Abstract
Tuberculosis is difficult to cure, requiring a minimum of 6 months of treatment with multiple antibiotics. Small numbers of organisms are able to tolerate the antibiotics and persist in the lungs of infected humans, but they still require some metabolic activity to survive. We studied the role of the hypoxia-induced Rv1894c gene in Mycobacterium tuberculosis virulence in guinea pigs, which develop hypoxic, necrotic granulomas histologically resembling those in humans and found this gene to be necessary for full bacillary growth and survival. We characterized the function of the encoded enzyme as a nitronate monooxygenase, which is needed to prevent the buildup of toxic products during hypoxic metabolism and is negatively regulated by the transcriptional repressor KstR. Future studies will focus on developing small-molecule inhibitors that target Rv1894c and its homologs, with the goal of killing persistent bacteria, thereby shortening the time needed to treat tuberculosis.
Keywords: Mycobacterium tuberculosis, persistence, latency, latent tuberculosis infection, dormancy, virulence, guinea pig, animal models, 2-nitropropane dioxygenase, nitroalkane, oxidation, detoxification, fatty acid synthesis, lipids, Pseudomonas aeruginosa
Global efforts to control the tuberculosis pandemic have been hampered by the ability of the causative agent, Mycobacterium tuberculosis, to persist despite robust immune responses and/or prolonged chemotherapy. After inhalation, the organisms are phagocytosed by alveolar macrophages, resulting in potent cell-mediated immune responses and formation of granulomas, which consist primarily of M. tuberculosis–infected macrophages and T cells [1, 2]. Between 6 and 8 weeks after infection in humans, these granulomas undergo caseous necrosis, resulting in the death of the majority of tubercle bacilli and destruction of surrounding host tissue [3]. Oxygen limitation is believed to be an important microenvironmental condition of these granulomas [4]. Exposure of M. tuberculosis to progressive hypoxia in vitro induces a dormant state in which the bacilli cease to divide and decrease metabolic activity [5], becoming phenotypically tolerant to isoniazid [6]. However, the adaptive mechanisms used by M. tuberculosis to persist indefinitely in the infected host remain to be elucidated.
The M. tuberculosis Rv1894c gene encodes a conserved hypothetical protein that is overexpressed during exposure to hypoxia [7–9] but is not a member of the hypoxia-induced DosR/DevR regulon [7, 8]. The TetR-type protein KstR, encoded by the Rv3574 gene, negatively regulates the Mycobacterium smegmatis Rv1894c homolog and also binds to the promoter region of the M. tuberculosis Rv1894c gene in vitro [10]. Previous high-throughput screening using defined M. tuberculosis mutant pools suggested that Rv1894c is essential for optimal survival under hypoxic conditions [11]. The encoded protein shows some similarity to the nitronate monooxygenase (NMO) family (EC 1.13.12.16) [12–14], which catalyzes oxidative denitrification of alkyl nitronates to their corresponding carbonyl compounds and nitrite [12, 13, 15–17]. Crystallization of the corresponding enzyme, PA1024, in Pseudomonas aeruginosa revealed that a ternary complex is formed with the cofactor flavin mononucleotide (FMN) and the substrate 2-nitropropane [17]. The authors proposed that the conserved histidine residue at position 152 most likely acts as the essential catalytic base, since it belongs to the highly conserved sequence motif IV and is correctly positioned in the structure. Several studies have shown that NMO activity can be inhibited specifically by superoxide dismutase (SOD), since significant alkyl nitronate oxidation results from nonenzymatic reactions with superoxide [18–20]. Database searches reveal the possibility of 4 additional NMO homologs in the M. tuberculosis genome, including Rv0021c, Rv1533, Rv2781c, and Rv3553.
In this study, we investigated the role of Rv1894c in M. tuberculosis virulence in guinea pig lungs. Unlike mice, guinea pigs infected with M. tuberculosis develop hypoxic granulomas [21, 22], which histologically resemble those in humans with respect to cellular composition, lesion architecture, and the presence of caseation necrosis [23], wherein M. tuberculosis persists extracellularly [24]. In addition, we experimentally tested the bioinformatic prediction of NMO activity for the Rv1894c protein and identified a conserved residue critical for enzymatic activity. Finally, since prior studies reported that KstR/Rv3574 binds to the Rv1894c promoter sequence [10], we studied Rv1894c gene expression in a kstR-deficient M. tuberculosis recombinant strain.
MATERIALS AND METHODS
Bacterial Strains and Growth Conditions
M. tuberculosis strains deficient in Rv1894c (Rv1894c::Tn) and kstR/Rv3574 (kstR::Tn) were generated by transposon mutagenesis of the wild-type CDC1551 strain [25]. The points of insertion of the himar1 transposon in these mutants are at nucleotide position 218 in Rv1894c, which is upstream of the putative catalytic histidine residue, and at nucleotide position 484 in Rv3574. To create the Rv1894c-complemented strain, the Rv1894c open reading frame, including 200-bp flanking sequences, was polymerase chain reaction amplified from CDC1551 genomic DNA, using forward primer Rv1894c-comp-L and reverse primer Rv1894c-comp-R (Table 1). The Gateway System (Invitrogen) was used to insert this fragment into the integrating plasmid pGS202 [26], which contains the hygromycin-selectable marker and integrates at the native attB site in M. tuberculosis. This plasmid was electroporated into the M. tuberculosis strain Rv1894c::Tn, generating the Rv1894c-complemented strain (Rv1894c::Tn Comp).
Table 1.
