Abstract
During infection, pathogenic bacteria manipulate the host cell in various ways to permit their own replication, propagation and escape from host immune responses. Post-translational modifications are unique mechanisms that allow cells to rapidly, locally, and specifically modify activity or interactions of key proteins. Some of these modifications, including phosphorylation and ubiquitylation1,2, can be induced by pathogens. However, the effects of pathogenic bacteria on SUMOylation, an essential post-translational modification in eukaryotic cells3 remain largely unknown. Here we show that Listeria monocytogenes infection leads to a decrease in the levels of cellular SUMO-conjugated proteins. This event is triggered by the bacterial virulence factor listeriolysin O (LLO) which induces a proteasome-independent degradation of Ubc9, an essential enzyme of the SUMOylation machinery. The effect of LLO on Ubc9 is dependent on the pore-forming capacity of the toxin and is shared by other bacterial pore-forming toxins like perfringolysin O (PFO) and pneumolysin (PLY). Ubc9 degradation was also observed in vivo in infected mice. Furthermore, we show that SUMO overexpression impairs bacterial infection. Together, our results reveal that Listeria, and probably other pathogens, dampen the host response to infection by preventing SUMOylation of key regulatory proteins.
Listeria monocytogenes is a facultative intracellular pathogen responsible for human listeriosis, a severe food-borne disease, and has emerged as a model for the study of host-pathogen interactions. This bacterium is able to cross the intestinal, maternofetal and blood brain barriers, to resist macrophage killing and to enter into normally non-phagocytic cells and replicate therein4. During infection, L. monocytogenes exploits an amazingly high number of mammalian cell functions to its own benefit. In particular, Listeria interferes with several signalling pathways and is able to regulate host protein activities by modifying their ubiquitylation or phosphorylation5-7. However, the impact of L. monocytogenes on SUMOylation, an essential post-translational modification, remains completely unknown.
SUMOylation is a reversible modification in which SUMO, an ubiquitin-like polypeptide of ~10 kDa, is covalently linked to target proteins. This conjugation results from the formation of an isopeptide bond between the C-terminal Gly residue of a mature SUMO moiety and a Lys side chain of the substrate protein3. The human genome encodes three functional SUMO isoforms that can be linked to distinct and overlapping sets of proteins8. Covalent linkage of SUMO to its substrate requires a set of different enzymes, in an analogous fashion to ubiquitylation. In humans, the SUMOylation machinery is composed of an E1 SUMO enzyme (the SAE1/SAE2 heterodimer), an E2 SUMO enzyme (Ubc9), and E3 SUMO enzymes that enhance SUMO conjugation of specific targets. In contrast to the ubiquitylation machinery in which several dozens E2 enzymes are found9, the E2 SUMO enzyme is unique in mammals and required for viability10. The SUMOylation level of cellular proteins is tightly regulated by SUMO-specific proteases that catalyze the deconjugation of SUMO from its substrates11. SUMOylation, as ubiquitylation, is essential for different cellular functions. Several hundreds of SUMO targets have now been identified, involved in transcription regulation, maintenance of genome integrity, intracellular transport, stress responses, protein stability and many other biological processes (for review, see 3,12).
Interestingly, some viruses interfere with the SUMOylation of host proteins13. We thus tested the hypothesis that pathogenic bacteria, as viruses, also alter host protein SUMOylation for the orchestration of the onset, establishment and/or persistence of the infectious process, and addressed this issue in the case of a L. monocytogenes infection.
To investigate whether L. monocytogenes was able to modify SUMOylation of host cell proteins upon infection, we compared the global pattern of proteins conjugated to SUMO1 or SUMO2/3 in uninfected cells with that of cells infected by L. monocytogenes or incubated with Listeria innocua, a non-pathogenic Listeria species. HeLa cells infected with L. monocytogenes displayed, after 3 hours of infection, a decrease in both SUMO1 and SUMO2/3-conjugated proteins of high molecular weight, compared to uninfected cells (Fig. 1a). This decrease was not observed with L. innocua and is thus specific to the pathogenic L. monocytogenes species. This global reduction in protein SUMOylation was confirmed by proteomic analysis of SUMO-conjugated proteins isolated from cells infected or not by L. monocytogenes, using the stable isotope labeling of amino acids in cell culture (SILAC) technique14 (Suppl. Fig 1 and Suppl. Table 1, 2). A decrease in SUMOylated proteins upon Listeria infection is of particular interest as it clearly differs from the reported increase in SUMOylation observed for cells subjected to various environmental stresses15,16.
