Abstract
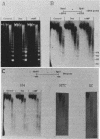
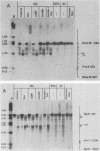
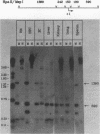
We used indirect end labeling to identify a series of five hypersensitive (HS) sites in the phosphoenolpyruvate carboxykinase (PEPCK) gene in H4IIE rat hepatoma cells. These sites were found at -4800 base pairs (bp) (site A), at -1300 bp (site B), over a broad domain between -400 and -30 bp (site C), at +4650 bp (site D), and at +6200 bp (site E). Sites A to D were detected only in cells capable of expressing the PEPCK gene, whereas site E was present in all of the cells examined thus far. The HS sites were present in H4IIE cells even when transcriptional activity was reduced to a minimum by treatment with insulin. Stimulation of transcription by a cyclic AMP analog to a 40-fold increase over the insulin-repressed level did not affect the main features of the HS sites. Furthermore, increased transcription did not disrupt the nucleosomal arrangement of the coding region of the gene, nor did it affect the immediate 5' region (site C), which is always nucleosome-free. In HTC cells, a rat hepatoma line that is hormonally responsive but unable to synthesize PEPCK mRNA, the four expression-specific HS sites were totally absent. Our experimental results also showed that, although there is a general correlation between lack of DNA methylation and transcriptional competence of the PEPCK gene, the role, if any, of methylation in the regulation of PEPCK gene activity is likely to be exerted at very specific sites.
Full text
PDF


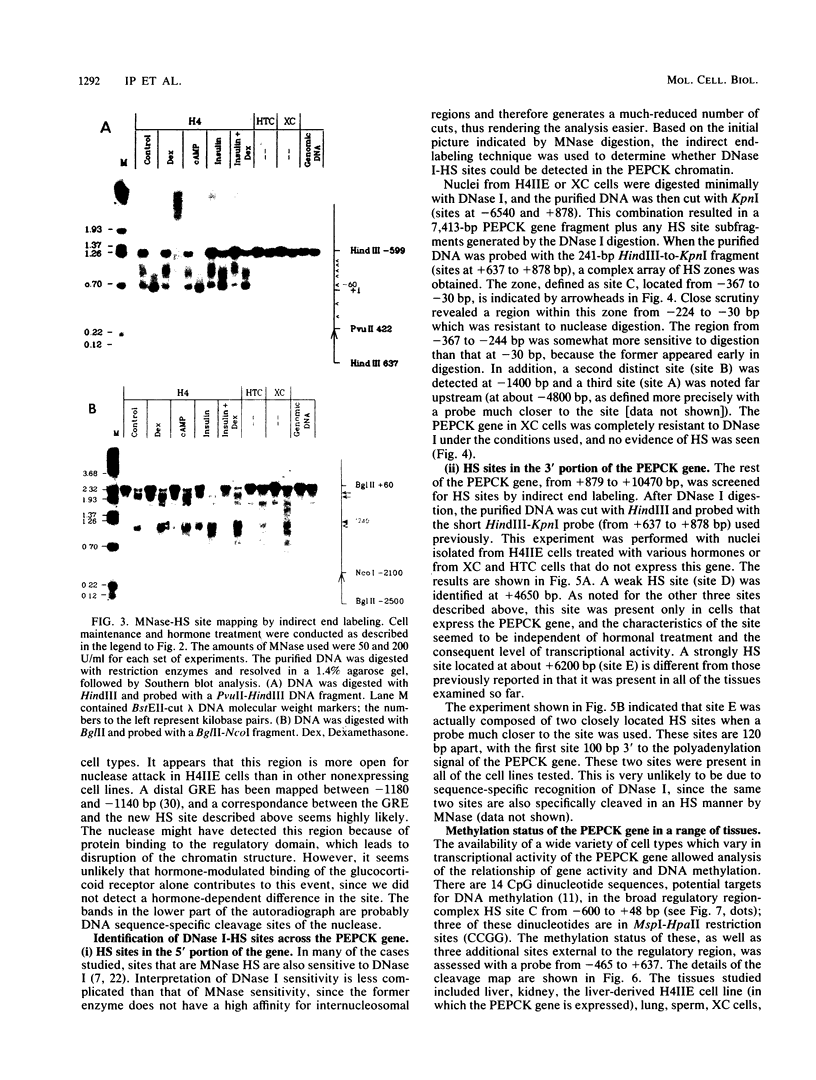
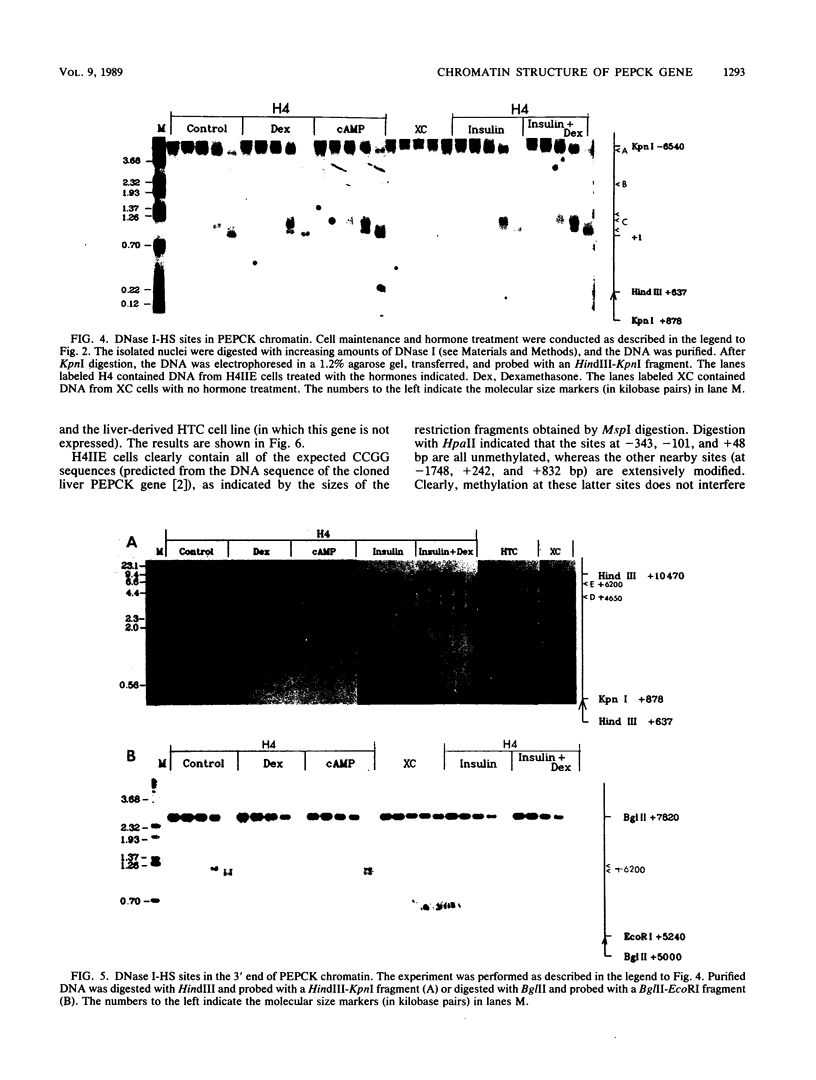
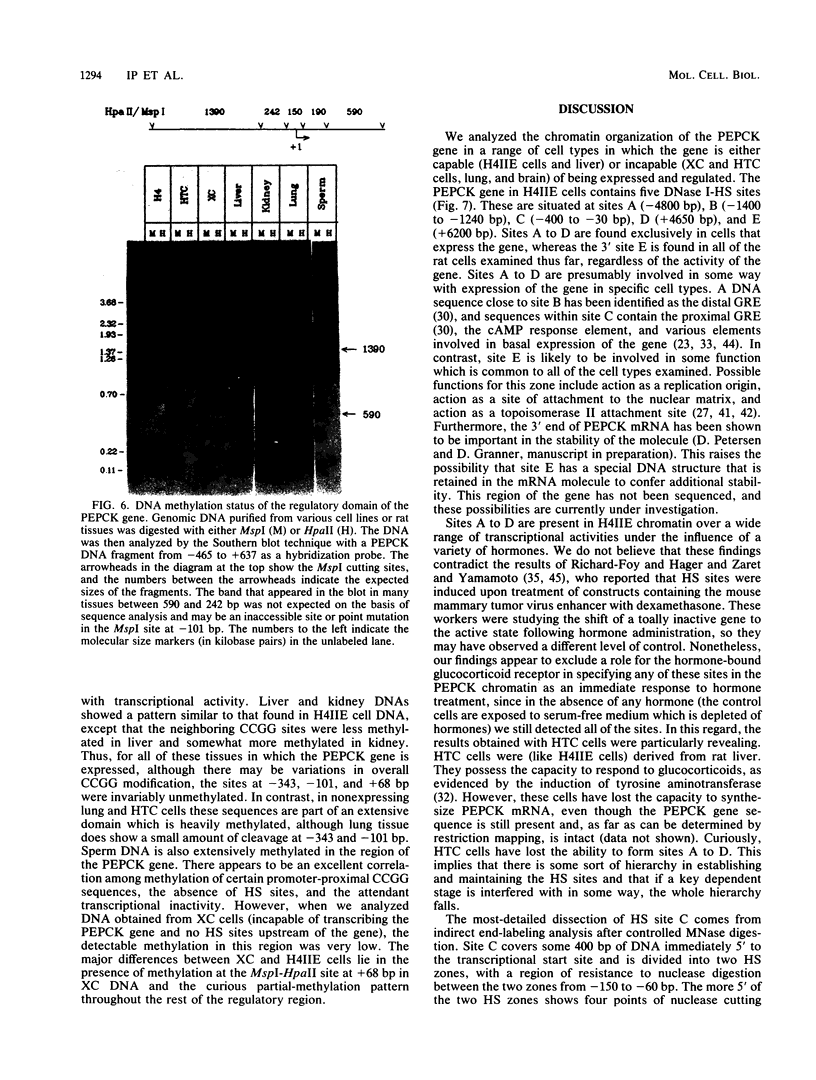



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allegra P., Sterner R., Clayton D. F., Allfrey V. G. Affinity chromatographic purification of nucleosomes containing transcriptionally active DNA sequences. J Mol Biol. 1987 Jul 20;196(2):379–388. doi: 10.1016/0022-2836(87)90698-x. [DOI] [PubMed] [Google Scholar]
- Beale E. G., Chrapkiewicz N. B., Scoble H. A., Metz R. J., Quick D. P., Noble R. L., Donelson J. E., Biemann K., Granner D. K. Rat hepatic cytosolic phosphoenolpyruvate carboxykinase (GTP). Structures of the protein, messenger RNA, and gene. J Biol Chem. 1985 Sep 5;260(19):10748–10760. [PubMed] [Google Scholar]
- Becker P. B., Ruppert S., Schütz G. Genomic footprinting reveals cell type-specific DNA binding of ubiquitous factors. Cell. 1987 Nov 6;51(3):435–443. doi: 10.1016/0092-8674(87)90639-8. [DOI] [PubMed] [Google Scholar]