Primers Used in This Study
| Primer Designation | Sequence | Source |
|---|---|---|
| Rv1894c-comp-L | GGGGACAAGTTTGTACAAAAAAGCAGGCTTTCTGCTCGGTGGGACAGCCGTCGT | This Study |
| Rv1894c-comp-R | GGGGACCACTTTGTACAAGAAAGCTGGGTTGCCGTCGAGAAGGCCTCGGACATC | This Study |
| Rv1894c-ATG-L | GGGGACAAGTTTGTACAAAAAAGCAGGCTCACACACTGCCATTTGCGACGAGCTCG | This Study |
| PA1024-ATG-L | GGGGACAAGTTTGTACAAAAAAGCAGGCTCAGGCGTGTTCAGGACCCGTTTCAC | This Study |
| PA1024-R | GGGGACCACTTTGTACAAGAAAGCTGGGTTGTCTCTTCGTCGTCAGTGGATAGA | This Study |
| H199A-F | GAGGCCGGCGGGGCCTGTGGCGAGGT | This Study |
| H199A-R | ACCTCGCCACAGGCCCCGCCGGCCTC | This Study |
| H152A-F | GAGTGTGCCGGCGCCCCGGGCGAGGA | This Study |
| H152A-R | TCCTCGCCCGGGGCGCCGGCACACTC | This Study |
| Rv1894c-L | ATGATCAAGCACATCCACGA | This Study |
| Rv1894c-R | GCTACCTCCTTGACGACCTG | This Study |
| Rv0021c-L | CCGAACTCGACATTCTCCAT | This Study |
| Rv0021c-R | GCCATACCGGTTATCACGTC | This Study |
| Rv1533-L | CGGTGGTCAAGGACAAGTTT | This Study |
| Rv1533-R | CCTGTCCGACGAAGTAGGTC | This Study |
| Rv2781c-L | TCCTATCGGAGCCAATCTGT | This Study |
| Rv2781c-R | AAGGTGAACGACACCACCTC | This Study |
| Rv3553-L | CGGCAAGTTTAGACGGATGT | This Study |
| Rv3553-R | CTCTTTGCACGACGGTAGGT | This Study |
| Rv3574-L | TTCCAGCGGCTGAACTTTAT | This Study |
| Rv3574-R | TACTGGTCCTCGGTTGGTTC | This Study |
Infection of Animals and Virulence Endpoints
Female Hartley guinea pigs (weight, 250–300 g; Charles River) were housed in a Biosafety level 3, specific-pathogen-free facility and fed water and chow ad libitum. All procedures followed protocols approved by the Institutional Animal Care and Use Committee at Johns Hopkins University. Separate groups of guinea pigs were aerosol infected with approximately 600 bacilli of wild-type M. tuberculosis CDC1551, Rv1894c::Tn, or Rv1894c::Tn Comp [27]. Mean log-transformed colony-forming units (CFUs) were compared by the Student t test.
Cloning, Expression, and Purification of Recombinant Native and Mutant Rv1894c and PA1024 Proteins
The Rv1894c coding sequence was PCR amplified using primers Rv1894c-ATG-L and Rv1894c-comp-R (Table 1). By use of the Gateway System (Invitrogen), this fragment was inserted into the protein expression plasmid pDEST17 (Invitrogen), incorporating an amino terminal His-tag into the Escherichia coli–expressed protein. The resulting plasmid, pHis-Rv1894c, was transformed into BL21-AI (Invitrogen). Soluble protein was batch bound to Ni-NTA (Qiagen) and purified according to the manufacturer's recommendation. The Pseudomonas aeruginosa gene PA1024, which encodes a protein with known NMO activity [17], was PCR amplified from a clinical isolate obtained from the Johns Hopkins Hospital Microbiology Laboratory, using primers PA1024-ATG-L and PA1024-R (Table 1); inserted into pDEST17 to generate pHis-PA1024; and expressed and purified as described above. The conserved putative catalytic histidine residue [17] of each of these 2 proteins was changed to alanine by site-directed mutagenesis with QuikChange II (Stratagene), using primers H199A-F and H199A-R (Table 1) to generate pHis-Rv1894c-H199A and primers H152A-F and H152A-R (Table 1) to generate pHis-PA1024-H152A. These modified proteins were expressed and purified as described above. Protein samples were dialyzed against 0.1 M potassium phosphate buffer (pH 7.0) for 16 hours and concentrated using Amicon Ultra Centrifugal 10 kDa Filters (Millipore), and protein concentration was determined by the Bradford assay [28].
Nitronate Monooxygenase Activity and Inhibition Assays
Enzyme activity was measured in time-course experiments at 37°C, using 10 mM 2-nitropropane (substrate). The nitrite (product) concentration was determined by the Griess reaction [29] and measured by absorbance at 540 nM in a Model 680 microplate reader (Bio-Rad). Inhibition by SOD was measured in a time-course experiment, as well as in inhibitor concentration-dependent assays at a single time point. In each experiment, the nitrite concentration was determined by comparison with a titration of known nitrite concentrations. Statistical analysis was performed by comparison of results from 3 independent protein purifications, using the Student t test.
Gene Expression Studies
Expression of Rv1894c, 4 other putative homologs (Rv0021c, Rv1533, Rv2781c, and Rv3553), and kstR were measured from wild-type CDC1551 and kstR::Tn. RNA was extracted from in vitro early log-phase cultures (OD600 = 0.5), late log-phase cultures (OD600 = 1.0), and early stationary cultures (OD600 = 3.0) from an equivalent number of bacilli. Purified RNA (250 ng) was used for first-strand complementary DNA (cDNA) synthesis, as per Superscript III (Invitrogen) protocols. Negative controls were prepared using identical procedures but excluding Superscript III enzyme. Gene expression levels were measured using primers listed in Table 1 and an iCycler 5.0 (Bio-Rad). Successful reactions contained a single peak by melt curves analysis. Synthesized cDNA was subjected to 3 technical replicates of PCR amplification, which were averaged to generate a single value for each biological replicate. The cycle threshold (CT) value obtained for each gene was normalized to a housekeeping gene, sigA. Statistical analysis was performed using 3 biological replicates of each sample at each time point.
RESULTS
Construction and Genotypic Characterization of an Rv1894c-Complemented Strain
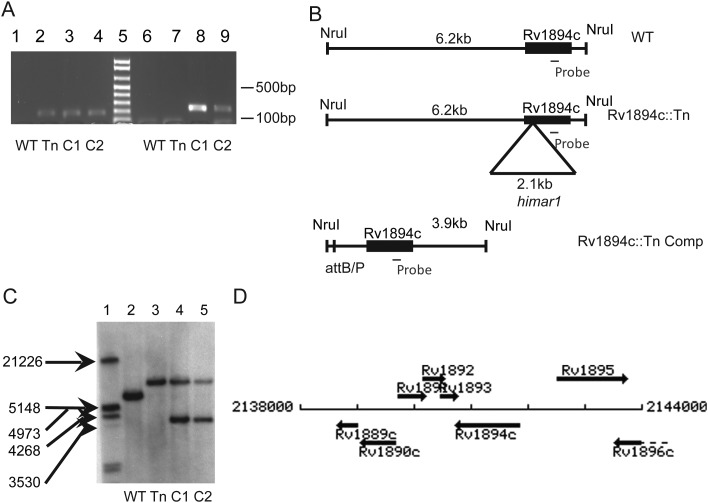
To confirm the mutant phenotype of Rv1894c::Tn, an Rv1894c-complemented strain (Rv1894c::Tn Comp) was constructed. PCR amplification of the kanamycin resistance marker linked to the himar1 transposon and the hygromycin resistance marker from the complementation construct revealed products of the correct size in Rv1894c::Tn and Rv1894c::Tn Comp strains, respectively, which were absent in the isogenic wild-type strain (Figure 1A). Single-copy Rv1894c integration at the proper location in Rv1894c::Tn Comp is expected to produce a 3.9-kb NruI restriction fragment, compared with a 6.2-kb NruI restriction fragment in the wild type, while maintaining the 8.3-kb NruI restriction fragment from Rv1894c::Tn (Figure 1B). These findings were further corroborated by Southern blot (Figure 1C), using a DNA probe recognizing the region of the Rv1894c coding sequence shown in Figure 1B. Rv1894c is expected to be transcribed independently because the neighboring genes are oriented in opposite directions (Figure 1D). No growth differences were noted between the 3 strains in nutrient-rich broth.