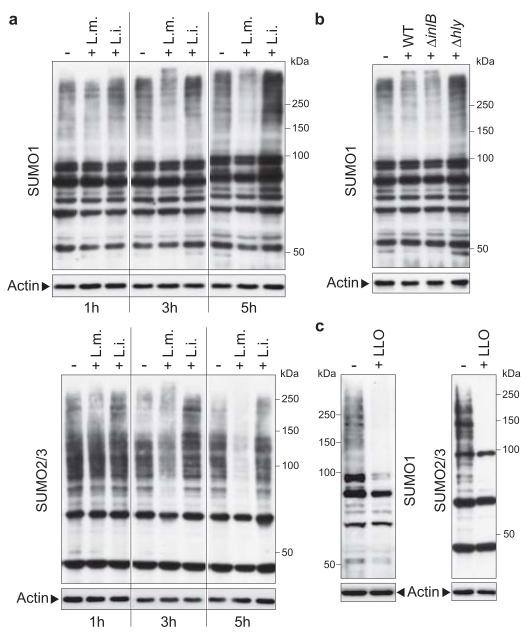
Figure 1. Decrease in SUMO-conjugated proteins upon Listeria infection.
a, Comparison of SUMO-conjugated protein patterns from uninfected HeLa cells (−) or cells infected for 1, 3 or 5 hours with Listeria monocytogenes (L. m.) or Listeria innocua (L. i.). Whole cell lysates were analysed by immunoblotting experiments using antibodies specific for SUMO1 (upper panel) or SUMO2/3 (lower panel) isoforms. b, Analysis of SUMO1-conjugated protein patterns from uninfected HeLa cells (−) or cells infected for 3 hours with wild type (WT), ΔinlB or Δhly L. monocytogenes. c, Analysis of SUMO1 and SUMO2/3-conjugated protein patterns from HeLa cells incubated with 3 nM LLO for 20 min.
A Listeria ΔinlB mutant impaired in entry into HeLa cells still induced a decrease in SUMO-conjugated proteins (Fig. 1b), suggesting that this decrease can be triggered by extracellular bacteria, and involves a surface or a secreted protein. We thus tested a Listeria Δhly mutant, defective for listeriolysin O (LLO), a secreted pore-forming toxin with a potent signalling activity, involved in the escape of Listeria from the internalization vacuole17. Strikingly, this mutant had no effect on host SUMOylated proteins (Fig. 1b), strongly suggesting that LLO plays a role in the decrease in SUMOylation observed with wild-type Listeria. We next analysed the global SUMOylation pattern of cells incubated with purified LLO alone, at a non-cytotoxic concentration7. LLO was sufficient to trigger a massive decrease in SUMOylated proteins in the host cell, both for SUMO1 and SUMO2/3 isoforms (Fig. 1c). This effect was not restricted to HeLa cells and also observed in Jeg3 cells (Suppl. Fig. 2).
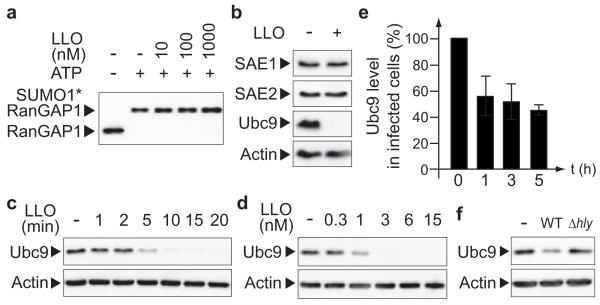
As many different SUMO targets were affected, we hypothesized that LLO could have an effect on the SUMOylation machinery. In vitro SUMOylation of RanGAP1, a well-characterized SUMO substrate18, was not affected by adding increasing amounts of LLO in the reaction, revealing that LLO has no direct inhibitory effect on E1 or E2 SUMO enzymes (Fig. 2a). We then quantified the levels of E1 or E2 SUMO enzymes in HeLa cells incubated with purified LLO. Strikingly, LLO induced a dramatic decrease in the level of Ubc9, in a dose-dependent manner, whereas the levels of SAE1 and SAE2 remained unaffected (Fig. 2b, d). The mRNA levels of Ubc9 were not affected upon LLO incubation (Suppl. Fig. 3). Moreover, LLO strongly reduced Ubc9 half-life (from >8 h to <10 min) and inhibition of cellular protein synthesis by cycloheximide did not prevent the LLO-triggered decrease in Ubc9 levels (Fig. 2b, c and Suppl. Fig. 3). Together, these results indicate that LLO does not affect Ubc9 translation but rather triggers its degradation. To our knowledge, LLO is the first bacterial factor inducing the degradation of the E2 SUMO enzyme. The decrease in Ubc9 level triggered by LLO was also observed during bacterial infection of HeLa and Jeg3 cells but was not observed in cells infected with a L. monocytogenes Δhly mutant (Fig. 2e, f and Suppl. Fig. 2).
