S. aureus and 23 (16.42%) were methicillin sensitive S. aureus; of 140 strains, 103 (73.57%) strains from persons with and 37 (26.42%) were without related infections; of 140 strains, 91 (65%) and 49 (35%) were from hospital- and community-acquired samples, respectively. In 140 strains, 118 (84.28%) with comorbidities and 22 (15.71%) without comorbidities cases were recorded; similarly, persons with prior antibiotic uses contributed 108 (77.14%) and without 32 (22.85%) positive strains. These binary data of surveillance were analyzed by a univariate analysis. It was evident that the prior antibiotic uses and comorbidities due to other ailments were the determinative factors in D-test positivity, corroborated by low P values, P=0.001 1 and 0.002 4, respectively. All isolates (278) were resistant to 17 antibiotics of nine groups, in varying degrees; the minimum of 28% resistance for vancomycin and the maximum of 97% resistance for gentamicin were recorded. Further, of 278 strains, only 42 (15.1%) strains were resistant constitutively to both antibiotics, erythromycin resistant and clindamycin resistant, while 45 (16.2%) strains were constitutively sensitive to both antibiotics (erythromycin sensitive and clindamycin sensitive). Further, of the rest 191 (68.7%) strains were with erythromycin resistant and clindamycin resistant, of which only 140 (50.35%) strains were D-test positives, while the rest 51 (18.34%) strains were D-test negatives.
Conclusions
In view of high prevalence of D-test positive S. aureus strains, and equally high prevalence of multidrug resistant strains both in community and hospital sectors, undertaking of D-test may be routinely conducted for suppurative infections.
Keywords: Antibiotics, Community-acquired, D-test, Erythromycin resistance, Hospital-acquired, Inducible clindamycin resistance, MRSA, MSSA, Staphylococcus aureus
1. Introduction
As a commensal, Staphylococcus aureus (S. aureus) colonizes asymptomatically nares of nose, skin and soft tissues of healthy individuals. But through bloodstream infection, a number of casual to serious ailments are caused, such as skin reactions, rhinitis, otitis media infection, mastitis, suppurative wounds, osteomyelitis, septic arthritis, urinary tract infections, and several life-threatening invasions, i.e. pneumonia, septicemia, bacteraemia, endocarditis and toxic-shock syndrome[1]; an infectious S. aureus brings a retinue of damnedest comorbidities, even fatality. Per se, S. aureus has been the most prevalent Gram-positive pathogen in India[2]. For the in vivo control, erythromycin has been in use since 3-4 decades and resistance to it by S. aureus has been reported since long[3]. Its invasive/insinuative nature is evident with its aggrandizement of resistance to multiple drugs, including vancomycin. And, methicillin resistant S. aureus (MRSA) was also found resistant to other preferred antibiotic, streptogramin B. Consequently, clindamycin, another wonted drug against Gram-positive pathogens was in use for S. aureus.
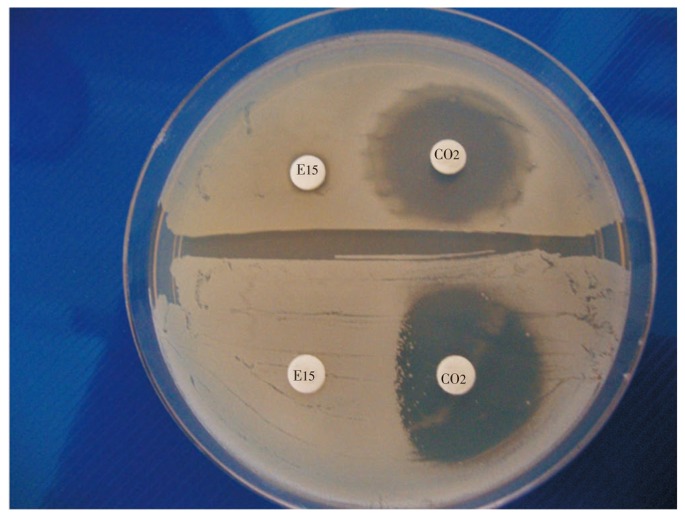
Surprisingly, inducible clindamycin resistance (Cd-r) of both methicillin sensitive S. aureus (MSSA) and MRSA, due to erythromycin resistance (Er-r) had been accentuated[4]. It was ascertained that, Cd-r mutants harbor the erm gene [Er-r gene that induces resistance to the macrolides, lincosamides and streptogramin B (MLSB) group, by a methylation at the 23s r-RNA subunit that leads to methylation,5]. There are two types of Er-r S. aureus strains, i.e., with and without the MLSB gene. In the presence of erythromycin, the strain with the MLSB gene induces resistance to clindamycin in the “Er-r, Cd-s” strain, conferring clindamycin resistance to the original Cd-s strain, eventually causing the well-known flattening of the clindamycin inhibition zone towards the erythromycin disc, so that the shape “D” is seen in the clindamycin zone or “D-test positivity” (Figure 1). Since, failure in the therapy with clindamycin used against S. aureus had been frequently met[4],[6], the D-test procedure is often recommended for checking the efficacy of the empiric use of clindamycin against isolated staphylococci in most hospitals to avoid the unbeknown pervasive error in the therapy, due to MLSB resistance. Admittedly, it is the standard procedure, being simple for checking the inducible erm mediated MLSB resistance in MRSA and other staphylococci. Moreover, inducible-MLSB S. aureus strains have been isolated independently with resistant patterns for a number of antibiotics in use, in diverse geographical zones[3]; their abundance have been reported up to the saturnine height of 94% of S. aureus isolates, a decade ago[7].