Figure 1.
Complementation of Rv1894c::Tn. A, Polymerase chain reaction analysis of genomic DNA. Lanes 1 and 6 contain wild-type genomic template (WT), lanes 2 and 7 contain Rv1894c::Tn genomic template (Tn), lanes 3 and 8 contain the first Rv1894c::Tn Comp candidate (C1), and lanes 4 and 9 contain the second Rv1894c::Tn Comp candidate (C2). Lanes 2–4 show the presence of the kanamycin cassette marking the himar1 insertion in the Rv1894c::Tn strain and both candidate complement strains. Lanes 8 and 9 show the presence of a region of the hygromycin gene marking the integration of the Rv1894c::Tn Comp plasmid into both candidate strains. Lane 5 is the molecular marker, showing 100-bp increments, beginning with 100 bp at the bottom. B, Diagram of expected sizes of genomic fragments binding to the probe-recognizing region of the Rv1894c coding sequence. C, Southern blot showing Rv1894c as a single-copy gene present in the correct fragment sizes in both wild-type (6.2 kb) and Rv1894c::Tn (8.3 kb) strains (lanes 2 and 3, respectively). In both of the complement candidate strains (lanes 4 and 5), the mutant allele (8.3 kb) and the complement allele (3.9 kb) are present. D, Map of genomic region surrounding Rv1894c.
Virulence in the Guinea Pig Aerosol Model
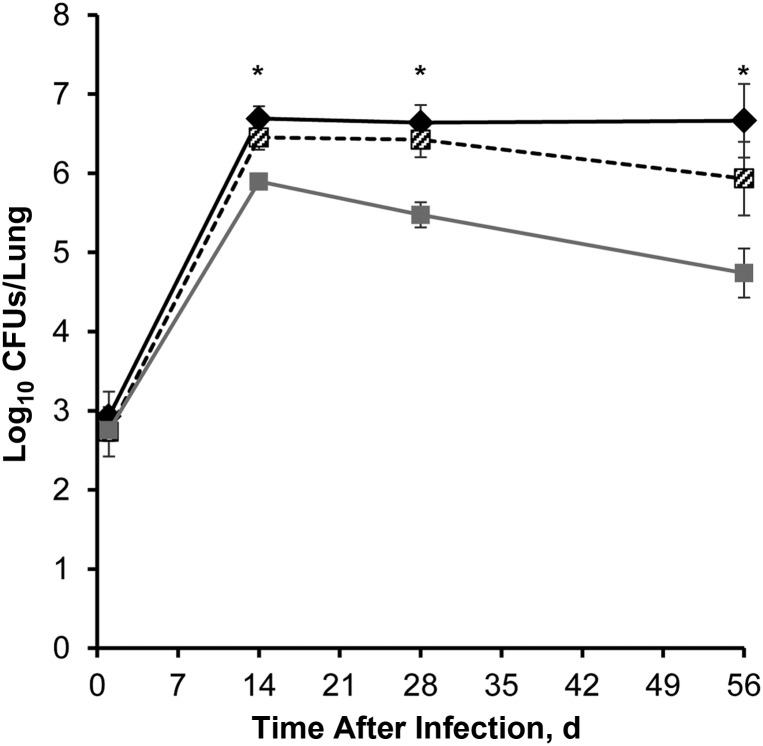
Following aerosol infection of guinea pigs with approximately 2.80 log10 bacilli of wild-type, Rv1894c::Tn, or Rv1894c::Tn Comp strains, lung bacillary counts increased to approximately 6.5 log10 in animals infected with either wild type or Rv1894c::Tn Comp after 14 days of infection and remained relatively stable and not significantly different from one another at day 28 (P = .240) or day 56 (P = .078; Figure 2). Conversely, initial growth of the Rv1894c::Tn mutant was mildly impaired in guinea pig lungs, peaking at 5.9 log10 CFUs/lung on day 14 after infection. Subsequently, the bacillary burden in Rv1894c::Tn mutant–infected guinea pigs dropped to 5.47 log10 CFUs/lung by day 28 after infection and finally declined to 4.74 log10 CFUs/lung by day 56 after infection. These lung CFU values were significantly lower than corresponding wild-type values at day 14 (P < .0006), day 28 (P < .003), and day 56 (P < .0012). The Rv1894c::Tn lung CFUs were significantly lower than corresponding Rv1894c::Tn Comp values at day 56 (P < .008).
Figure 2.
Reduced initial growth and long-term survival of the Rv1894c::Tn strain in the lungs of guinea pigs after aerosol infection. Black diamonds refer to wild-type Mycobacterium tuberculosis infection. Black and white crosshatched boxes refer to Rv1894c::Tn Comp infection. Grey boxes refer to Rv1894c::Tn infection. The asterisks indicate significant differences between colony-forming units (CFUs) recovered from wild-type– and mutant–infected lungs (P < .0006 for day 14, P < .003 for day 28, and P < .002 for day 56). The CFUs recovered from wild-type– and complement-infected lungs were not significantly different at any time point.