Figure 2. L. monocytogenes induces a specific degradation of Ubc9.
a, In vitro RanGAP1 SUMOylation assay in presence of increasing amounts of LLO. b, Immunoblot analysis of HeLa cells incubated with 3 nM LLO for 20 min. c, Kinetics of Ubc9 degradation in HeLa cells incubated with 3 nM LLO. d, Dose-dependent degradation of Ubc9 in HeLa cells incubated with LLO for 20 min. e, Quantification of Ubc9 levels in cell extracts from HeLa cells infected with L. monocytogenes (n=3; error bars, s.d.). f, Immunoblot analysis of HeLa cells infected for 3 hours with wild-type (WT) or Δhly L. monocytogenes.
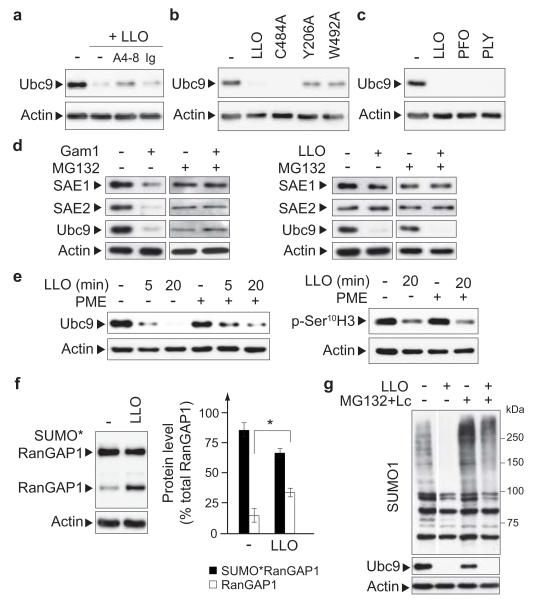
To determine whether LLO pore-formation is required to trigger Ubc9 degradation, we first preincubated LLO with A4-8, a monoclonal antibody preventing the binding of the toxin to cellular membranes19. This preincubation impaired the LLO-dependent decrease in Ubc9 levels in HeLa cells extracts, indicating that binding of the toxin to membranes is required for Ubc9 targeting (Fig. 3a). We then monitored the effect of three different LLO variants able to bind cellular membranes but affected in their haemolytic activities (Suppl. Table 3), i.e. LLOC484A and LLOW492A, which carry substitutions in the undecapeptide sequence conserved among cholesterol-dependent pore-forming toxins20, and LLOY206A, designed by homology to PFOY181A, a mutant of PFO unable to form pores21. Analysis of HeLa cells incubated with these three LLO mutants revealed a correlation between the hemolytic activities of the mutants and degradation of Ubc9 (Fig. 3b). Together, these data reveal that both binding of LLO to cellular membranes and pore formation are required to trigger Ubc9 degradation.
Figure 3. Characterization of LLO-mediated Ubc9 degradation.
a, Immunoblot analysis of HeLa cells incubated with either LLO or LLO preincubated with A4-8 or control antibody (Ig). b, Immunoblot analysis of HeLa cells incubated with LLO or LLO variants. c, Immunoblot analysis of HeLa cells incubated with LLO, PFO or PLY. d, Immunoblot analysis of HeLa cells either expressing Gam1 (left panel) or incubated with LLO (right panel), in the presence of DMSO or MG132. e, Immunoblot analysis of HeLa cells pretreated with DMSO or PME and incubated with LLO. f, Analysis of RanGAP1 and SUMO1*RanGAP1 levels in HeLa cells incubated with LLO (n=4; error bars, s.d.; *, P<0.01). g, Immunoblot analysis of HeLa cells pretreated with DMSO or MG132+Lactacystin (MG132+Lc) and incubated with LLO. For all LLO treatments, cells were incubated for 20 min with 3 nM of the toxin.
To determine whether the effect of LLO on Ubc9 is a property shared by other pore-forming toxins, we analysed the effect of PFO and PLY, two toxins of the same family as LLO, encoded by two other bacterial pathogens (Clostridium perfringens and Streptococcus pneumoniae, respectively). Interestingly, as LLO, PFO and PLY triggered a degradation of Ubc9 (Fig. 3c). Thus, other pathogenic bacteria, including extracellular pathogens, can target the SUMOylation machinery with mechanisms similar to that used by Listeria.
