Abstract
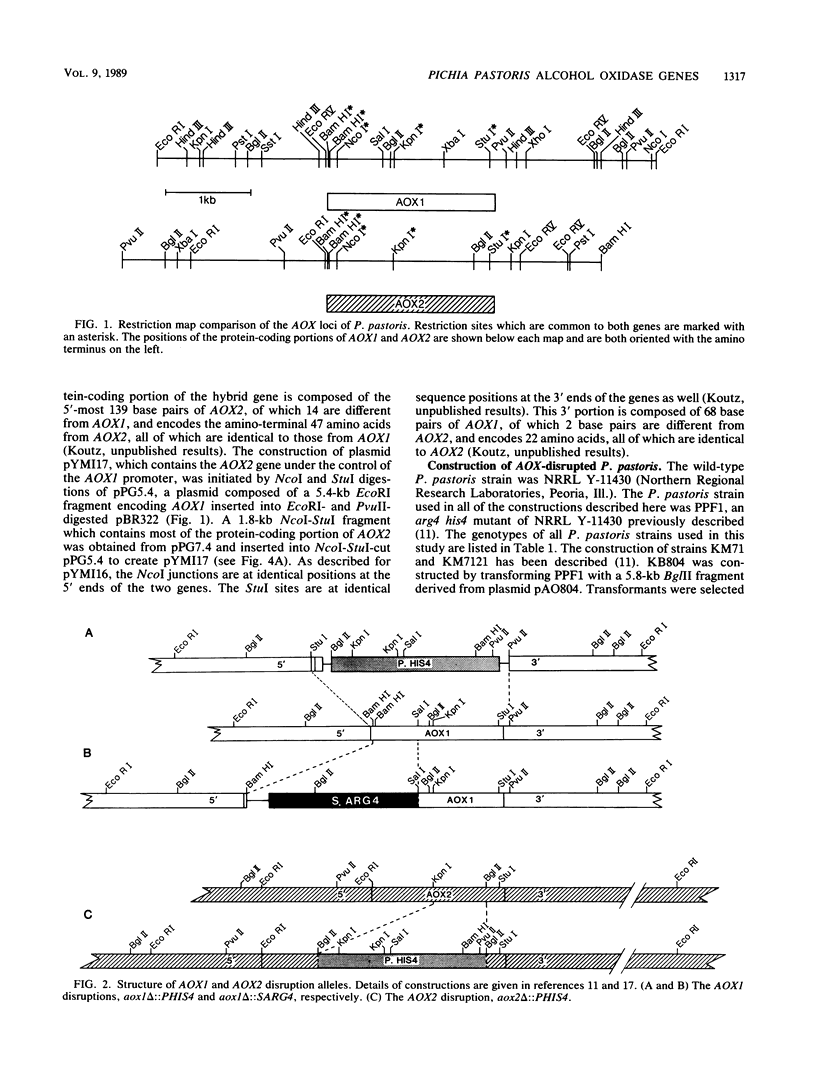
In Pichia pastoris, alcohol oxidase (AOX) is the first enzyme in the methanol utilization pathway and is encoded by two genes, AOX1 and AOX2. The DNA and predicted amino acid sequences of the protein-coding portions of the genes are closely homologous, whereas flanking sequences share no homology. The functional roles of AOX1 and AOX2 in the metabolism of methanol were examined. Studies of strains with disrupted AOX genes revealed that AOX1 was the major source of methanol-oxidizing activity in methanol-grown P. pastoris. The results of two types of experiments each suggested that the difference in AOX activity contributed by the two genes was a consequence of sequences located 5' of the protein-coding portions of the genes. First, the coding portion of AOX2 was able to functionally substitute for that of AOX1 when placed under the control of AOX1 regulatory sequences. Second, when labeled oligonucleotide probes specific for the 5' nontranslated region of each gene were used, it was apparent that the steady-state level of AOX1 mRNA was much higher than that of AOX2. Except for the difference in the amount of mRNA present, the two genes appeared to be regulated in the same manner. A physiological reason for the existence of AOX2 was sought but was not apparent.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abovich N., Rosbash M. Two genes for ribosomal protein 51 of Saccharomyces cerevisiae complement and contribute to the ribosomes. Mol Cell Biol. 1984 Sep;4(9):1871–1879. doi: 10.1128/mcb.4.9.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajwa W., Rudolph H., Hinnen A. PHO5 upstream sequences confer phosphate control on the constitutive PHO3 gene. Yeast. 1987 Mar;3(1):33–42. doi: 10.1002/yea.320030106. [DOI] [PubMed] [Google Scholar]
- Basson M. E., Thorsness M., Rine J. Saccharomyces cerevisiae contains two functional genes encoding 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5563–5567. doi: 10.1073/pnas.83.15.5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beacham I. R., Schweitzer B. W., Warrick H. M., Carbon J. The nucleotide sequence of the yeast ARG4 gene. Gene. 1984 Sep;29(3):271–279. doi: 10.1016/0378-1119(84)90056-8. [DOI] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. The primary structure of the Saccharomyces cerevisiae gene for alcohol dehydrogenase. J Biol Chem. 1982 Mar 25;257(6):3018–3025. [PubMed] [Google Scholar]
- Cohen R., Yokoi T., Holland J. P., Pepper A. E., Holland M. J. Transcription of the constitutively expressed yeast enolase gene ENO1 is mediated by positive and negative cis-acting regulatory sequences. Mol Cell Biol. 1987 Aug;7(8):2753–2761. doi: 10.1128/mcb.7.8.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cregg J. M., Barringer K. J., Hessler A. Y., Madden K. R. Pichia pastoris as a host system for transformations. Mol Cell Biol. 1985 Dec;5(12):3376–3385. doi: 10.1128/mcb.5.12.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumsky M. G., Trueblood C. E., Ko C., Poyton R. O. Structural analysis of two genes encoding divergent forms of yeast cytochrome c oxidase subunit V. Mol Cell Biol. 1987 Oct;7(10):3511–3519. doi: 10.1128/mcb.7.10.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis C. L. Identification of new genes involved in the regulation of yeast alcohol dehydrogenase II. Genetics. 1984 Dec;108(4):833–844. doi: 10.1093/genetics/108.