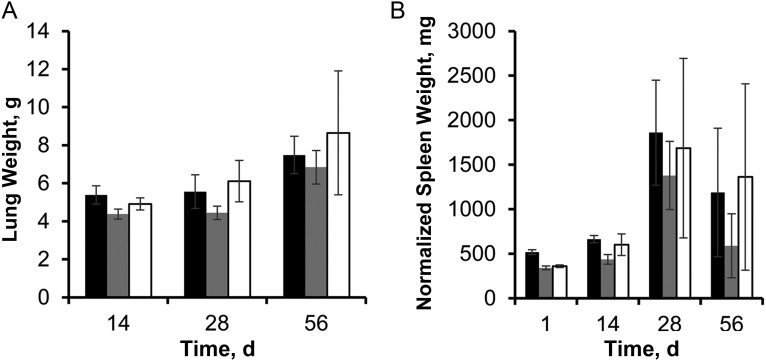
Examination of guinea pig organs showed minimal differences between the groups at days 28 and 56, with a statistically insignificant trend toward lower mean lung weight (Figure 3A) and lower normalized mean spleen weight (Figure 3B) in the Rv1894c::Tn-infected guinea pigs. Histological examination of lung samples revealed peribronchiolar inflammation and multiple well-circumscribed granulomas comprising primarily lymphocytes and histiocytes with central necrosis, which did not differ significantly in size or number between groups (data not shown). Ziehl-Neelsen staining revealed the presence of multiple acid-fast bacilli in each group, primarily in the periphery of the necrotic cores of granulomas (data not shown).
Figure 3.
Effect of Rv1894c deficiency on guinea pig pathology. A, Guinea pig lung weights of lungs infected with Mycobacterium tuberculosis. B, Guinea pig spleen weights, normalized to mean body weight on day 1 divided by body weight of individual animal at time of sacrifice. Black bars represent wild type; grey bars represent Rv1894c::Tn; and white bars represent Rv1894c::Tn Comp.
NMO Activity and Inhibition
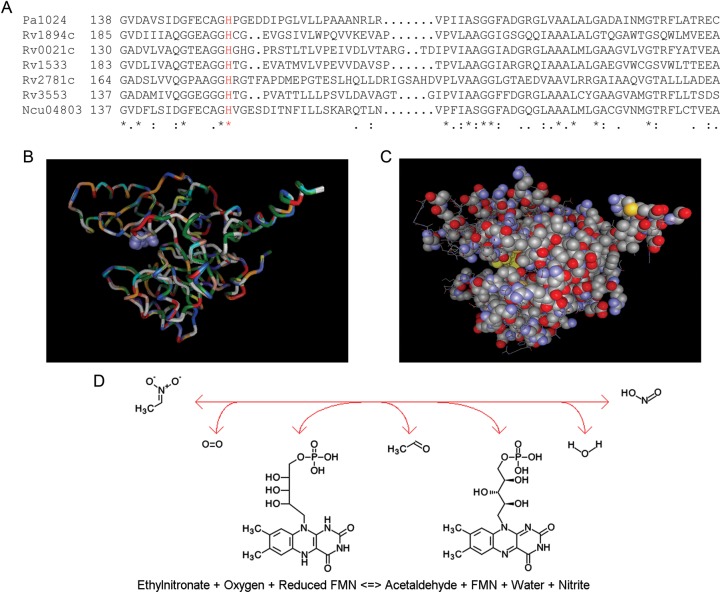
The amino acid sequences of the known NMO proteins from P. aeruginosa and Neurospora crassa were aligned with the 5 putative NMO proteins from M. tuberculosis, revealing strict conservation of the proposed catalytic histidine residue and strong homology throughout the NMO domain (Figure 4A). A FATCAT comparison of PA1024 and Rv1894c yielded a raw score of 848.61 (P < .01) for similarity of the structure pairs [30]. The structure alignment of the 2 proteins revealed 315 equivalent positions out of 376, with a root mean square deviation of 0.64 and no twists in the structure required. This is summarized graphically in Figure 4B, where the predicted secondary structure of Rv1894c is superimposed on the known structure of PA1024, and the conserved histidine residue is shown in space-fill. The equivalent positions between PA1024 and Rv1894c reconstitute the structure of PA1024, including the cofactor (FMN) and substrate (2-nitropropane) binding sites (Figure 4C). On the basis of these modeling data, we predicted that M. tuberculosis Rv1894c encodes an NMO that is capable of converting an ethylnitronate to an acetaldehyde and nitrite (Figure 4D).
Figure 4.
The proposed nitronate monooxygenase family shows amino acid sequence and structural similarity. A, Clustal X multiple alignment of PA1024 from Pseudomonas aeruginosa with 5 nitronate monooxygenase (NMO) proteins from Mycobacterium tuberculosis and with NCU04803 from Neurospora crassa. Segment of whole protein alignment showing one area of conservation near the proposed catalytic histidine residue (shown in red). B, Overlaid alignment of alignment of PA1024 and Rv1894c, using FATCAT, predicting strong structural similarity. The color scheme shows each amino acid as a different color, as defined by Vector-NTI 3D Molecule Viewer software. The histidine in the catalytic cleft is shown in space-fill. C, View of the PA1024 structure, showing conserved residues from Clustal X paired alignment with Rv1894c in space-fill. Also shown in this image is the space-fill view in yellow of the flavin cofactor and the 2-nitropropane substrate, both near the center of the image. D, Enzymatic reaction, from left to right, catalyzed by NMO.
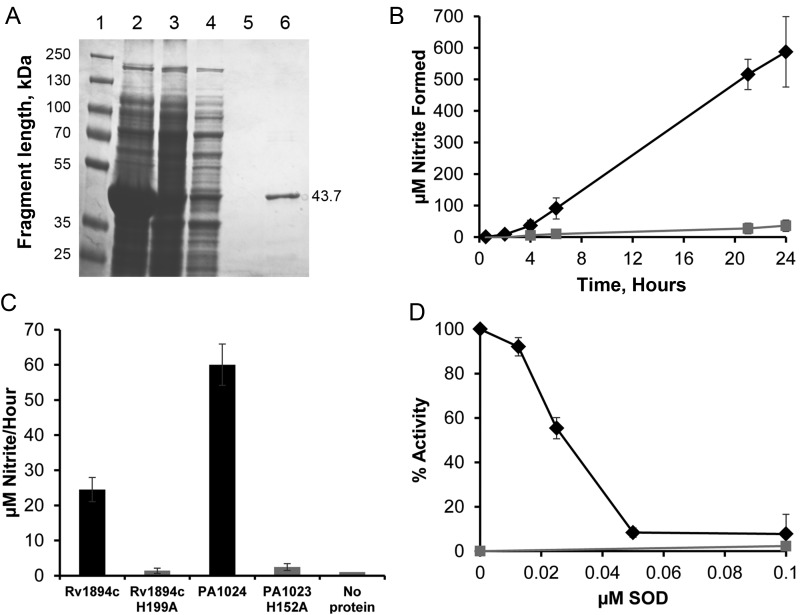
Recombinant native Rv1894c protein and Rv1894c mutant protein containing an alanine substitution at the putative histidine catalytic site in residue 199 (Rv1894c H199A) were purified from E. coli by affinity chromatography, using a 6-His tag (Figure 5A). Copurification of a flavin molecule with both the wild-type and mutant protein was inferred from the yellow color that was specifically eluted in the protein fraction and measured as absorption peaks at 370 and 445 nM in the visible light range (Supplementary Figure 1). Activity of Rv1894c and Rv1894c H199A to convert the substrate (2-nitropropane) to the product (nitrite) was measured (Figure 5B). After 6 hours of incubation at 37°C, recombinant Rv1894c protein converted 91.0 ± 33.5 µM nitrite from 10 mM 2-nitropropane. This amount increased to 515.7 ± 48.3 µM nitrite after 21 hours of incubation, further increasing to 587.4 ± 111.6 µM nitrite after 24 hours. Each experiment was performed in triplicate, using independent protein purifications.