To identify the mechanisms underlying Ubc9 degradation, we used different pharmacological inhibitors. Inhibition of proteasome activity by MG132 has no effect on the level of Ubc9, which still decreased in HeLa cells upon incubation with LLO (Fig. 3d). As a control, we verified that HeLa cells expressing Gam1, a viral protein triggering the proteasomal degradation of SAE1, SAE2 and Ubc922,23, displayed a reduction in levels of E1 and E2 SUMO enzymes compared to control cells, whereas MG132 treated cells did not (Fig. 3d). Together, these data indicate that LLO-induced Ubc9 degradation is proteasome-independent and highlight differences in the mechanism of action of LLO and Gam1, although both proteins target the host cell SUMOylation machinery. Previous reports have shown that LLO can activate different signalling pathways via triggering of calcium influx from the extracellular medium and activation of MAPK17,24,25. We showed that LLO-mediated Ubc9 degradation is independent of the calcium influx triggered by LLO and independent of p38 or ERK activities (Suppl. Fig. 4). In contrast, Pepstatin-A-Methyl-Ester (PME), a cell permeable aspartyl-protease inhibitor, was able to partially impair Ubc9 decrease (Fig. 3e). The effect of PME on Ubc9 degradation is specific as it had no effect on histone H3 Ser10 dephosphorylation, another LLO-induced modification7. Together, our results strongly suggest that the LLO-mediated Ubc9 degradation is induced through a non classical pathway, which involves an aspartyl-protease remaining to be identified.
We then focused on the downstream effects of Ubc9 degradation. We hypothesized that degradation of Ubc9 is a key event that not only triggers a blockade of SUMOylation but also leads to the deSUMOylation of SUMO-conjugated proteins due to the activity of deSUMOylases. Consistently, whereas the total level of RanGAP1 protein (i.e. SUMO*RanGAP1+RanGAP1 in Fig. 3f) remains mostly unchanged upon LLO treatment in HeLa cells, the quantity of unSUMOylated RanGAP1 significantly increases, in agreement with a deSUMOylation of this target (Fig. 3f). However, one cannot exclude that the observed decrease in some SUMO-conjugated proteins might also be due to degradation, rather than deSUMOylation. Indeed, inhibition of the proteasome partially abrogates the decrease in SUMO-conjugated proteins triggered by LLO (~50% inhibition, Fig. 3g). We thus conclude that LLO triggers both a global deSUMOylation event and also degradation of some SUMO-conjugated proteins.
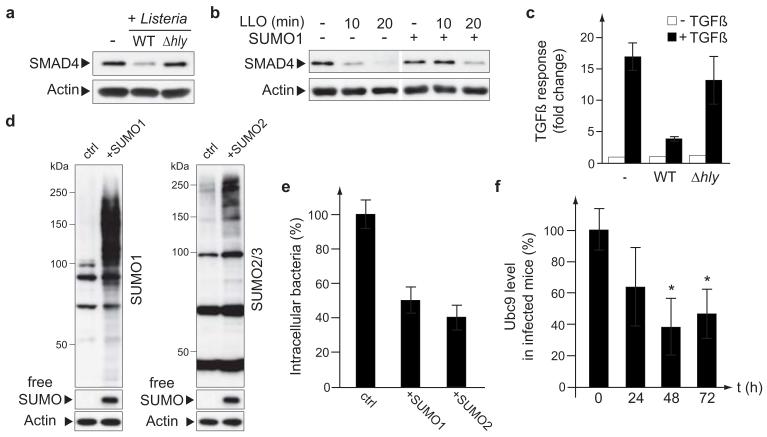
To investigate further the downstream effects of LLO-mediated decrease in SUMO-conjugated host proteins, we selected as a read-out the TGFβ response of the cell, a pathway regulated by SUMOylation. Indeed, SUMOylation stabilizes SMAD4, a central intracellular signal transducer for TGFβ signalling26 and regulates the TGFβ receptor TβR127. We first observed that infection with wild-type L. monocytogenes, as well as incubation with LLO, leads to a decrease in SMAD4 level, in agreement with the reported role of SUMOylation in SMAD4 stabilization (Fig. 4a, b). This decrease in SMAD4 level was not observed in cells infected with a Listeria Δhly mutant (Fig. 4a). Overexpression of SUMO1 in HeLa cells counteracts the decrease in SMAD4 induced by LLO, thus confirming that this decrease is linked to the effect of the toxin on the SUMOylation of host proteins (Fig. 4b). We then quantified the capacity of cells infected by Listeria to respond to TGFβ by using a reporter plasmid encoding luciferase under the control of SMAD transcriptional response elements. TGFβ response of infected 293FT cells was strongly impaired compared to non infected cells or cells infected with a Listeria Δhly mutant (Fig. 4c). Together, these results suggest that loss of SUMO-conjugated proteins upon L. monocytogenes infection affects the TGFβ response of the host cell. However, we cannot rule out that other pathways affected by LLO might participate to the observed impairment of TGF-β response in infected cells. Of note, TGFβ is known to play a role in pathogen infections28 and, in particular, it has been proposed to have a protective role in host resistance against L. monocytogenes infection29. Impairment of TGFβ signalling might thus be beneficial for Listeria to replicate and disseminate in its host.