Figure 1. D-shape flattening of clindamycin sensitive zone of S. aureus induced by erythromycin resistance.
Both bacterial strains plated were erythromycin resistant, but the strain with MLSB gene had D-shape flattening or D-test positivity, while the other strain without MLSB gene had no D-shape zone from clindamycin toward the erythromycin disc.
This fixated study characterizes the prevalence of S. aureus in samples from in-house patients, hospitalized in wards, cabins, intensive care units, and neonatal intensive care units for 2 or more d, taken as hospital-acquired (HA), and samples from patients who regularly/intermittently visit outpatient department, taken as community-acquired (CA). Further, since clindamycin is frequently used empirically before results of cultures of clinical samples could be obtained for patients with aerobic-anaerobic infection from intra-abdominal sepsis, aspiration pneumonia, soft tissue infections, cellulites and post-surgical wounds, etc.[8], it was a deliberate attempt of surveillance in probing to the occurrence of D-test positive S. aureus strains, in a resource-limited setting. Obviously, a post-hoc analysis on the cause of failure in to-do-away-with the multidrug resistant strains of this pathogen by an empiric treatment with any member of the MLSB group, specifically the clindamycin would be a clinical misdemeanour. A heedful univariate analysis of the bivalence of D-test results with several hospital factors such as, sex, presence of comorbidities, etc., vindicates this study. Further, an antibiogram of a spectrum of 278 isolates of S. aureus with 17 antibiotics was obtained that gave an idea on the prevalence of the insidious infection-dynamics and the associated shenanigans of this notorious super-bug of health domain, for a benefit of apothecary in dove-tailing suitable drugs and to decrease unwarranted increases in the growing cost of hospital care, in face of the intimidating erythromycin-induced MLSB resistance.
2. Materials and methods
2.1. Isolation and antibiotic susceptibility
The study was conducted for a period of 6 months (April to September 2011) and a total of 278 strains of S. aureus were isolated from different clinical samples from HA and CA sources of Institute of Medical Science & Sum Hospital. Isolated strains were identified by using the standard microbiological procedures[9]. The MSSA strain, Microbial Type Culture Collections strain number 7443 was used as the reference control. This strain and all isolated strains were subjected to antibiotic sensitivity test, by the disc diffusion method, detailed previously[10].
2.2. Detection of MRSA
For the cefoxitin disc diffusion test, a 0.5 McFarland standard equivalent suspension of a test isolate was plated for lawn culture on a Muller-Hinton agar plate; a cefoxitin disc 30 µg/disc was placed on the lawn-center. Plates were incubated at 37 °C for 18 h and inhibition-zone diameters were measured; a value ≥19 mm was recorded as methicillin resistant and a value, ≤20 mm was considered as methicillin sensitive[11]. For the chromogenic agar media test, pure clinical isolates of S. aureus were streaked onto MRSA-agar, the Hichrome-MeReSa agar (HiMedia, Mumbai), and were incubated for 24 h at 37 °C; MRSA strains had blue colonies and MSSA strains had white colonies[12].
2.3. D-test
Isolates that were “Er-r, Cd-s” were tested for inducible Cd-r, by susceptibility to clindamycin 2 µg/disc and erythromycin 15 µg/disc levels along with the reference strain, according to CLSI criteria[13]. Erythromycin and clindamycin discs (HiMedia, Mumbai) were placed (17±2) mm apart (edge to edge) on a Muller-Hinton agar plate, incubated at 37 °C for 18 h and D-test positivity was identified by the flattening of clindamycin zone between erythromycin and clindamycin discs. Any isolate with “Er-r, Cd-r” was considered as constitutive MLSB resistant strain[14].