Figure 5.
Recombinant Rv1894c protein functions as a nitronate monooxygenase (NMO). A, Coomassie-stained sodium dodecyl sulfate polyacrylamide gel electrophoresis showing purification of Rv1894c from Escherichia coli. Lane 1 is a size standard. Lane 2 is a crude extract from lysed E. coli. Lane 3 is the cleared lysate soluble fraction. Lane 4 is the column flow through. Lane 5 is the final wash of the column. Lane 6 is the eluted protein of the correct size, 43.7 kDa. B, Enzyme assay of NMO activity of Rv1894c, using 2-nitropropane as substrate. Black diamonds represent reactions containing Rv1894c. Grey squares represent a control reaction containing no protein. Conversion from absorbance at 540 nM to micromoles of nitrite was performed using a standard curve of known nitrite concentration. C, Enzyme assay of NMO activity of Rv1894c and PA1024 containing site-directed mutations of the proposed catalytic histidine residues (H199A and H152A, respectively) compared to the native proteins. Each bar represents the rate of nitrite conversion from 3 independent purifications of each protein, and the error bars are SDs. D, Inhibition of NMO activity by superoxide dismutase (SOD). Rv1894c protein (black diamonds) and a no-protein control (grey boxes) were incubated with substrate and increasing concentrations of SOD. Nitrite production was measured after several hours of incubation. Each time point was performed in triplicate with independent protein purifications. Error bars represent SDs.
Replacement of the proposed catalytic histidine residue with alanine (H199A) abrogated enzymatic function. Thus, after 24 hours the Rv1894c H199A mutant protein converted only 35.9 ± 17.9 µM nitrite. The substrate conversion rate was calculated and compared to rates determined for the control protein derived from P. aeruginosa, PA1024, and the corresponding mutant protein, PA1024 H152A (Figure 5C). This yielded reaction rates of 24.5 ± 3.5 µM nitrite/hour for Rv1894c, 1.4 ± 0.8 µM nitrite/hour for Rv1894c H199A, 60.0 ± 5.9 µM nitrite/hour for PA1024, and 2.5 ± 1.0 µM nitrite/hour for PA1024 H199A. In both cases, the wild-type protein showed statistically significantly more activity than the mutant protein (Rv1894c, P < .001; PA1024, P < .001), and the mutant protein activities did not differ significantly from those of the protein-negative control.
Enzyme inhibition by SOD was measured using 200 ng of purified protein, incubated at 37°C with 10 mM 2-nitropropane, in the presence of SOD at 2-fold increasing concentrations ranging from 0.0125 µM to 0.1 µM and was performed in triplicate using independent protein purifications. The half maximal inhibitory concentration was determined to be 0.028 µM (Figure 5D), which is similar to the concentration shown to inhibit other members of this enzyme family [16, 18–20].
Expression of Rv1894c Remains Constant Throughout the Growth Cycle and Is Repressed by KstR
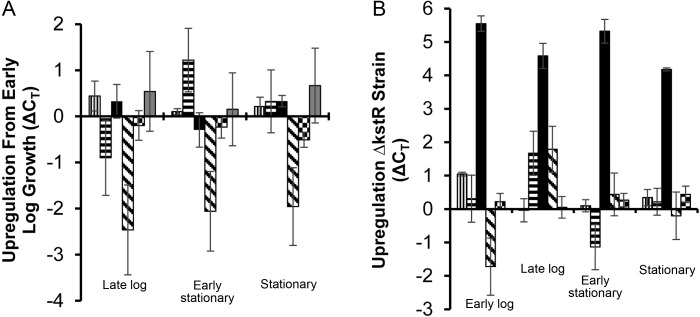
To determine whether Rv1894c is induced in conditions independent of hypoxia, we studied its gene expression during different phases of axenic growth in nutrient-rich broth. As shown in Figure 6A, expression of Rv1894c remained relatively constant throughout the growth cycle, including during the transition to stationary phase. We also examined expression of Rv3574 (kstR), the repressor of Rv1894c, and found no significant changes in expression throughout the growth cycle.
Figure 6.
Expression of Rv1894c in Mycobacterium tuberculosis is regulated by KstR but is growth-phase independent. A, Gene expression of the 5 nitronate monooxygenase (NMO) homologs as well as the expression level of the repressor kstR (Rv3574) in wild-type M. tuberculosis grown in 7H9 were compared to normalized sigA expression levels at different growth stages. Each value is shown relative to the levels of expression measured at the early log phase of growth. B, Cycle threshold (CT) values of the 5 NMO homolog expression levels were compared between wild type and a kstR::Tn strain at different growth stages. Rv0021c values are represented by a bar with vertical lines. Rv1533 values are represented by a bar with horizontal lines. Rv1894c values are represented by a solid black bar. Rv2781c values are represented by a bar with diagonal lines. Rv3553 values are represented by a bar with a checkerboard pattern. Rv3574 (kstR) values are represented by a solid grey bar. Each experimental time point was performed in triplicate. Error bars represent SDs.