Figure 4. Roles of SUMOylation in Listeria infection.
a, Immunoblot analysis of HeLa cells infected for 5 hours with L. monocytogenes. b, Immunoblot analysis of HeLa cells transfected with a SUMO1 expression vector or a control plasmid and treated with 3 nM LLO. c, Luciferase activities, in response to hTGF-β1, in 293FT cells transfected with a SMAD reporter plasmid and infected with wild-type (WT) or Δhly L. monocytogenes (n=3; error bars, s.d.). d, Immunoblot analysis of HeLa cells transfected with an expression vector for SUMO1, SUMO2 or the corresponding empty vector as a control. e, Number of intracellular bacteria recovered from HeLa cells transfected as in d and infected with L. monocytogenes for 7 hours (n=3; error bars, s.d.). f, Quantification of Ubc9 levels in liver homogenates from C57Bl/6J mice infected with L. monocytogenes (n=8 animals per group; error bars, s.d.; *, P<0.01).
To characterize the putative role of SUMOylation during infection, we increase the level of SUMOylated proteins in HeLa cells by overexpressing SUMO1 or SUMO2 (Fig. 4d). We infected these cells with L. monocytogenes and the number of intracellular bacteria was determined 7 hours after infection. This number was significantly reduced in cells overexpressing SUMO1 or SUMO2 compared to control cells (Fig. 4e). Thus, induction of an increase in SUMOylation is detrimental for bacterial invasion and/or replication. Conversely, Ubc9 degradation triggered by L. monocytogenes is probably important to promote bacterial infection by lowering SUMOylation of different host proteins.
To examine the relevance of our in vitro data during infection, we tested whether Ubc9 degradation observed in tissue cultured cells was also detectable in vivo. Quantification of Ubc9 levels in livers of C57Bl/6J mice infected with L. monocytogenes showed a significant reduction at 48h and 72h after infection (Fig. 4f). Thus, Ubc9 degradation occurs in vivo in organs infected by L. monocytogenes.
In conclusion, we report here a previously unknown strategy used by Listeria to promote its infection capacity. By targeting an essential enzyme of the SUMO pathway, L. monocytogenes has developed an efficient strategy to modify simultaneously activity of many different proteins of the host cell. That toxins produced by other pathogens also induced Ubc9 degradation highlights that inhibition of SUMOylation by pathogens may be a widespread phenomenon.
METHODS
Plasmids
Cignal SMAD reporter plasmid encodes the firefly luciferase gene under the control of a minimal CMV promoter and tandem repeats of the SMAD transcriptional response element (SABiosciences Corporation). pSG5-SUMO1HisHA (BUG2863) and pSG5-SUMO2HisHA (BUG2864) plasmids encode mature SUMO isoforms with N-terminal His6- and HA tags. pSG9-Gam1wt plasmid encodes the adenoviral Gam1 protein (kind gift of S. Chiocca22). Plasmids used for purification of His-tagged LLO mutants were derived from pET29b-LLO-His630 with punctual mutations inserted by PCR mutagenesis (pET29b-LLO C484A-His6, BUG2662; pET29b-LLO Y206A-His6, BUG2860; pET29b-LLO W492A-His6, BUG2664)
Bacterial strains
Listeria strains were grown in brain heart infusion (BHI) broth (Difco) or BHI agar plates. Strains used in this study were Listeria innocua (BUG499), Listeria monocytogenes wild-type strain EGD (BUG600) and L. monocytogenes isogenic EGD Δhly (BUG2132, Hamon et al., in preparation) and ΔinlB (BUG104731) deletion mutants.