- Bellard M., Dretzen G., Bellard F., Oudet P., Chambon P. Disruption of the typical chromatin structure in a 2500 base-pair region at the 5' end of the actively transcribed ovalbumin gene. EMBO J. 1982;1(2):223–230. doi: 10.1002/j.1460-2075.1982.tb01151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benvenisty N., Mencher D., Meyuhas O., Razin A., Reshef L. Sequential changes in DNA methylation patterns of the rat phosphoenolpyruvate carboxykinase gene during development. Proc Natl Acad Sci U S A. 1985 Jan;82(2):267–271. doi: 10.1073/pnas.82.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch J. B., Weintraub H. Temporal order of chromatin structural changes associated with activation of the major chicken vitellogenin gene. Cell. 1983 May;33(1):65–76. doi: 10.1016/0092-8674(83)90335-5. [DOI] [PubMed] [Google Scholar]
- Cartwright I. L., Elgin S. C. Nucleosomal instability and induction of new upstream protein-DNA associations accompany activation of four small heat shock protein genes in Drosophila melanogaster. Mol Cell Biol. 1986 Mar;6(3):779–791. doi: 10.1128/mcb.6.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H. DNA methylation and gene activity. Cell. 1988 Apr 8;53(1):3–4. doi: 10.1016/0092-8674(88)90479-5. [DOI] [PubMed] [Google Scholar]
- Cochet-Meilhac M., Nuret P., Courvalin J. C., Chambon P. Animal DNA-dependent RNA polymerases. 12. Determination of the cellular number of RNA polymerase B molecules. Biochim Biophys Acta. 1974 Jun 27;353(2):185–192. doi: 10.1016/0005-2787(74)90183-x. [DOI] [PubMed] [Google Scholar]
- Cohen R. B., Sheffery M. Nucleosome disruption precedes transcription and is largely limited to the transcribed domain of globin genes in murine erythroleukemia cells. J Mol Biol. 1985 Mar 5;182(1):109–129. doi: 10.1016/0022-2836(85)90031-2. [DOI] [PubMed] [Google Scholar]
- Doerfler W. DNA methylation and gene activity. Annu Rev Biochem. 1983;52:93–124. doi: 10.1146/annurev.bi.52.070183.000521. [DOI] [PubMed] [Google Scholar]
- Eissenberg J. C., Cartwright I. L., Thomas G. H., Elgin S. C. Selected topics in chromatin structure. Annu Rev Genet. 1985;19:485–536. doi: 10.1146/annurev.ge.19.120185.002413. [DOI] [PubMed] [Google Scholar]