Next, we studied the gene expression patterns of the 4putative Rv1894c homologs (Rv0021c, Rv1533, Rv2781c, and Rv3553) during growth in nutrient-rich broth. Expression of Rv2781c was repressed during the transition from logarithmic growth into stationary phase. Thus, in late log phase, early stationary phase, and stationary phase, Rv2781c expression was downregulated by 2.46 ± 0.98 cycles, 2.06 ± 0.86 cycles, and 1.96 ± 0.84 cycles, respectively, relative to its expression during log-phase growth. Expression of Rv1533 was repressed by 0.9 ± 0.82 cycles during late log phase but then induced in early stationary phase by 1.22 ± 0.69 cycles relative to its expression at early log phase. The expression levels of Rv0021c and Rv3553 remained relatively constant throughout the growth cycle.
Previous studies have reported that Rv1894c expression might be negatively regulated by KstR (Rv3574) [10]. We hypothesized that KstR deficiency would result in derepression of Rv1894c expression during bacillary growth in nutrient-rich broth. Consistent with this hypothesis, Rv1894c expression was upregulated in a kstR/Rv3574-deficient transposon mutant (kstR::Tn) relative to the isogenic wild-type strain during all phases of the growth cycle (Figure 6B). Thus, relative to that of the parental wild type, expression of Rv1894c in kstR::Tn was induced by 5.55 ± 0.23 cycles, 4.58 ± 0.38 cycles, 5.32 ± 0.36 cycles, and 4.18 ± 0.04 cycles in mid-log phase, late log phase, early stationary phase, and late stationary phase, respectively. Gene expression of the 4 putative Rv1894c homologs was not derepressed in kstR::Tn.
DISCUSSION
M. tuberculosis is a highly adapted pathogen, with the ability to persist for prolonged periods within oxygen-limited, necrotic granulomas of the infected human host. Identification of metabolic and regulatory pathways essential for M. tuberculosis persistence during hypoxia may provide insight into novel strategies to eradicate this infection. Previously, we have shown in a high-throughput screen that the M. tuberculosis gene Rv1894c is required for bacillary survival during progressive hypoxia in vitro [11]. In the current study, we have determined that Rv1894c encodes a flavin-dependent protein with NMO activity, which is required for optimal growth during acute infection, as well as for persistence in guinea pig lungs. On the basis of these findings, we propose its annotation as nmo1.
As in the case of other NMOs [13], M. tuberculosis Nmo1 showed time-dependent oxidative denitrification of the alkyl nitronate substrate and concentration-dependent inhibition by SOD. On the basis of the absorption spectra (Supplementary Figure 1) and because FMN was copurified in the crystal structure of PA1024, we conclude that FMN was copurified with our protein. However, both of these proteins were purified from E. coli, so the physiological flavin cofactor of Rv1894c remains to be determined. Bacterial NMO have been best characterized in P. aeruginosa [12, 13, 15–17]. Confirming the predictions of Ha et al [17], we have determined that the conserved histidine residue at position 199 in M. tuberculosis Nmo1, corresponding to H152 in P. aeruginosa PA1024, is critical for enzymatic function and is strictly conserved among NMO from other bacteria and fungi (Figure 4A). These enzymes have been shown to play an important role in oxidizing various alkyl nitronates that are known to be toxic or mutagenic [31–33]. In plants and fungi, propionate-3-nitronate is a natural metabolite that is detoxified by fungal NMOs [14]. The hypoxic induction of Rv1894c [7, 8], which requires molecular oxygen for NMO activity [13], seems counterintuitive, although it is likely that an increased concentration of enzyme is required to successfully compete for limiting oxygen. Recent evidence suggests that a hypoxia-induced NMO in Neisseria gonorrhoeae is involved in unsaturated fatty acid synthesis under anaerobic conditions [34]. While the physiological Nmo1 substrate remains unknown, we hypothesize that it is not host immune derived, but rather a product of bacterial metabolism, since the attenuated phenotype of the Rv1894c-deficient mutant was also observed in the progressive hypoxia model in vitro [11]. The nitrate assimilation pathway, which is responsible for reducing nitrate to nitrite and then to ammonium, is important for M. tuberculosis metabolism and survival during carbon and oxygen limitation [35]. It is possible that reduced nitrite production as a result of Rv1894c/Nmo1 deficiency could contribute to the phenotype of Rv1894c::Tn during progressive hypoxia and in guinea pig lungs.
We found that Rv1894c/nmo1 expression was unchanged during axenic growth of wild-type M. tuberculosis in nutrient-rich broth, and an Rv1894c/nmo1-deficient mutant showed equivalent growth relative to the isogenic wild-type and complement strains. In contrast, Rv1894c is upregulated during hypoxia [7, 8] and in a mouse hypoxic granuloma model [9]. Consistent with an important role for this gene in M. tuberculosis adaptation to microaerophilic conditions, we have shown previously in a high-throughput screen that an Rv1894c-deficient mutant shows impaired survival during progressive hypoxia in vitro, in a mouse hypoxic granuloma model, and in guinea pig lungs but not in the lungs of BALB/c mice [11], which do not develop hypoxic lesions following infection [36, 37]. In the current study, the requirement of Rv1894c for M. tuberculosis persistence in guinea pig lungs was confirmed by individual infection with Rv1894c::Tn and near complete restoration of the wild-type phenotype following complementation with the native gene. Similar to our findings, Isabella and Clark recently found that N. gonorrhoeae deficient in the hypoxia-induced NMO (NGO1024, UfaA) exhibited impaired anaerobic growth [34], suggesting that this protein family may have widespread importance in many facultative and obligate anaerobes. Interestingly, Rv1894c::Tn showed a mild growth defect during the acute phase of infection in guinea pig lungs, prior to the development of necrotic granulomas. This phenotype may be attributable to local hypoxia at the site of infection in the absence of tissue necrosis, a defective nitrate assimilation pathway in the setting of carbon limitation, or other unknown factors. In any case, it appears that differences in lung CFUs between the mutant and control groups prior to immune containment were not significant enough to result in discernible differences in gross pathology or histology.
Unlike many other hypoxia-regulated genes, nmo1 expression is not DosR-dependent [38]. The negative regulator KstR/Rv3574 is known to bind the promoter sequence of Rv1894c [10]. The KstR regulon includes many genes involved in lipid metabolism and transport, including 10 different fad genes and the mce4 operon [10], which are important for M. tuberculosis survival in mice and macrophages [39–42], and kstR itself has been shown to be necessary for survival in mice [39] and macrophages [42]. We found that Rv1894c/nmo1 expression was derepressed throughout the growth cycle as a result of kstR deficiency. Increased nmo1expression in hypoxia-exposed M. tuberculosis may be due to such derepression, since kstR expression is significantly downregulated during prolonged hypoxia [43]. Alternatively, hypoxia-induced upregulation of Rv1894c/nmo1 may be due to transcriptional activation by unidentified positive regulators. In support of the existence of additional regulators, the genes Rv1894/nmo1 and kstR/Rv3574 were found to be concurrently induced in palmitic acid [44] and during infection of mice [39] and macrophages [42].