Cell culture, transfections and infections
HeLa and Jeg3 cells were cultivated in MEM-glutamax medium and 293FT cells in DMEM-glutamax medium (Invitrogen). Both media were supplemented with 10% FCS, MEM non essential aminoacids and 1 mM sodium pyruvate (Invitrogen). For transfections, HeLa cells were seeded at 1.25×105 cells per 4 cm2 well and transfected the day after with 1.5 μg of DNA, using Lipofectamine LTX reagent (Invitrogen). 293FT cells were seeded at 1.2×105 cells per 2 cm2 well and transfected the day after with 250 ng of DNA, using GeneJuice transfection reagent (Novagen). For bacterial infection and LLO treatment, HeLa and Jeg3 cells were seeded at 5×105 cells per 10 cm2 well the day before treatment. Cells were then serum-starved for 2 hours before infection or addition of LLO.
For infections, exponential-phase bacteria grown in BHI at 37°C were washed twice in PBS and added to cells at a multiplicity of infection (MOI) of 50. After 1 hour of infection, cells were washed and incubated with 10 μg/ml gentamicin (Euromedex) to kill extracellular bacteria. For quantification of intracellular bacteria, infected cells were lysed 7 hours after infection with PBS-Triton X-100 0.2% (Sigma) and the number of viable bacteria released from the cells was assessed by plating on BHI agar plates. Each experiment was done in duplicate.
For LLO, PFO and PLY treatments, purified proteins were added directly in the culture medium as indicated in the text. PFO and PLY were used at the same haemolytic titer as LLO. For inhibition of LLO binding to cellular membranes, 0.1 mg/mL LLO were pre-incubated for 45 minutes at 4°C either with 0.5 mg/mL A4-8 antibody19 or with 0.5 mg/mL D21-1-4 control antibody19 before addition to HeLa cells.
For analysis of Gam1 expressing cells, HeLa cells were transfected with pSG9-Gam1wt or a control plasmid and treated since the beginning of the transfection with 10 μM MG132 (Calbiochem) or DMSO. 24 hours after transfection, cells were lysed in Laemmli buffer and analyzed by immunoblotting experiments. In parallel, HeLa cells pretreated in the same conditions with MG132 or DMSO were incubated with 3 nM LLO for 20 min before analysis by immunoblotting experiment.
For analysis of SUMO1-conjugated protein degradation, HeLa cells were pretreated for 5 hours with DMSO or with 20 μM MG132+20 μM Lactacystin (Sigma-Aldrich) and then incubated for 20 min with 3 nM LLO.
Inhibition of aspartyl-proteases was done by incubating cells for 1 hour with 200 μM Pepstatin-A Methyl Ester (Sigma-Aldrich). Cells were then washed twice before incubation with LLO.
For in vivo infection, 106 bacteria were injected intravenously in C57Bl/6J mice. Livers of mice were then collected, homogenized and centrifuged 10 min at 13000 rpm. 1 volume of liver supernatant was then added to 5 volumes of Laemmli buffer (0.125 M Tris pH 6.8, 4% SDS, 20% glycerol, 100 mM DTT), sonicated and loaded on SDS-PAGE for immunoblotting experiments.
Immunoblotting and LLO purification
Cells infected or incubated with purified LLO were lysed directly in Laemmli buffer, sonicated and loaded on SDS-polyacrylamide gels. Primary antibodies used for immunoblot analysis were: mouse anti-SAE1 (ab56957, Abcam), mouse anti-Uba2/SAE2 (611602, BD Biosciences), mouse anti-Ubc9 (610748, BD Biosciences), rabbit anti-SUMO132, rabbit anti-SUMO2 (51-9100, Zymed laboratories), mouse anti-SMAD4 (sc-7966, Santa Cruz Biotechnology), rabbit anti-RanGAP1 (R0155, Sigma), rabbit anti-phospho-Ser10-H3 (05-817, Upstate), mouse anti-actin (A5441, Sigma-Aldrich) and mouse anti-GST (05-782, Millipore). A rabbit polyclonal antibody anti-LLO (R176) was obtained by immunizing rabbits with purified recombinant LLO, followed by affinity purification of the immune serum. Anti-mouse and anti-rabbit HRP-conjugated secondary antibodies are from AbCys. All displayed imunoblots are representative of at least 3 independent experiments. Quantification of immunoblots was performed using a G:Box Gel Documentation system (Syngene). Protein levels were normalized using actin levels as an internal control. P values were calculated using a two-tailed two-sample equal variance Student’s t-test. A P value of less than 0.01 was determined to be statistically significant.
