Abstract
The application of high-throughput, massively parallel sequencing technologies to hematologic malignancies over the past several years has provided novel insights into disease initiation, progression, and response to therapy. Here, we describe how these new DNA sequencing technologies have been applied to hematolymphoid malignancies. With further improvements in the sequencing and analysis methods as well as integration of the resulting data with clinical information, we expect these technologies will facilitate more precise and tailored treatment for patients with hematologic neoplasms.
Keywords: next-generation sequencing, high-throughput sequencing, massively parallel sequencing, sequencing technologies, genome sequencing, exome sequencing, hematologic
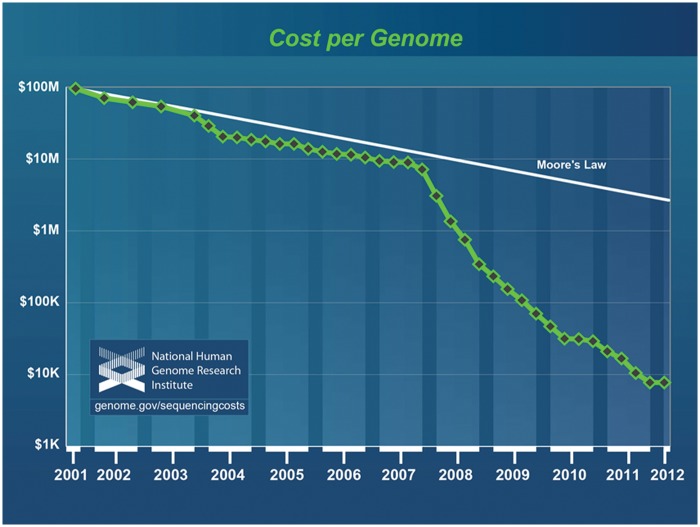
The introduction of high-throughput, massively parallel DNA sequencing technologies is rapidly providing new insights into hematology, oncology, clinical genetics, and many other disease areas. These sequencing data have the potential to facilitate more precision in our practice of medicine. The rise of these technologies has been fueled by dramatic reductions in the sequencing cost (Figure 1); US$3000 genomes have already become a reality, and the much-discussed US$1000 genome will likely be achieved soon. Sequencing is particularly well suited to the analysis of cancer genomes and, not surprisingly, a number of hematologic malignancies have been sequenced and analyzed in the last few years.
Figure 1.
Sequencing cost per genome according to the National Human Genome Research Institute (NHGRI). This graph illustrates the production costs of sequencing a human genome since 2001. The marked reduction in cost first observed in early 2008 corresponds to the time when genome centers adopted next-generation sequencing technologies. Sequencing costs are compared with hypothetical data reflecting Moore’s law, a long-term trend in the computer industry whereby the number of transistors on a computer chip (and hence computing power) doubles approximately every 2 years. Technologies that keep pace with Moore’s law are considered to be improving rapidly, making it a common reference for comparison. The figure is used by courtesy of the National Human Genome Research Institute (Wetterstrand KA, DNA sequencing costs: data from the NHGRI Large-Scale Genome Sequencing Program; available at: http://www.genome.gov/sequencingcosts. Accessed 31 May 2012).
The first complete cancer genome, obtained from a patient with acute myeloid leukemia (AML), was published 4 years ago [Ley et al. 2008]. In the past year we have seen a dramatic increase in the number of hematolymphoid genomes and exomes published. These new data sets are now yielding many unexpected and novel insights into the genetics underlying myeloid and lymphoid neoplasms. A common approach of such studies is to identify, catalog, and interpret somatic mutations with the idea that this knowledge will be useful for medical interpretation. Despite the remarkable reduction of sequencing costs, however, the reliable clinical interpretation of these genomic variants remains a major challenge to the widespread adoption of high-throughput sequencing in hematology and other areas of medicine. To determine the clinical utility of these genetic variants, we ultimately need to correlate them with specific clinical information from a very large number of patients with neoplastic hematologic disorders. We believe the dividends will be plentiful, leading to improved understanding of the relevance of somatic mutations with respect to diagnosis, prognosis, and management, ultimately enabling more precise and tailored cancer treatment.