The functional redundancy suggested by the presence of 4 additional putative NMOs (Rv0021c, Rv1533, Rv2781c, and Rv3553) highlights the potential importance of this pathway. These proteins are each predicted to contain an NMO domain, with strong conservation at the regions that interact with the flavin cofactor and at the catalytic histidine residue. Consistent with overlapping protein function, none of these genes appear to be essential in vitro [45] or in mice [39]. They are not upregulated in hypoxia-exposed M. tuberculosis [7, 8, 43, 46–48], and our data reveal they are not derepressed in the absence of KstR, suggesting divergent regulation relative to Rv1894c/nmo1.
In conclusion, we have identified a novel enzyme family with NMO activity in M. tuberculosis, of which Rv1894c is the prototype. We have identified the catalytic residue critical for enzyme activity and demonstrated that Rv1894c/nmo1 is required for long-term M. tuberculosis persistence in guinea pig lungs. Future studies will focus on the development of small-molecule NMO inhibitors [49] with the goal of shortening treatment duration by targeting hypoxia-exposed, persistent bacilli [50].
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Financial support. This work was supported by the Potts Memorial Foundation (to L. G. K.) and the National Institutes of Health (R01-AI083125 and R01-HL106786 to P. C. K.).
Disclaimer. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Flynn JL, Chan J. Tuberculosis: latency and reactivation. Infect Immun. 2001;69:4195–201. doi: 10.1128/IAI.69.7.4195-4201.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaplan G, Post FA, Moreira AL, et al. Mycobacterium tuberculosis growth at the cavity surface: a microenvironment with failed immunity. Infect Immun. 2003;71:7099–108. doi: 10.1128/IAI.71.12.7099-7108.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grosset J. Mycobacterium tuberculosis in the extracellular compartment: an underestimated adversary. Antimicrob Agents Chemother. 2003;47:833–6. doi: 10.1128/AAC.47.3.833-836.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haapanen JH, Kass I, Gensini G, Middlebrook G. Studies on the gaseous content of tuberculous cavities. Am Rev Respir Dis. 1959;80:1–5. doi: 10.1164/arrd.1959.80.1P1.1. [DOI] [PubMed] [Google Scholar]
- 5.Wayne LG, Hayes LG. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun. 1996;64:2062–9. doi: 10.1128/iai.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wayne LG, Sramek HA. Metronidazole is bactericidal to dormant cells of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1994;38:2054–8. doi: 10.1128/aac.38.9.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park HD, Guinn KM, Harrell MI, et al. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol Microbiol. 2003;48:833–43. doi: 10.1046/j.1365-2958.2003.03474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sherman DR, Voskuil M, Schnappinger D, Liao R, Harrell MI, Schoolnik GK. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha -crystallin. Proc Natl Acad Sci U S A. 2001;98:7534–9. doi: 10.1073/pnas.121172498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karakousis PC, Yoshimatsu T, Lamichhane G, et al. Dormancy phenotype displayed by extracellular Mycobacterium tuberculosis within artificial granulomas in mice. J Exp Med. 2004;200:647–57. doi: 10.1084/jem.20040646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kendall SL, Withers M, Soffair CN, et al. A highly conserved transcriptional repressor controls a large regulon involved in lipid degradation in Mycobacterium smegmatis and Mycobacterium tuberculosis. Mol Microbiol. 2007;65:684–99. doi: 10.1111/j.1365-2958.2007.05827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klinkenberg LG, Sutherland LA, Bishai WR, Karakousis PC. Metronidazole lacks activity against Mycobacterium tuberculosis in an in vivo hypoxic granuloma model of latency. J Infect Dis. 2008;198:275–83. doi: 10.1086/589515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francis K, Gadda G. Kinetic evidence for an anion binding pocket in the active site of nitronate monooxygenase. Bioorganic Chemistry. 2009;37:167–72. doi: 10.1016/j.bioorg.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 13.Gadda G, Francis K. Nitronate monooxygenase, a model for anionic flavin semiquinone intermediates in oxidative catalysis. Archives of Biochemistry and Biophysics. 2010;493:53–61. doi: 10.1016/j.abb.2009.06.018. [DOI] [PubMed] [Google Scholar]
- 14.Francis K. On the biochemistry, mechanism and physiological role of fungal nitronate monooygenase [PhD dissertation] Atlanta, GA: Georgia State University; 2011. p. 269. [Google Scholar]
- 15.Francis K, Russell B, Gadda G. Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane dioxygenase. J Biol Chem. 2005;280:5195–204. doi: 10.1074/jbc.M411249200. [DOI] [PubMed] [Google Scholar]
- 16.Gorlatova N, Tchorzewski M, Kurihara T, Soda K, Esaki N. Purification, characterization, and mechanism of a flavin mononucleotide-dependent 2-nitropropane dioxygenase from Neurospora crassa. Applied and Environmental Microbiology. 1998;64:1029–33. doi: 10.1128/aem.64.3.1029-1033.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ha JY, Min JY, Lee SK, et al. Crystal structure of 2-nitropropane dioxygenase complexed with FMN and substrate. Identification of the catalytic base. J Biol Chem. 2006;281:18660–7. doi: 10.1074/jbc.M601658200. [DOI] [PubMed] [Google Scholar]
- 18.Kido T, Soda K. Properties of 2-nitropropane dioxygenase of Hansenula mrakii. Formation and participation of superoxide. J Biol Chem. 1978;253:226–32. [PubMed] [Google Scholar]
- 19.Kido T, Soda K. Oxidation of anionic nitroalkanes by flavoenzymes, and participation of superoxide anion in the catalysis. Archives of Biochemistry and Biophysics. 1984;234:468–75. doi: 10.1016/0003-9861(84)90294-7. [DOI] [PubMed] [Google Scholar]
- 20.Kuo CF, Fridovich I. Free-radical chain oxidation of 2-nitropropane initiated and propagated by superoxide. Biochem J. 1986;237:505–10. doi: 10.1042/bj2370505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Via LE, Lin PL, Ray SM, et al. Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and nonhuman primates. Infect Immun. 2008;76:2333–40. doi: 10.1128/IAI.01515-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lenaerts AJ, Hoff D, Aly S, et al. Location of persisting mycobacteria in a Guinea pig model of tuberculosis revealed by r207910. Antimicrob Agents Chemother. 2007;51:3338–45. doi: 10.1128/AAC.00276-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flynn J, Chan J. Animal models of tuberculosis. In: Rom W, Garay S, editors. Tuberculosis. 2nd ed. Philadelphia: Lippincott Williams & Wilkins; 2004. pp. 237–50. [Google Scholar]