LLO mutant proteins were purified as described30. Purified PFO and PLY were kindly provided by Timothy Mitchell (University of Glasgow, Glasgow, U.K.).
In vitro SUMOylation assays
In vitro SUMOylation assays were conducted using BioMol SUMOylation kit. 250 nM GST-RanGAP1 SUMO target was incubated with 70 nM SAE1/SAE2, 350 nM Ubc9 and 700 nM SUMO1 proteins in reaction buffer. MgATP was added to the reaction mix together with various amount of purified LLO (ranging from 10 to 1000 nM). After 1 hour at 30°C, reactions were stopped by adding 1 volume of Laemmli buffer. SUMOylation products were analysed by immunoblotting experiments using an anti-GST antibody.
Luciferase assays
293FT cells were transfected with a DNA mix containing the SMAD reporter plasmid and a plasmid encoding Renilla luciferase (ratio 40:1, SABiosciences Corporation). 24 hours after transfection, cells were infected with L. monocytogenes (MOI of 50). 6 hours after infection, cells were incubated with recombinant human TGFβ1 (Peprotech, 50 ng/ml) for another 18h. Cells were then harvested and luciferase activities were quantitated on a Tristar LB941 luminometer (Berthold technologies) using Dual Luciferase Assay (Promega). Firefly luciferase activities were normalized using Renilla luciferase activities as an internal control.
Supplementary Material
ACKNOWLEDGMENTS
We thank Véronique Villiers for technical assistance. Work in P.C. laboratory received financial support from Institut Pasteur, Inserm, INRA, and ERC (Advanced Grant 233348). The authors further acknowledge support by research grants from the Fund for Scientific Research – Flanders (Belgium) (project number G.0042.07), the Concerted Research Actions (project BOF07/GOA/012) from the Ghent University and the Inter University Attraction Poles (IUAP06). D.R. is supported by a fellowship from the Association pour la Recherche sur le Cancer, F.I. is a Research Assistant of the Research Foundation – Flanders (FWO-Vlaanderen), P.C. is an international research scholar of the Howard Hughes Medical Institute.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature/com/nature.
SUPPLEMENTARY INFORMATION is linked to the online version of the paper at www.nature.com/nature.
AUTHOR INFORMATION
Reprints and permissions information is available at www.nature.com/reprints.
REFERENCES
- 1.Hamon MA, Cossart P. Histone modifications and chromatin remodeling during bacterial infections. Cell Host Microbe. 2008;4:100–109. doi: 10.1016/j.chom.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 2.Rytkonen A, Holden DW. Bacterial interference of ubiquitination and deubiquitination. Cell Host Microbe. 2007;1:13–22. doi: 10.1016/j.chom.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol. Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 4.Cossart P, Toledo-Arana A. Listeria monocytogenes, a unique model in infection biology: an overview. Microbes Infect. 2008;10:1041–1050. doi: 10.1016/j.micinf.2008.07.043. [DOI] [PubMed] [Google Scholar]
- 5.Veiga E, Cossart P. Listeria hijacks the clathrin-dependent endocytic machinery to invade mammalian cells. Nat. Cell Biol. 2005;7:894–900. doi: 10.1038/ncb1292. [DOI] [PubMed] [Google Scholar]
- 6.Bonazzi M, Veiga E, Cerda JP, Cossart P. Successive post-translational modifications of E-cadherin are required for InlA-mediated internalisation of Listeria monocytogenes. Cell Microbiol. 2008;10:2208–22. doi: 10.1111/j.1462-5822.2008.01200.x. [DOI] [PubMed] [Google Scholar]
- 7.Hamon MA, et al. Histone modifications induced by a family of bacterial toxins. Proc. Natl Acad. Sci USA. 2007;104:13467–13472. doi: 10.1073/pnas.0702729104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vertegaal AC, et al. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol. Cell Proteomics. 2006;5:2298–2310. doi: 10.1074/mcp.M600212-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nacerddine K, et al. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev. Cell. 2005;9:769–779. doi: 10.1016/j.devcel.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 11.Hay RT. SUMO-specific proteases: a twist in the tail. Trends Cell Biol. 2007;17:370–376. doi: 10.1016/j.tcb.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 12.Zhao J. Sumoylation regulates diverse biological processes. Cell Mol. Life Sci. 2007;64:3017–3033. doi: 10.1007/s00018-007-7137-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boggio R, Chiocca S. Viruses and sumoylation: recent highlights. Curr. Opin. Microbiol. 2006;9:430–436. doi: 10.1016/j.mib.2006.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gevaert K, et al. Stable isotopic labeling in proteomics. Proteomics. 2008;8:4873–4885. doi: 10.1002/pmic.200800421. [DOI] [PubMed] [Google Scholar]