For decades, Sanger sequencing, based on a chain-terminator method, was the primary method for determining the sequence of DNA [Sanger et al. 1977]. Post-Sanger technologies, which are collectively referred to as next-generation sequencing (NGS) technologies, provide a significant increase in sequencing throughput largely via parallelization, automation and computerization of the sequencing methods [reviewed by Metzker, 2010; Mardis, 2011]. These technological leaps have made it possible to obtain the sequence of an entire human genome in the timeframe of days to weeks using only a single sequencing instrument.
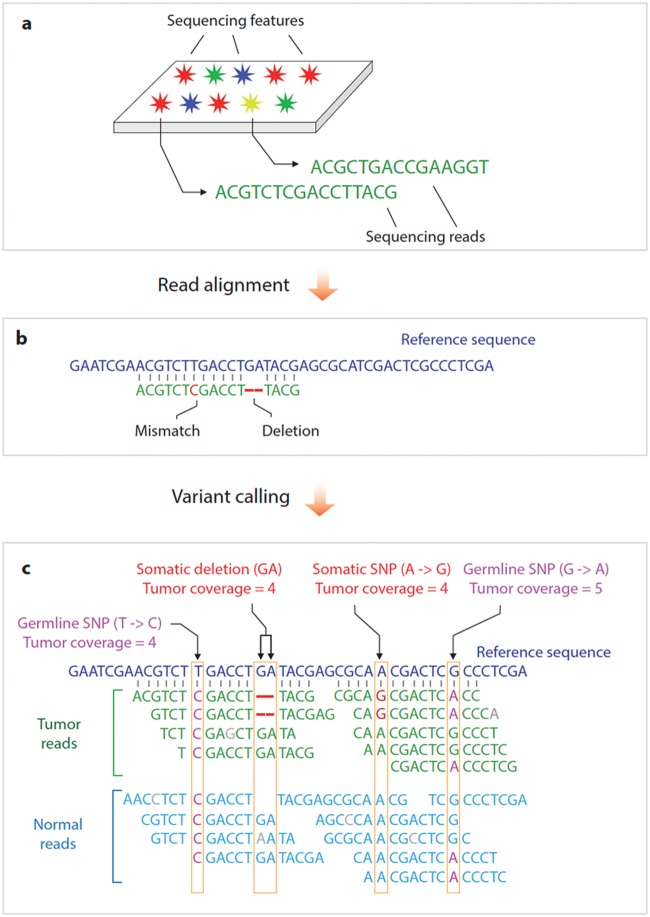
Although multiple sequencing chemistries and platforms are currently available (and many more are in development), they generally share a common work flow (Figure 2). In the process of DNA sequencing, a distinguishable signal (such as fluorescence or change in electrical current) is produced by a chemical sequencing reaction (such as DNA polymerization). This signal distinguishes the four nucleotides, progressively generating sequencing reads. The sequencing is massively parallel, concurrently producing up to billions of individual reads approximately 30–600 bases in length.
Figure 2.
Next-generation sequencing work flow. The work flow can generally be divided into at least three steps. (a) Up to billions of sequencing reads are generated in parallel using one of multiple different sequencing chemistries. (b) These sequence reads are then aligned to a reference genome in order to establish the genomic location of every read. (c) Variant calling algorithms are used to evaluate whether the number of reads and associated quality metrics provide support for the presence of a nucleotide change relative to the reference sequence at a specified confidence level. This illustration shows both a single-nucleotide variant (single-nucleotide polymorphism [SNP]) and a deletion. Parallel analysis of a matched normal specimen allows these variants to be interpreted as either germline, in which case variant reads are observed in tumor and normal reads, or somatic, in which case the variant reads are observed only in tumor reads. Other variant-calling algorithms can be used to detect copy-number changes or structural variants.