- Emerson B. M., Lewis C. D., Felsenfeld G. Interaction of specific nuclear factors with the nuclease-hypersensitive region of the chicken adult beta-globin gene: nature of the binding domain. Cell. 1985 May;41(1):21–30. doi: 10.1016/0092-8674(85)90057-1. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Garel A., Axel R. Selective digestion of transcriptionally active ovalbumin genes from oviduct nuclei. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3966–3970. doi: 10.1073/pnas.73.11.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garel A., Zolan M., Axel R. Genes transcribed at diverse rates have a similar conformation in chromatin. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4867–4871. doi: 10.1073/pnas.74.11.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross D. S., Garrard W. T. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- Hebbes T. R., Thorne A. W., Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988 May;7(5):1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemenway C., Robins D. M. DNase I-hypersensitive sites associated with expression and hormonal regulation of mouse C4 and Slp genes. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4816–4820. doi: 10.1073/pnas.84.14.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson P. D., Felsenfeld G. A method for mapping intranuclear protein-DNA interactions and its application to a nuclease hypersensitive site. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2296–2300. doi: 10.1073/pnas.82.8.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene M. A., Corces V., Lowenhaupt K., Elgin S. C. DNase I hypersensitive sites in Drosophila chromatin occur at the 5' ends of regions of transcription. Proc Natl Acad Sci U S A. 1981 Jan;78(1):143–146. doi: 10.1073/pnas.78.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Wilson B., Fortier C., Blackhart B. D., McCarthy B. J. DNase I- and micrococcal nuclease-hypersensitive sites in the human apolipoprotein B gene are tissue specific. Mol Cell Biol. 1988 Jan;8(1):71–80. doi: 10.1128/mcb.8.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson M. A., Quinn P. G., Granner D. K. Multihormonal regulation of phosphoenolpyruvate carboxykinase-chloramphenicol acetyltransferase fusion genes. Insulin's effects oppose those of cAMP and dexamethasone. J Biol Chem. 1987 Nov 5;262(31):14917–14920. [PubMed] [Google Scholar]