- 15.Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 16.Golebiowski F, et al. System-wide changes to SUMO modifications in response to heat shock. Sci Signal. 2009;2:ra24. doi: 10.1126/scisignal.2000282. [DOI] [PubMed] [Google Scholar]
- 17.Schnupf P, Portnoy DA. Listeriolysin O: a phagosome-specific lysin. Microbes Infect. 2007;9:1176–1187. doi: 10.1016/j.micinf.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 18.Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nato F, et al. Production and characterization of neutralizing and nonneutralizing monoclonal antibodies against listeriolysin O. Infect. Immun. 1991;59:4641–4646. doi: 10.1128/iai.59.12.4641-4646.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michel E, Reich KA, Favier R, Berche P, Cossart P. Attenuated mutants of the intracellular bacterium Listeria monocytogenes obtained by single amino acid substitutions in listeriolysin O. Mol. Microbiol. 1990;4:2167–2178. doi: 10.1111/j.1365-2958.1990.tb00578.x. [DOI] [PubMed] [Google Scholar]
- 21.Hotze EM, et al. Monomer-monomer interactions drive the prepore to pore conversion of a beta-barrel-forming cholesterol-dependent cytolysin. J. Biol. Chem. 2002;277:11597–11605. doi: 10.1074/jbc.M111039200. [DOI] [PubMed] [Google Scholar]
- 22.Boggio R, Colombo R, Hay RT, Draetta GF, Chiocca S. A mechanism for inhibiting the SUMO pathway. Mol. Cell. 2004;16:549–561. doi: 10.1016/j.molcel.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 23.Boggio R, Passafaro A, Chiocca S. Targeting SUMO E1 to ubiquitin ligases: a viral strategy to counteract sumoylation. J. Biol. Chem. 2007;282:15376–15382. doi: 10.1074/jbc.M700889200. [DOI] [PubMed] [Google Scholar]
- 24.Dramsi S, Cossart P. Listeriolysin O-mediated calcium influx potentiates entry of Listeria monocytogenes into the human Hep-2 epithelial cell line. Infect. Immun. 2003;71:3614–3618. doi: 10.1128/IAI.71.6.3614-3618.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang P, Rosenshine I, Cossart P, Finlay BB. Listeriolysin O activates mitogen-activated protein kinase in eucaryotic cells. Infect. Immun. 1996;64:2359–2361. doi: 10.1128/iai.64.6.2359-2361.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin X, Liang M, Liang YY, Brunicardi FC, Feng XH. SUMO-1/Ubc9 promotes nuclear accumulation and metabolic stability of tumor suppressor Smad4. J. Biol. Chem. 2003;278:31043–31048. doi: 10.1074/jbc.C300112200. [DOI] [PubMed] [Google Scholar]
- 27.Kang JS, Saunier EF, Akhurst RJ, Derynck R. The type I TGF-beta receptor is covalently modified and regulated by sumoylation. Nat. Cell Biol. 2008;10:654–664. doi: 10.1038/ncb1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fitzpatrick DR, Bielefeldt-Ohmann H. Transforming growth factor beta in infectious disease: always there for the host and the pathogen. Trends Microbiol. 1999;7:232–236. doi: 10.1016/s0966-842x(99)01498-5. [DOI] [PubMed] [Google Scholar]
- 29.Nakane A, et al. Transforming growth factor beta is protective in host resistance against Listeria monocytogenes infection in mice. Infect. Immun. 1996;64:3901–3904. doi: 10.1128/iai.64.9.3901-3904.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glomski IJ, Gedde MM, Tsang AW, Swanson JA, Portnoy DA. The Listeria monocytogenes hemolysin has an acidic pH optimum to compartmentalize activity and prevent damage to infected host cells. J. Cell Biol. 2002;156:1029–1038. doi: 10.1083/jcb.200201081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dramsi S, et al. Entry of Listeria monocytogenes into hepatocytes requires expression of inIB, a surface protein of the internalin multigene family. Mol. Microbiol. 1995;16:251–261. doi: 10.1111/j.1365-2958.1995.tb02297.x. [DOI] [PubMed] [Google Scholar]
- 32.Bischof O, et al. The E3 SUMO ligase PIASy is a regulator of cellular senescence and apoptosis. Mol. Cell. 2006;22:783–794. doi: 10.1016/j.molcel.2006.05.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.