The analysis of sequencing data begins with alignment of the sequencing reads (also known as ‘mapping’) to a reference genome in order to establish the genomic location of every read. The specific algorithm that is then employed depends on the overall goal of the analysis. Typical analysis of a cancer genome involves the detection of somatic and sometimes germline variants (a process known as ‘variant calling’). Such variants may include single-nucleotide changes, small insertions and deletions, large-scale copy-number variants (CNVs), and structural variants (SVs) such as translocations or inversions. Analysis is often complicated by false-positive and false-negative calls resulting from systematic biases and random errors in the sequencing data as well as algorithmic artifacts. The detected variants are then evaluated for potential functional and clinical impact in a process known as ‘variant annotation’ and ‘variant interpretation’. Additional confirmation of significant variants by an independent method is often required to rule out any potential artifacts.
We will now examine how NGS technologies have been applied to neoplastic hematologic disorders and the special considerations associated with the analysis of such cases. Whole-genome sequencing (WGS) is an increasingly common application of NGS that provides a comprehensive view of the neoplastic genome. The goal of such analysis is typically to identify and interpret somatic variants by comparing the sequence of the neoplastic population with the matching normal (or germline) counterpart, such as skin or an uninvolved blood cell lineage. The comprehensive nature of such data sets is attractive; however, significant computational difficulties are associated with the underlying analyses, primarily due to the very large amount of data to be analyzed. The compressed reads from a standard-coverage genome use approximately 250 GB of hard-drive space. The Medicine Now exhibition of the Wellcome Collection (London, UK) provides another illustration of the size of the human genome. This printed version of a human genome occupies more than 100 volumes; each volume has 1000 pages with a standard size font. Cancer genomes are generally sequenced to a minimum of 30-fold coverage, indicating that, on average, each base in the genome is represented in at least 30 sequence reads. Therefore, the read data from a single cancer genome fill 3000 of the above volumes, which will form a stack higher than a 50-story building. An additional challenge of working with genome-scale data is that certain relatively large portions of the genome are still difficult to examine accurately because of the ambiguity of the underlying sequences originating from centromeric, telomeric, and other highly repetitive regions of the genome.
A more specialized approach is to focus only on specific and the most informative parts of the genome using an approach known as ‘targeted sequencing’. Such analysis will typically screen for somatic mutation hotspots of cancer genes or detect recurrent gene fusions using PCR or hybridization-based DNA capture methods [Sulonen et al. 2011]. The number of specific gene targets can vary from a single selected genic region up to the entire set of protein-coding regions (in a technique known as exome sequencing). The benefit of targeted approaches is that one can achieve very significant sequencing coverage and consequently lower the detection limit for mutations within the targeted regions. Exome sequencing focuses on the most functionally relevant parts of the genome, the 1–2% that includes protein-coding regions, and generally achieves substantially higher sequence coverage compared with WGS. This higher coverage allows analysis of specimens with lower purity and compromised quality, which is a frequent issue with cancer specimens. A major drawback of targeted resequencing is that certain portions of the exome are difficult to capture, sequence, and analyze [Parla et al. 2011], especially those with particularly high or low G/C content [Clark et al. 2011].
High-throughput sequencing is well suited to screening the repertoire of immunoglobulin or T-cell receptor rearrangements, thus detecting clonal lymphocyte populations in neoplastic lymphoid disorders [reviewed by Benichou et al. 2012]. Owing to the large number of reads from a single sequencing run, this approach provides highly comprehensive evaluation of immunoglobulin or T-cell receptor rearrangements in the specimen, detecting cellular clones even at the lowest end of the frequency spectrum. Tracking of tumor clones in lymphoid tumors such as chronic lymphocytic leukemia (CLL) is one area where this technique could provide immediate value, with the ability to definitively detect and measure minimal residual disease, thereby providing a potential metric for management and clinical decision making [Boyd et al. 2009].