3. Results
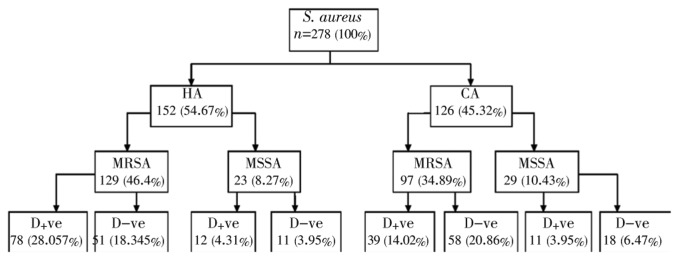
A total of 278 strains of S. aureus were isolated from clinical samples, pus, different swabs, urine, body fluids and blood, in the cited order of prevalence, both in HA and CA sources (Table 1). Of 278 (100%) strains, 152 (54.67%) isolates were from HA and 126 (45.32%) from CA samples. Of 152 HA isolates, 129 (46.4%) strains were MRSA and 23 (8.27%) were MSSA, whereas of the total 126 (45.32%) CA strains, 97 (34.89%) were MRSA and 29 (10.43%) were MSSA (Table 1 and Figure 2).
Table 1. Occurrence of inducible clindamycin resistant isolates as D-test positives in total S. aureus strains in different clinical samples.
| Source samples | Hospital acquired |
Community acquired |
||
| MRSA | MSSA | MRSA | MSSA | |
| Pus | 41 (32) | 6 (4) | 33 (14) | 9 (3) |
| Wound swabs | 32 (18) | 4 (2) | 24 (7) | 5 (3) |
| Skin swabs | 21 (13) | 4 (2) | 17 (8) | 4 (2) |
| Nasal swabs | 16 (7) | 3 (2) | 12 (4) | 4 (2) |
| Urine | 9 (4) | 3 (2) | 5 (3) | 3 (-) |
| Body fluids | 7 (4) | 2 (-) | 3 (2) | 3 (-) |
| Blood | 3 (-) | 1 (-) | 3 (1) | 1 (1) |
| Total | 129 (78) | 23 (12) | 97 (39) | 29 (11) |
Numbers in parenthesis represents D-test positives strains, the total being 140, out of the grand total number of 278 strains.
Figure 2. An account of D-test positive and negative colonies of S. aureus with respect to other variables.
Of the total 278 strains, 140 (50.35%) were D-test positive and 138 (49.64%) were D-test negative. D-test positivity/negativity had concern to other variable factors, sensitivity to methicillin, source of strains, sex, other associated ailments, other infections, and prior antibiotic use. Thus, the following bivalents with respect to both D-test positive and negative strains were monitored (Table 2): 1. MRSA/MSSA; 2. HA/CA sources; 3. Males/females; 4. Presence/absence of associated comorbidities (presence/absence of diabetes-related ailments any other problems like cardiac complaints, noteworthy diseases, etc.); 5. Presence/absence of related infections; and 6. Their prior antibiotic uses within >90 d (Table 2). The P-value for two pairs of data, D-test positive/negative for MRSA/MSSA pair was 0.324 2, which signified that there was no statistically respectable difference of D-test positivity between MRSA/MSSA pair (for 140 strains of D-test positives and 138 negatives); similarly, the D-test positive/negative cases related to sex were not significant at the level. On the contrary, for the rest 4 pairs of bivalents as mentioned above (numbers 2, 4, 5 and 6), P-values were statistically significant, signifying there were statistically respectable differences (Table 2). Further, for both the bivalent data for “prior antibiotic use”, and “presence or absence of comorbidities” with P=0.001 1 and 0.002 4, respectively, for D-test positive/negative cases confirm the difference statistically. In other words, the distribution of MLSB gene in samples from patients “with prior antibiotic use” and “presence of comorbidities” had determinative roles in D-test positivity.
Table 2. Univariate analysis of D-test positive and D-test negative isolates of S. aureus*.
| Variables | D-test positive | D-test negative | P-value | Odds ratio | Range(%95 CI) | |
| Strains | MRSA | 117 | 109 | 0.324 2 | 1.353 4 | 0.7381-2.4816 |
| MSSA | 23 | 29 | ||||
| Sources | HA | 91 | 67 | 0.005 0 | 1.968 0 | 1.2157-3.1859 |
| CA | 49 | 71 | ||||
| Sex | Male | 87 | 102 | 0.033 0 | 0.740 3 | 0.4546-1.2056 |
| Female | 53 | 46 | ||||
| Comorbidity | Present | 118 | 96 | 0.002 4 | 2.346 6 | 1.3113-4.1993 |
| Absent | 22 | 42 | ||||
| Related infections | Present | 103 | 116 | 0.028 1 | 1.007 0 | 0.6019-1.6877 |
| Absent | 37 | 42 | ||||
| use >90 d Prior antibiotic | Present | 108 | 82 | 0.001 1 | 3.539 6 | 2.153-5.8191 |
| Absent | 32 | 86 | ||||
*See text for detailed information on variables and for abbreviations.