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas M. G., McCammon M. T., Vassarotti A. Targeting proteins into mitochondria. Microbiol Rev. 1986 Jun;50(2):166–178. doi: 10.1128/mr.50.2.166-178.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder R. T., Loh E. Y., Davis R. W. RNA from the yeast transposable element Ty1 has both ends in the direct repeats, a structure similar to retrovirus RNA. Proc Natl Acad Sci U S A. 1983 May;80(9):2432–2436. doi: 10.1073/pnas.80.9.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis S. B., Brust P. F., Koutz P. J., Waters A. F., Harpold M. M., Gingeras T. R. Isolation of alcohol oxidase and two other methanol regulatable genes from the yeast Pichia pastoris. Mol Cell Biol. 1985 May;5(5):1111–1121. doi: 10.1128/mcb.5.5.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S. G., Keller G. A., Subramani S. Identification of a peroxisomal targeting signal at the carboxy terminus of firefly luciferase. J Cell Biol. 1987 Dec;105(6 Pt 2):2923–2931. doi: 10.1083/jcb.105.6.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland J. P., Holland M. J. Structural comparison of two nontandemly repeated yeast glyceraldehyde-3-phosphate dehydrogenase genes. J Biol Chem. 1980 Mar 25;255(6):2596–2605. [PubMed] [Google Scholar]
- Kalderon D., Roberts B. L., Richardson W. D., Smith A. E. A short amino acid sequence able to specify nuclear location. Cell. 1984 Dec;39(3 Pt 2):499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Kim K. S., Rosenkrantz M. S., Guarente L. Saccharomyces cerevisiae contains two functional citrate synthase genes. Mol Cell Biol. 1986 Jun;6(6):1936–1942. doi: 10.1128/mcb.6.6.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laz T. M., Pietras D. F., Sherman F. Differential regulation of the duplicated isocytochrome c genes in yeast. Proc Natl Acad Sci U S A. 1984 Jul;81(14):4475–4479. doi: 10.1073/pnas.81.14.4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarow P. B., Fujiki Y. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- Ledeboer A. M., Edens L., Maat J., Visser C., Bos J. W., Verrips C. T., Janowicz Z., Eckart M., Roggenkamp R., Hollenberg C. P. Molecular cloning and characterization of a gene coding for methanol oxidase in Hansenula polymorpha. Nucleic Acids Res. 1985 May 10;13(9):3063–3082. doi: 10.1093/nar/13.9.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Botstein D. Effects of null mutations in the hexokinase genes of Saccharomyces cerevisiae on catabolite repression. Mol Cell Biol. 1986 Nov;6(11):4046–4052. doi: 10.1128/mcb.6.11.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlister L., Holland M. J. Differential expression of the three yeast glyceraldehyde-3-phosphate dehydrogenase genes. J Biol Chem. 1985 Dec 5;260(28):15019–15027. [PubMed] [Google Scholar]
- McAlister L., Holland M. J. Isolation and characterization of yeast strains carrying mutations in the glyceraldehyde-3-phosphate dehydrogenase genes. J Biol Chem. 1985 Dec 5;260(28):15013–15018. [PubMed] [Google Scholar]
- McAlister L., Holland M. J. Targeted deletion of a yeast enolase structural gene. Identification and isolation of yeast enolase isozymes. J Biol Chem. 1982 Jun 25;257(12):7181–7188. [PubMed] [Google Scholar]
- Niederacher D., Entian K. D. Isolation and characterization of the regulatory HEX2 gene necessary for glucose repression in yeast. Mol Gen Genet. 1987 Mar;206(3):505–509. doi: 10.1007/BF00428892. [DOI] [PubMed] [Google Scholar]