For the remainder of this perspective, we will illustrate how NGS has been utilized for the analysis of hematologic neoplasms and discuss some key seminal papers pioneering such applications. In 2008, Timothy Ley and coworkers described the sequencing of a cytogenetically normal AML and paired normal genome [Ley et al. 2008], the first publication describing the sequencing of a complete cancer genome. It represented a monumental effort to sequence and analyze the tumor genome, requiring 132 instrument runs for the cancer–normal pair. This study successfully demonstrated how WGS could be used to identify acquired (or somatic) mutations and interpret them in the context of disease biology. The authors identified 10 genes with nonsynonymous somatic mutations in the coding regions. Of these mutations, two were previously known to be recurrent mutations and relevant to AML biology: the FLT3 internal tandem duplication mutation and the NPM1 exon 12 insertion mutation. The other eight genes were not previously implicated in AML pathogenesis, and recurrent somatic mutations were not identified upon screening of 187 additional AML cases. The clinical significance of these mutations is unclear at present. One possible explanation is that such somatic mutations represent very rare events that contribute to AML pathogenesis by conferring growth or survival advantages (i.e. ‘driver’ mutations). An alternative explanation is that some or all of these variants represent neutral acquired mutations that have been carried along during the disease course (commonly referred to as ‘passenger’ mutations). To further comprehensively characterize the entire spectrum of AML-associated variants, one needs to both sequence many more additional AML cases and carry out further functional studies on promising variants to fully understand their clinical and biological significance. This example illustrates the general difficulty of distinguishing driver from passenger mutations that are identified by large-scale NGS.
There are two other important points from this seminal paper. First, to identify somatic mutations one needs to also sequence a paired nonneoplastic specimen from the same patient to accurately identify somatic variants. DNA from a skin biopsy provides one source of the germline DNA state for that patient, as demonstrated by Ley and colleagues, who found that 98% of the variants identified in the AML specimen represented germline alleles [Ley et al. 2008]. In addition, sequencing of a matched normal specimen allows identification of systematic sequencing and mapping errors. Consequently, we expect that sequencing of a matched normal specimen will be beneficial for the foreseeable future. Second, additional relevant variants emerge upon further improvement of sequencing technologies and bioinformatics tools. Just two years later, the same group reported the discovery of a novel recurrent mutation in the DNA methyltransferase gene DNMT3A upon re-examination of this same case with improved sequence coverage, sequencing techniques, and data analysis methods [Ley et al. 2010]. DNMT3A mutations were present in 20% of AML cases and associated with poor overall survival independently of age and FLT3 or NPM1 mutational status. The clinical utility of DNMT3A mutations is further supported by a recent study by Patel and coworkers, which indicates that these mutations are predictive of improved survival in response to high-dose induction chemotherapy in patients under 60 years of age [Patel et al. 2012]. This is an excellent example of using clinical outcome data to inform results from the WGS, ultimately improving patient care. Recent studies have also utilized NGS to describe patterns of clonal architecture and tumor evolution in AML, which also has the potential to guide therapeutic options [Ding et al. 2012; Walter et al. 2012].
The next set of papers describes exome sequencing of a large number of CLL cases to identify genes important for CLL pathogenesis. Quesada and coworkers sequenced exomes of matched CLL and normal cells from 105 individuals [Quesada et al. 2011]. Within this cohort, 78 genes contained recurrent somatic mutations resulting in protein-coding changes. Mutations in any specific gene occurred only in a small percentage of cases, however: approximately 10% for NOTCH1 and the splicing factor SF3B1 and <5% for the remaining genes.