The univariate analysis of surveillance data revealed that MRSA detection had 1.353 4 times more risk factor or vulnerability to express the MLSB gene along with the acquiring of Er-r factor than MSSA prevalence in causing D-test positivity. Similarly, there was 1.968 0 times more chance of prevalence of the MLSB gene in Er-r S. aureus from HA samples to have often D-test positivity than those from CA samples. Patients with other comorbidities had been recorded to have 2.346 6 more chance than patients without any comorbidity for positivity. And patients with a history of prior antibiotic uses had the highest value of 3.359 6 more chance than patients without any such history for positivity. On the other hand, males were found to have 0.740 3 times less chance in acquiring inducible Cd-r than females, in this surveillance. However, patients with or without other related infections had an equal chance of acquiring inducible Cd-r (Table 2).
Resistance at a minimum of 36% and 13% for daptomycin, 34% and 28% for vancomycin, and the maximum of 97% and 95% for gentamicin and 95% and 86% for oxacillin were recorded for HA and CA S. aureus isolates, respectively. Further, it was clear that resistant values of isolates to erythromycin were 83% and 67%, and those were independently resistant to clindamycin by 76% and 81% at CA and HA isolates, respectively (Table 3).
Table 3. Percentage of resistance of S. aureus to 17 antibiotics of various groups with both hospital acquired and community acquired strains (n=278).
| Antibiotic group | Antibiotics (µg/disc) | HA isolates (%) | CA isolates (%) |
| Aminoglycosides | Amikacin 30 | 89 | 74 |
| Gentamicin 10 | 97 | 95 | |
| β-lactams | Amoxyclav 30 | 85 | 76 |
| Ampicillin 10 | 88 | 68 | |
| Oxacillin 1 | 95 | 86 | |
| Penicillin 10 | 36 | 56 | |
| Fluoroquinolone | Gatifloxacin 05 | 78 | 67 |
| Glycopeptides | Teicoplanin 10 | 80 | 59 |
| Vancomycin 30 | 28 | 34 | |
| Lincosamide | Clindamycin 2 | 81 | 76 |
| Lipopeptide | Daptomycin 30 | 36 | 13 |
| Macrolides | Azithromycin 15 | 72 | 54 |
| Erythromycin 15 | 83 | 67 | |
| Sulfonamide | Co-trimoxazole 5 | 78 | 49 |
| Stand-alone antibiotics | Chloramphenicol 30 | 72 | 61 |
| Linezolid 30 | 67 | 37 | |
| Tetracycline 30 | 45 | 34 |
These multidrug resistant isolates were at such a high abundance that antibiotics, gentamicin and gatifloxacin were excluded deliberately for S. aureus, from antimicrobial stewardship programme.
On separate lawn cultures of all isolates, two discs-erythromycin 15 µg/disc and clindamycin 2 µg/disc were used for checking the susceptibility pattern. Of 278 (100%) strains, 42 (15.1%) were constitutively resistant to both antibiotics, while 45 (16.1%) more strains were constitutively sensitive to both. The rest 191 (68.7%) strains were expected to be D-test positive, but only 140 (50.35%) strains had positivity, while the rest 51 (18.34%) strains were negatives (Table 4). Thus, this study had data of constitutive resistant pattern for both (Cd-r, Er-r) and constitutive susceptibility pattern for both (Er-s, Cd-s). These phenotypes in the routine isolation procedure were isolated, which helped to assess the prevalence of D-test positives among MSSA and MRSA isolates. It was found that the double constitutive resistance of MSSA was totally absent in both HA and CA samples; on the other hand, double constitutive sensitive phenotype (Er-s, Cd-s) were 4 and 8 isolates in total from MSSA isolates, whereas this phenotype occurred as 7 and 26 from MRSA isolates, in HA and CA cohorts, respectively (of total D-test positives). This clearly indicated that negligible fractions of both constitutive sensitive and resistant phenotypes were prevalent, which were unsuitable for checking D-test positivity (Table 4).