- 24.Hoff DR, Ryan GJ, Driver ER, et al. Location of intra- and extracellular M. tuberculosis populations in lungs of mice and guinea pigs during disease progression and after drug treatment. PLoS One. 2011;6:e17550. doi: 10.1371/journal.pone.0017550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamichhane G, Zignol M, Blades NJ, et al. A postgenomic method for predicting essential genes at subsaturation levels of mutagenesis: application to Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2003;100:7213–8. doi: 10.1073/pnas.1231432100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta R, Lavollay M, Mainardi JL, Arthur M, Bishai WR, Lamichhane G. The Mycobacterium tuberculosis protein LdtMt2 is a nonclassical transpeptidase required for virulence and resistance to amoxicillin. Nature Medicine. 2010;16:466–9. doi: 10.1038/nm.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klinkenberg LG, Lee JH, Bishai WR, Karakousis PC. The stringent response is required for full virulence of Mycobacterium tuberculosis in guinea pigs. J Infect Dis. 2010;202:1397–404. doi: 10.1086/656524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry. 1976;72:248–54. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 29.Griess P. Bemerkungen zu der abhandlung der H.H. Weselsky und Benedikt “Ueber einige azoverbindungen.”. Chemische Berichte. 1879;12:426–8. [Google Scholar]
- 30.Ye Y, Godzik A. Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics. 2003;19(Suppl 2):ii246–55. doi: 10.1093/bioinformatics/btg1086. [DOI] [PubMed] [Google Scholar]
- 31.Hornfeldt CS, Rabe WH., 3rd Nitroethane poisoning from an artificial fingernail remover. Journal of Toxicology Clinical Toxicology. 1994;32:321–4. doi: 10.3109/15563659409017967. [DOI] [PubMed] [Google Scholar]
- 32.Kohl C, Gescher A. Denitrification of the genotoxicant 2-nitropropane: relationship to its mechanism of toxicity. Xenobiotica. 1997;27:843–52. doi: 10.1080/004982597240208. [DOI] [PubMed] [Google Scholar]
- 33.Kohl C, Mynett K, Davies JE, Gescher A, Chipman JK. Propane 2-nitronate is the major genotoxic form of 2-nitropropane. Mutation Research. 1994;321:65–72. doi: 10.1016/0165-1218(94)90121-x. [DOI] [PubMed] [Google Scholar]
- 34.Isabella VM, Clark VL. Identification of a conserved protein involved in anaerobic unsaturated fatty acid synthesis in Neiserria gonorrhoeae: implications for facultative and obligate anaerobes that lack FabA. Mol Microbiol. 2011;82:489–501. doi: 10.1111/j.1365-2958.2011.07826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malm S, Tiffert Y, Micklinghoff J, et al. The roles of the nitrate reductase NarGHJI, the nitrite reductase NirBD and the response regulator GlnR in nitrate assimilation of Mycobacterium tuberculosis. Microbiology. 2009;155:1332–9. doi: 10.1099/mic.0.023275-0. [DOI] [PubMed] [Google Scholar]
- 36.Aly S, Wagner K, Keller C, et al. Oxygen status of lung granulomas in Mycobacterium tuberculosis-infected mice. J Pathol. 2006;210:298–305. doi: 10.1002/path.2055. [DOI] [PubMed] [Google Scholar]
- 37.Tsai MC, Chakravarty S, Zhu G, et al. Characterization of the tuberculous granuloma in murine and human lungs: cellular composition and relative tissue oxygen tension. Cell Microbiol. 2006;8:218–32. doi: 10.1111/j.1462-5822.2005.00612.x. [DOI] [PubMed] [Google Scholar]
- 38.Voskuil MI, Schnappinger D, Visconti KC, et al. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J Exp Med. 2003;198:705–13. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci U S A. 2003;100:12989–94. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joshi SM, Pandey AK, Capite N, Fortune SM, Rubin EJ, Sassetti CM. Characterization of mycobacterial virulence genes through genetic interaction mapping. Proc Natl Acad Sci U S A. 2006;103:11760–5. doi: 10.1073/pnas.0603179103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosas-Magallanes V, Stadthagen-Gomez G, Rauzier J, et al. Signature-tagged transposon mutagenesis identifies novel Mycobacterium tuberculosis genes involved in the parasitism of human macrophages. Infect Immun. 2007;75:504–7. doi: 10.1128/IAI.00058-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rengarajan J, Bloom BR, Rubin EJ. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc Natl Acad Sci U S A. 2005;102:8327–32. doi: 10.1073/pnas.0503272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Voskuil MI, Visconti KC, Schoolnik GK. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis (Edinb) 2004;84:218–27. doi: 10.1016/j.tube.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 44.Schnappinger D, Ehrt S, Voskuil MI, et al. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J Exp Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 46.Bacon J, James BW, Wernisch L, et al. The influence of reduced oxygen availability on pathogenicity and gene expression in Mycobacterium tuberculosis. Tuberculosis (Edinb) 2004;84:205–17. doi: 10.1016/j.tube.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 47.Kendall SL, Movahedzadeh F, Rison SC, et al. The Mycobacterium tuberculosis dosRS two-component system is induced by multiple stresses. Tuberculosis (Edinb) 2004;84:247–55. doi: 10.1016/j.tube.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 48.Muttucumaru DG, Roberts G, Hinds J, Stabler RA, Parish T. Gene expression profile of Mycobacterium tuberculosis in a non-replicating state. Tuberculosis (Edinb) 2004;84:239–46. doi: 10.1016/j.tube.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 49.Ramesh KV, Akhila BN, Deshmukh S. Molecular modeling of 2-nitropropane dioxygenase domain of Mycobacterium tuberculosis H37Rv and docking of herbal ligands. Indian Journal of Biochemistry & Biophysics. 2011;48:164–9. [PubMed] [Google Scholar]
- 50.Mc DW. Inapparent infection: relation of latent and dormant infections to microbial persistence. Public Health Reports. 1959;74:485–99. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.