Wang and coworkers independently performed exome sequencing of CLL–normal pairs from 88 individuals with CLL and WGS of an additional 3 CLL cases [Wang et al. 2011]. They observed an average of 20 nonsynonymous somatic mutations per patient, with a range of 2–76. This is similar to what has been reported in exome sequencing studies of other hematolymphoid malignancies, but generally lower than what is observed in solid tumors [Greenman et al. 2007; Ley et al. 2008; Mardis et al. 2009; Chapman et al. 2011]. Importantly, Wang and coworkers identified nine genes that were mutated at a significantly higher rate than background, suggesting a causative relationship. Four of these genes, TP53, ATM, MYD88, and NOTCH1, have been previously implicated in CLL biology. Independently of Quesada and coworkers, their analysis also identified recurrent mutations in the splicing factor gene SF3B1. The other four novel genes, FBXW7, DDX3X, MAPK1, and ZMYM3, function in a similar set of pathways to the above genes. These nine genes function within five pathways: DNA damage repair and cell-cycle control, Notch signaling, inflammatory pathways, Wnt signaling, and RNA splicing and processing. Furthermore, numerous other genes within these pathways were mutated in these CLL exomes, even though the mutation rate of these genes did not reach statistical significance because of their lower mutation rate. These observations suggest that although a large number of different genes may be mutated in an individual CLL case, a common set of pathways is typically impaired or activated by rare variants. Sequencing an even larger cohort of cases may therefore be necessary to uncover a complete set of causative CLL variants and identify pathways relevant to CLL biology.
The identification of recurrent somatic mutations in the splicing factor SF3B1 by both groups was a novel and unexpected finding, implicating RNA splicing and processing in the pathogenesis of CLL. Another exciting observation is that the involvement of RNA splicing, in particular SF3B1, is not exclusive to CLL. Two other groups, also using exome sequencing, observed alteration of genes involved in RNA splicing and processing in myelodysplastic syndromes (MDS). Papaemmanuil and coworkers reported that SF3B1 somatic mutations are found in approximately 20% of patients with MDS and approximately 65% of MDS cases with ring sideroblasts [Papaemmanuil et al. 2011]. Mutations in SF3B1 were also present at a lower level (<5%) in other myeloid malignancies and a wide variety of other tumor types. Independently, Yoshida and coworkers observed variants in SF3B1 as well as other components of the splicing machinery in the overwhelming majority of MDS cases with ring sideroblasts (approximately 85%) and also in MDS without ring sideroblasts (approximately 45%) or chronic myelomonocytic leukemia (55%) [Yoshida et al. 2011]. Such concordance among unrelated studies both reinforces the relevance of SF3B1 mutations and illustrates the utility of sequencing in the identification of novel important genes and pathways relevant to cancer.
An important aspect of genome, exome, and to a lesser extent more targeted sequencing that must be considered prior to performing these studies is the risk of discovering clinically relevant germline mutations unrelated to the hematologic malignancy being studied. Currently, there are no national or international guidelines for disclosing the results of such genomic studies to research participants. However, a recent survey of institutional review board professionals indicated that if research results are to be returned to the research participant, he/she should be given the option to receive or not receive results as part of an informed consent process, and that the return of validated, medically actionable results was generally encouraged [Dressler et al. 2012]. If these research results are not be returned to the patient, this should be clearly indicated in the informed consent process and document. We suggest that, optimally, transparent informed consent should accompany both research and clinical genomic testing, but the extent of such pretest counseling, who should perform the counseling and consent, and other related issues are still being actively examined.
Since the publication of the first cancer genome just 4 years ago, many groups have utilized NGS to further elucidate the biology of hematologic malignancies. These efforts have been remarkably successful and uncovered novel and important disease genes with clear clinical relevance. We expect this trend to accelerate in the coming years, providing more comprehensive characterization of causative variants and their biological role. Likewise, we expect further integration of genomic sequencing studies with complementary data, including RNA sequencing and epigenetic analysis. Sequencing technologies and analysis methods have continued to improve rapidly, and this has enabled studies that were unimaginable until today. In the coming years, we anticipate major efforts to establish the clinical relevance of the variants that will be discovered by more comprehensive sequencing efforts. Accelerating the inclusion of genomic data into prospective clinical trials and combining it with current classification and prognostic scoring systems should allow further refinement of clinical care. Over time, these efforts should ultimately facilitate precision medicine, whereby therapy is guided by the genetic makeup of the patient and their malignancy.
Acknowledgments
We would like to thank Dr Andrew Fire for critical reading of this manuscript.
Footnotes
Funding: This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Conflict of interest statement: Dr Merker is co-inventor for a patent involving the measurement and monitoring of cell clonality using massively parallel sequencing.