Table 4. Patterns of sensitivity and resistance to antibiotics in strains of S. aureus to erythromycin and clindamycin during D-test.
| Strains | Hospital acquired |
Community acquired |
Total | ||
| MRSA | MSSA | MRSA | MSSA | ||
| Er-r + Cd-r Constitutive resistant | 25 | 0 | 17 | 0 | 42 |
| Er-s + Cd-sV Constitutive sensitive | 7 | 4 | 26 | 8 | 45 |
| Er-r + Cd-s D-test negative | 18 | 7 | 21 | 5 | 51 |
| Er-r + Cd-s D-test positive | 78 | 12 | 39 | 11 | 140 |
Er-r: erythromycin resistant; Er-s: erythromycin sensitive; Cd-r: clindamycin resistant; Cd-s: clindamycin sensitive. Both Er-r and Cd-r strains were taken as of constitutive resistance; both Er-s and Cd-s strains were taken as of constitutive sensitive. Total number of D-test positives=140; total number of S. aureus strains=278.
4. Discussion
During the unifying assessment of 278 strains resistant to 17 antibiotics, the infection-dynamics of this iconic notorious pathogen was discernible with the minimum of 28% resistance to vancomycin and the maximum of 97% resistance to gentamicin. Indeed, occurrence of high percentage of resistance to daptomycin at 36% in HA samples is of high clinical concern in this study. The most striking situation was that S. aureus strains have emerged with concomitant resistance to many commonly used antibiotics of groups seen here, also as seen elsewhere[1]. Surprisingly, the imperiling value of ∼30% epidemiological prevalence of vancomycin resistant S. aureus in this hospital is a matter of concern; these could be due to errors in manual method of determining antibiotic susceptibility pattern in a resource limited settings with the absence of an automated technique, the use of vancomycin in empiric therapy and overall, the absence of a stringent antibiotic policy in local hospitals, to state contemplatively. Moreover, in a European country, of 750 clinically isolated S. aureus strains, 38% D-test positives were obtained in CA and 67% in HA-MRSA isolates; but the D-test positive figure for HA-MSSA was 63.6%; further, MRSA isolates were often found resistant to cephalosporins, cefems and other β-lactams, ampicillin-sulbactam, amoxyclav, ticarcillin-clavulanic acid, piperacillin-tazobactam and the carbapenem, imipenem[15]. According to our survey, the percentage of D-test positivity in CA isolates was lower than that of HA strains.
Strains that were Er-r when plated with Cd-s were expected to have D-test positivity, but out of the total 191 (Er-r, Cd-s) isolates, only 140 strains were D-test positive. Thus, a cohort of 51 Er-r strains was unable to induce D-like flattening of clindamycin-inhibition zone, due to the absence of MLSB gene. From the analysis of D-test positivity with variable factors, MRSA/MSSA, sex, absence/presence of comorbidities, etc., it was evident that the distribution pattern of MLSB gene was not universal among all Er-r isolates that could be the cause of 140 D-test positives only, among 191 Er-r strains. It is imperative that some other mechanism is also involved in Er-r, at least with 51 stains herein that could be the active efflux mechanisms to evade antibiotics of the MLSB group by an intrinsic gene[5]. Moreover, in this study the two types of phenotypes, D and D+, basing on the size of the clear zone around the erythromycin disc less than 6 mm for former and more than 8 mm for the later as described[16], were not detected in this study.
Among 244 clinical isolates of staphylococci reported from Karnataka, India, 13.1% strains had inducible clindamycin resistance with the MLSB phenotype; among them, 10 isolates were MRSA (38.4% of the total MRSA), 16 were MSSA (12.9% of the total MSSA) and 6 were “coagulase-negative staphylococci” or CONS, i.e., 6.3% of the total isolated CONS[17]. In another laboratory from Karnataka, 10% isolates had inducible clindamycin resistance, 9% had constitutive resistance and 8% had MS phenotype. Inducible resistance and constitutive resistance were found to be higher in MRSA as compared to MSSA (20%, 16% and 6%, 6%, respectively)[18]. The prevalence of MLSB strains both in CA and HA S. aureus isolates, as well as the prevalence of CA-MRSA strains were identified as clinical predictors of both CA-MRSA and MLSB, in Albama, USA[3]. Among 402 S. aureus isolates, the prevalence of MLSB was 52%, of which 50% of MRSA and 60% of MSSA isolates were MLSB; CA-MRSA were 14% of all isolates and had a lower prevalence of MLSB than HA-MRSA: 33% versus 55%, respectively[3]. A total of 159 staphylococcal isolates from burn patients in the Tripoli Burn Center were tested for inducible clindamycin resistance, which was detected in 66.2% of 65 MRSA isolates and in none of 55 MSSA, 10 methicillin-resistant CONS and 29 methicllin-sensitive CONS isolates[19]. It was reported that 88.6% MRSA isolates were Er-r and 52.3% were Cd-r in Iran; values of resistance in MSSA strains to erythromycin and clindamycin were 22% and 11.4%, respectively. Inducible clindamycin resistance was detected in 20.5% MRSA isolates; but, 52.3% of MRSA isolates and 7.3% of MSSA had constitutive MLSB phenotype[20].