- McGinnis W., Shermoen A. W., Heemskerk J., Beckendorf S. K. DNA sequence changes in an upstream DNase I-hypersensitive region are correlated with reduced gene expression. Proc Natl Acad Sci U S A. 1983 Feb;80(4):1063–1067. doi: 10.1073/pnas.80.4.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahon J. L., Venetianer A., Sala-Trepat J. M. Specific sets of DNase I-hypersensitive sites are associated with the potential and overt expression of the rat albumin and alpha-fetoprotein genes. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2135–2139. doi: 10.1073/pnas.84.8.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palen T. E., Cech T. R. Chromatin structure at the replication origins and transcription-initiation regions of the ribosomal RNA genes of Tetrahymena. Cell. 1984 Apr;36(4):933–942. doi: 10.1016/0092-8674(84)90043-6. [DOI] [PubMed] [Google Scholar]
- Parslow T. G., Granner D. K. Chromatin changes accompany immunoglobulin kappa gene activation: a potential control region within the gene. Nature. 1982 Sep 30;299(5882):449–451. doi: 10.1038/299449a0. [DOI] [PubMed] [Google Scholar]
- Pederson D. S., Thoma F., Simpson R. T. Core particle, fiber, and transcriptionally active chromatin structure. Annu Rev Cell Biol. 1986;2:117–147. doi: 10.1146/annurev.cb.02.110186.001001. [DOI] [PubMed] [Google Scholar]
- Petersen D. D., Magnuson M. A., Granner D. K. Location and characterization of two widely separated glucocorticoid response elements in the phosphoenolpyruvate carboxykinase gene. Mol Cell Biol. 1988 Jan;8(1):96–104. doi: 10.1128/mcb.8.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piette J., Kryszke M. H., Yaniv M. Specific interaction of cellular factors with the B enhancer of polyoma virus. EMBO J. 1985 Oct;4(10):2675–2685. doi: 10.1002/j.1460-2075.1985.tb03987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plesko M. M., Hargrove J. L., Granner D. K., Chalkley R. Inhibition by sodium butyrate of enzyme induction by glucocorticoids and dibutyryl cyclic AMP. A role for the rapid form of histone acetylation. J Biol Chem. 1983 Nov 25;258(22):13738–13744. [PubMed] [Google Scholar]
- Quinn P. G., Wong T. W., Magnuson M. A., Shabb J. B., Granner D. K. Identification of basal and cyclic AMP regulatory elements in the promoter of the phosphoenolpyruvate carboxykinase gene. Mol Cell Biol. 1988 Aug;8(8):3467–3475. doi: 10.1128/mcb.8.8.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen J. A., Gilboa E. Significance of DNase I-hypersensitive sites in the long terminal repeats of a Moloney murine leukemia virus vector. J Virol. 1987 May;61(5):1368–1374. doi: 10.1128/jvi.61.5.1368-1374.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard-Foy H., Hager G. L. Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J. 1987 Aug;6(8):2321–2328. doi: 10.1002/j.1460-2075.1987.tb02507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha E., Davie J. R., van Holde K. E., Weintraub H. Differential salt fractionation of active and inactive genomic domains in chicken erythrocyte. J Biol Chem. 1984 Jul 10;259(13):8558–8563. [PubMed] [Google Scholar]