- Roggenkamp R., Janowicz Z., Stanikowski B., Hollenberg C. P. Biosynthesis and regulation of the peroxisomal methanol oxidase from the methylotrophic yeast Hansenula polymorpha. Mol Gen Genet. 1984;194(3):489–493. doi: 10.1007/BF00425563. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Russell D. W., Smith M., Williamson V. M., Young E. T. Nucleotide sequence of the yeast alcohol dehydrogenase II gene. J Biol Chem. 1983 Feb 25;258(4):2674–2682. [PubMed] [Google Scholar]
- Sahm H., Wagner F. Microbial assimilation of methanol. The ethanol- and methanol-oxidizing enzymes of the yeast Candida boidinii. Eur J Biochem. 1973 Jul 2;36(1):250–256. doi: 10.1111/j.1432-1033.1973.tb02907.x. [DOI] [PubMed] [Google Scholar]
- Schatz G., Butow R. A. How are proteins imported into mitochondria? Cell. 1983 Feb;32(2):316–318. doi: 10.1016/0092-8674(83)90450-6. [DOI] [PubMed] [Google Scholar]
- Schatz P. J., Georges G. E., Solomon F., Botstein D. Insertions of up to 17 amino acids into a region of alpha-tubulin do not disrupt function in vivo. Mol Cell Biol. 1987 Oct;7(10):3799–3805. doi: 10.1128/mcb.7.10.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz P. J., Pillus L., Grisafi P., Solomon F., Botstein D. Two functional alpha-tubulin genes of the yeast Saccharomyces cerevisiae encode divergent proteins. Mol Cell Biol. 1986 Nov;6(11):3711–3721. doi: 10.1128/mcb.6.11.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz P. J., Solomon F., Botstein D. Genetically essential and nonessential alpha-tubulin genes specify functionally interchangeable proteins. Mol Cell Biol. 1986 Nov;6(11):3722–3733. doi: 10.1128/mcb.6.11.3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tait-Kamradt A. G., Turner K. J., Kramer R. A., Elliott Q. D., Bostian S. J., Thill G. P., Rogers D. T., Bostian K. A. Reciprocal regulation of the tandemly duplicated PHO5/PHO3 gene cluster within the acid phosphatase multigene family of Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jun;6(6):1855–1865. doi: 10.1128/mcb.6.6.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueblood C. E., Poyton R. O. Differential effectiveness of yeast cytochrome c oxidase subunit genes results from differences in expression not function. Mol Cell Biol. 1987 Oct;7(10):3520–3526. doi: 10.1128/mcb.7.10.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschopp J. F., Brust P. F., Cregg J. M., Stillman C. A., Gingeras T. R. Expression of the lacZ gene from two methanol-regulated promoters in Pichia pastoris. Nucleic Acids Res. 1987 May 11;15(9):3859–3876. doi: 10.1093/nar/15.9.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschumper G., Carbon J. Delta sequences and double symmetry in a yeast chromosomal replicator region. J Mol Biol. 1982 Apr 5;156(2):293–307. doi: 10.1016/0022-2836(82)90330-8. [DOI] [PubMed] [Google Scholar]
- Veenhuis M., Van Dijken J. P., Harder W. The significance of peroxisomes in the metabolism of one-carbon compounds in yeasts. Adv Microb Physiol. 1983;24:1–82. doi: 10.1016/s0065-2911(08)60384-7. [DOI] [PubMed] [Google Scholar]
- Young E. T., Pilgrim D. Isolation and DNA sequence of ADH3, a nuclear gene encoding the mitochondrial isozyme of alcohol dehydrogenase in Saccharomyces cerevisiae. Mol Cell Biol. 1985 Nov;5(11):3024–3034. doi: 10.1128/mcb.5.11.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]