The round zones due to erythromycin and clindamycin radiating out from each disc partially were observed. Erythromycin molecules reach the outer region of clindamycin zone prior to clindamycin molecules. The presence of the MLSB genotype in the lawn of Er-r stain led a methylase translation permitting the growth in this region, despite the diffusion of an inhibitory concentration of clindamycin. The D-test positives render clinical difficulty, due to the failure of clindamycin treatment of MRSA; eventually the D-test becomes an implicit trust. In vitro testing of isolates with MLSB genotype demonstrates clindamycin susceptibility. But macrolide inducible DNA sequence that preceded the erm (methylase) open reading undergoes mutation, substitution or deletion that generate a readily translatable (now the constitutive MLSB) stretch of DNA; its secondary m-RNA structure was recorded to be about of 1-2 million base pairs[21].
In the realm of imagination, origin of multidrug resistance in S. aureus has many possibilities: 1. The development of exquisite clonal nexuses of S. aureus is fast due to the genome simplicity[21]. 2. “Positive selection pressure”, the accepted/viable concept of evolution could be valid, not least because of the availability of antibiotics and their degraded toxic products readily in nature, such as in untreated hospital and community drains, but an altered influx potential in disallowing an antibiotic through plasma membrane, could often be the mechanism involved for resistance, as exemplified elsewhere[22]. 3. Genetic recombination mechanisms-conjugation and transformation should occur more readily than expected in untreated hospital sewage system, because all sorts of bacteria with grading levels of antibiotic resistance are physically together, and DNA from lysed cells would be readily available for uptake by living cells, trickling genome improvement as discussed[10]. In developing sections, the scientific disposition of hospital waste should be expected to be in a developing state, unwittingly-giving space for pathogen spreads. 4. Horizontal transfer/suffusion of drug-resistant pathogens to both community and hospital settings is expected because of the accumulated grime from crowding of patients and their attendants in resource limited hospital settings; a priory, slum areas of developing zones of developing countries might be conducive to pathogen spread in community. When, a bacterial strain musters a set of drug-resistant characters as an armamentarium against antibiotics of present time in an individual patient, it acts as a doppelgänger during improvement in all strains of the species in the patient-body, as if with a snowball decent time-to-time. Thus, spread of the novel strain in community is the aftermath. Such events occur continually and independently with each pathogen. This is the mechanism of transformation of the harmless commensal S. aureus to the ghoulish, intractable, perilous and wily superbug MRSA[23].
Binary outcomes of surveillance data elucidated that “prior antibiotic uses” and “comorbidities due to other ailments” were determinative factors of D-test positivity. It could be identified here that inducible MLSB strain was widespread in hospital sectors, so D-test protocol need to be included as the routine diagnostic procedure for suppurative ailments of incoming patients. But, in the community sector, there was less Cd-r S. aureus strains. This study indicated negligible fractions of constitutive “sensitive” and “resistant” phenotypes that were pejorative for D-test positivity. In view of the high prevalence of D-test positive S. aureus strains both in community and hospital sectors, undertaking of D-test may be routinely conducted.
Acknowledgments
D Dubey is supported by an INSPIRE Fellowship from DST, Govt. of India, New Delhi. We are grateful to Dr. DK Roy, Dean, IMS & Sum Hospital, for extended facilities. This work, in part, was supported by a MRP in Botany, ‘Alternative drug search from ethno-medicinal plants of Odisha against multidrug resistant bacteria’, by UGC (New Delhi) and by another research scheme from CSIR (New Delhi), no. 21 (0859)/11/EMR-II, awarded to RN Padhy. We thank Prof. Dr. MR Nayak, Honourable President, SOA University, for encouragements.