- Rose S. M., Garrard W. T. Differentiation-dependent chromatin alterations precede and accompany transcription of immunoglobulin light chain genes. J Biol Chem. 1984 Jul 10;259(13):8534–8544. [PubMed] [Google Scholar]
- Sasaki K., Cripe T. P., Koch S. R., Andreone T. L., Petersen D. D., Beale E. G., Granner D. K. Multihormonal regulation of phosphoenolpyruvate carboxykinase gene transcription. The dominant role of insulin. J Biol Chem. 1984 Dec 25;259(24):15242–15251. [PubMed] [Google Scholar]
- Sasaki K., Granner D. K. Regulation of phosphoenolpyruvate carboxykinase gene transcription by insulin and cAMP: reciprocal actions on initiation and elongation. Proc Natl Acad Sci U S A. 1988 May;85(9):2954–2958. doi: 10.1073/pnas.85.9.2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. L., Xu Y. Z., Bellard M., Chambon P. Digestion of the chicken beta-globin gene chromatin with micrococcal nuclease reveals the presence of an altered nucleosomal array characterized by an atypical ladder of DNA fragments. EMBO J. 1986 Feb;5(2):293–300. doi: 10.1002/j.1460-2075.1986.tb04212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F. Protein-DNA interactions and nuclease-sensitive regions determine nucleosome positions on yeast plasmid chromatin. J Mol Biol. 1986 Jul 20;190(2):177–190. doi: 10.1016/0022-2836(86)90291-3. [DOI] [PubMed] [Google Scholar]
- Villeponteau B., Lundell M., Martinson H. Torsional stress promotes the DNAase I sensitivity of active genes. Cell. 1984 Dec;39(3 Pt 2):469–478. doi: 10.1016/0092-8674(84)90454-9. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Wynshaw-Boris A., Lugo T. G., Short J. M., Fournier R. E., Hanson R. W. Identification of a cAMP regulatory region in the gene for rat cytosolic phosphoenolpyruvate carboxykinase (GTP). Use of chimeric genes transfected into hepatoma cells. J Biol Chem. 1984 Oct 10;259(19):12161–12169. [PubMed] [Google Scholar]
- Zaret K. S., Yamamoto K. R. Reversible and persistent changes in chromatin structure accompany activation of a glucocorticoid-dependent enhancer element. Cell. 1984 Aug;38(1):29–38. doi: 10.1016/0092-8674(84)90523-3. [DOI] [PubMed] [Google Scholar]