Contributor Information
Jason D. Merker, Department of Pathology, Stanford University School of Medicine, Stanford, CA 94304, USA
Anton Valouev, Department of Preventive Medicine, Division of Bioinformatics, University of Southern California Keck School of Medicine, Los Angeles, CA, USA.
Jason Gotlib, Department of Medicine (Hematology), Stanford University School of Medicine, Stanford Cancer Center, Stanford, CA, USA.
References
- Benichou J., Ben-Hamo R., Louzoun Y., Efroni S. (2012) Rep-Seq: uncovering the immunological repertoire through next-generation sequencing. Immunology 135: 183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd S., Marshall E., Merker J., Maniar J., Zhang L., Sahaf B., et al. (2009) Measurement and clinical monitoring of human lymphocyte clonality by massively parallel V-D-J pyrosequencing. Sci Transl Med 1: 12ra23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman M., Lawrence M., Keats J., Cibulskis K., Sougnez C., Schinzel A., et al. (2011) Initial genome sequencing and analysis of multiple myeloma. Nature 471: 467–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M., Chen R., Lam H., Karczewski K., Euskirchen G., Butte A., et al. (2011) Performance comparison of exome DNA sequencing technologies. Nat Biotechnol 29: 908–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Ley T., Larson D., Miller C., Koboldt D., Welch J., et al. (2012) Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481: 506–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressler L., Smolek S., Ponsaran R., Markey J., Starks H., Gerson N., et al. (2012) Irb Perspectives on the return of individual results from genomic research. Genet Med 14: 215–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman C., Stephens P., Smith R., Dalgliesh G., Hunter C., Bignell G., et al. (2007) Patterns of somatic mutation in human cancer genomes. Nature 446: 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley T., Ding L., Walter M., Mclellan M., Lamprecht T., Larson D., et al. (2010) Dnmt3a mutations in acute myeloid leukemia. N Engl J Med 363: 2424–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley T., Mardis E., Ding L., Fulton B., McLellan M., Chen K., et al. (2008) DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature 456: 66–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis E. (2011) A decade’s perspective on DNA sequencing technology. Nature 470: 198–203 [DOI] [PubMed] [Google Scholar]
- Mardis E., Ding L., Dooling D., Larson D., McLellan M., Chen K., et al. (2009) Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med 361: 1058–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzker M. (2010) Sequencing technologies - the next generation. Nat Rev Genet 11: 31–46 [DOI] [PubMed] [Google Scholar]
- Papaemmanuil E., Cazzola M., Boultwood J., Malcovati L., Vyas P., Bowen D., et al. (2011) Somatic Sf3b1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med 365: 1384–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parla J., Iossifov I., Grabill I., Spector M., Kramer M., McCombie W. (2011) A comparative analysis of exome capture. Genome Biol 12: R97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel J., Gonen M., Figueroa M., Fernandez H., Sun Z., Racevskis J., et al. (2012) Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med 366: 1079–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V., Conde L., Villamor N., Ordonez G., Jares P., Bassaganyas L., et al. (2011) Exome sequencing identifies recurrent mutations of the splicing factor Sf3b1 gene in chronic lymphocytic leukemia. Nat Genet 44: 47–52 [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74: 5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulonen A., Ellonen P., Almusa H., Lepisto M., Eldfors S., Hannula S., et al. (2011) Comparison of solution-based exome capture methods for next generation sequencing. Genome Biol 12: R94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M., Shen D., Ding L., Shao J., Koboldt D., Chen K., et al. (2012) Clonal architecture of secondary acute myeloid leukemia. N Engl J Med 366: 1090–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Lawrence M., Wan Y., Stojanov P., Sougnez C., Stevenson K., et al. (2011) Sf3b1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med 365: 2497–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Sanada M., Shiraishi Y., Nowak D., Nagata Y., Yamamoto R., et al. (2011) Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478: 64–69 [DOI] [PubMed] [Google Scholar]