Comments
Background
An infectious S. aureus brings a chain of comorbidities, even fatality, and S. aureus has been the most prevalent Gram-positive pathogen. For the in vivo control, erythromycin has been in use since 3-4 decades and resistance to it by S. aureus had been reported since long. Its invasive nature is evident with its attainment of resistance to multiple drugs, including vancomycin. And, methicillin resistant S. aureus (MRSA) was too found resistant to the other preferred antibiotic, streptogramin B. Consequently, clindamycin, another mostly used drug against Gram-positive pathogens was in use for S. aureus. Surprisingly, inducible clindamycin resistance (Cd-r) of both methicillin sensitive S. aureus (MSSA) and MRSA, due to erythromycin resistance (Er-r) had been known. It was ascertained that, Cd-r mutants harbor the erm gene [Er-r gene that induces resistance to the MLSB group (macrolides, lincosamides and streptogramin B), by a methylation at the 23s r-RNA subunit that leads to methylation. In the presence of erythromycin, the strain with the MLSB gene induces resistance to clindamycin in the “Er-r, Cd-s” strain, conferring clindamycin resistance to the original Cd-s strain, eventually causing the well-known flattening of the clindamycin inhibition zone towards the erythromycin disc, so that the shape “D” is seen in the clindamycin zone or “D-test positivity”.
Research frontiers
Since, failure in the therapy with clindamycin used against S. aureus had been frequently met, the D-test procedure is often recommended for checking the efficacy of the empiric use of clindamycin against isolated staphylococci in most hospitals to avoid the unbeknown pervasive error in the therapy, due to MLSB resistance.
Related reports
Inducible-MLSB S. aureus strains have been isolated independently with resistant patterns for a number of antibiotics in use, in diverse geographical zones; their abundance have been reported up to the height of 94% of S. aureus isolates (Patel et al. 2006; Sibbery et al. 2002; Jorgensen et al. 2004).
Innovations and breakthroughs
It could be identified in this study that inducible MLSB strain was widespread in hospital sectors, so D-test protocol need be included as the routine diagnostic procedure for suppurative ailments of incoming patients. But, in community sector, there was less Cd-r S. aureus strains. This study indicated negligible fractions of constitutive “sensitive” and “resistant” phenotypes that were pejorative for D-test positivity. Indeed, occurrence of high percentage of resistance for daptomycin at 36% in hospital aquired samples are of high clinical concern, in this study. The most striking situation was that S. aureus strains have emerged with concomitant resistance to many commonly used antibiotics seen here.
Applications
In view of high prevalence of D-test positive S. aureus strains both in community and hospital sectors, undertaking of D-test may be routinely conducted to prevent mis-matches out of empiric use of clindamycin against this super-bug of health domain, for wound sites.
Peer review
This is a good study in which the authors investigated the infection dynamics of S. aureus isolates from a hospital. The results are interesting. This study is significant for the hospital managers, decision makers, physicians and students to avoid cross infections in hospitals. It is also ringing alarm bells for the necessities in the solutions of nosocomial infection.
Footnotes
Foundation Project: Financially supported by the research scheme from CSIR (New Delhi), No. 21 (0859)/11/EMR-II.
Conflict of interest statement: We declare that we have no conflict of interest.
References
- 1.DeLeo FR, Chambers HF. Reemergence of antibiotic-resistant Staphylococcus aureus in the genomics era. J Clin Invest. 2009;119:2464–2474. doi: 10.1172/JCI38226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nadig S, Namburi P, Raghunath D, Arakere G. Genotyping of methicillin-resistant Staphylococcus aureus isolates from Indian hospitals. Curr Sci. 2006;91:1364–1369. [Google Scholar]
- 3.Patel M, Waites KB, Moser SA, Cloud GA, Hoesley CJ. Prevalence of inducible clindamycin resistance among community and hospital-associated Staphylococcus aureus isolates. J Clin Microbiol. 2006;44:2481–2484. doi: 10.1128/JCM.02582-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woods CR. Macrolide-inducible resistance to clindamycin and the D-test. Pediatr Infect Dis J. 2009;28:1115–1118. doi: 10.1097/INF.0b013e3181c35cc5. [DOI] [PubMed] [Google Scholar]
- 5.Leclercq R. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin Infect Dis. 2002;34:482–492. doi: 10.1086/324626. [DOI] [PubMed] [Google Scholar]
- 6.Siberry GK, Tekle T, Carroll K, Dick J. Failure of clindamycin treatment of methicillin- resistant Staphylococcus aureus expressing inducible clindamycin resistance in vitro. Clin Infect Dis. 2003;37:1257–1260. doi: 10.1086/377501. [DOI] [PubMed] [Google Scholar]
- 7.Sattler CA, Mason EO, Kaplan SL. Prospective comparison of risk factors and demographic and clinical characteristics of community-acquired, methicillin-resistant versus methicillin-susceptible Staphylococcus aureus infection in children. Pediatr Infect Dis J. 2002;21:910–916. doi: 10.1097/00006454-200210000-00005. [DOI] [PubMed] [Google Scholar]
- 8.Khawcharoenporn T, Tice A. Empiric outpatient therapy with trimethoprim- sulfamethoxazole, cephalexin, or clindamycin for cellulitis. Am J Med. 2010;123:942–950. doi: 10.1016/j.amjmed.2010.05.020. [DOI] [PubMed] [Google Scholar]
- 9.Forbes BA, Sahm DF, Weissfeld AS, editors. Bailey and Scott's diagnostic microbiology. 12 th ed. St. Louis: Mosby Elsevier; 2007. [Google Scholar]
- 10.Sahu MC, Dubey D, Rath S, Debata NK, Padhy RN. Multidrug resistance of Pseudomonas aeruginosa as known from surveillance of nosocomial and community infections in an Indian teaching hospital. J Publ Health. 2012;20:413–423. [Google Scholar]
- 11.Dubey D, Padhy RN. Surveillance of multidrug resistance of two Gram-positive pathogenic bacteria in a teaching hospital and in vitro efficacy of 30 ethnomedicinal plants used by an aborigine of India. Asian Pac J Trop Dis. 2012;2:273–281. [Google Scholar]
- 12.Datta P, Gulati N, Singla N, Vasdeva HR, Bala K, Chander J, et al. Evaluation of various methods for the detection of meticillin-resistant Staphylococcus aureus strains and susceptibility patterns. J Med Microbiol. 2011;60:1613–1616. doi: 10.1099/jmm.0.032219-0. [DOI] [PubMed] [Google Scholar]
- 13.Clinical and Laboratory Standards Institute Performance standard for antimicrobial susceptibility testing: twenty-first informational supplement. 2011. Document M200-S21; Wayne, PA: CLSI.
- 14.Jorgensen JH, Crawford SA, McElmeel ML, Fiebelkorn KR. Detection of inducible clindamycin resistance of staphylococci in conjunction with performance of automated broth susceptibility testing. J Clin Microbiol. 2004;42:1800–1802. doi: 10.1128/JCM.42.4.1800-1802.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farrell D, Shackloth J, Cossins L, Quintana A, Hogan P. High prevalence of inducible clindamycin resistance among community-associated methicillin resistant Staphylococcus aureus in Europe. Int J Antimicrob Agent. 2007;29(Suppl):S520. [Google Scholar]
- 16.Steward CD, Raney PM, Morrell AK, Williams PP, McDougal LK, Jevitt L, et al. Testing for induction of clindamycin resistance in erythromycin-resistant isolates of Staphylococcus aureus. J Clin Microbiol. 2005;43:1716–1721. doi: 10.1128/JCM.43.4.1716-1721.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ciraj AM, Vinod P, Sreejith G, Rajani K. Inducible clindamycin resistance among clinical isolates of staphylococci. Indian J Pathol Microbiol. 2009;52:49–51. doi: 10.4103/0377-4929.44963. [DOI] [PubMed] [Google Scholar]
- 18.Prabhu K, Rao S, Rao V. Inducible clindamycin resistance in Staphylococcus aureus isolated from clinical samples. J Lab Physic. 2011;3:25–27. doi: 10.4103/0974-2727.78558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zorgani A, Shawerf O, Tawil K, El-Turki E, Ghenghesh KS. Inducible clindamycin resistance among staphylococci isolated from burn patients. Lybian J Med. 2009;4:104–106. doi: 10.4176/090128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seifi N, Kahani N, Askari E, Mahdipour S, Nasab M. Inducible clindamycin resistance in Staphylococcus aureus isolates recovered from Mashhad, Iran. Iranian J Microbiol. 2012;4:82–86. [PMC free article] [PubMed] [Google Scholar]
- 21.Daurel C, Huet C, Dhalluin A, Bes M, J Etienne J, Leclercq R. Differences in potential for selection of clindamycin-resistant mutants between inducible erm(A) and erm(C) Staphylococcus aureus genes. J Clin Microbiol. 2008;46:546–550. doi: 10.1128/JCM.01925-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zechini B, Versace I. Inhibitors of multidrug resistant efflux systems in bacteria. Rec Pat Antiinfect Drug Discov. 2009;4:37–50. doi: 10.2174/157489109787236256. [DOI] [PubMed] [Google Scholar]
- 23.Dubey D, Rath S, Sahu MC, Pattnaik L, Debata NK, Padhy RN. Surveillance of infection status of drug resistant Staphylococcus aureus in an Indian teaching hospital. Asian Pac J Trop Dis. 2012 in press. [Google Scholar]