SUMMARY
In neurons, dendritic protein synthesis is required for many forms of long-term synaptic plasticity. The population of mRNAs that are localized to dendrites, however, remains sparsely identified. Here, we use deep sequencing to identify the mRNAs resident in the synaptic neuropil in the hippocampus. Analysis of a neuropil data set yielded a list of 8,379 transcripts of which 2,550 are localized in dendrites and/or axons. Using a fluorescent barcode strategy to label individual mRNAs, we show that their relative abundance in the neuropil varies over 3 orders of magnitude. High-resolution in situ hybridization validated the presence of mRNAs in both cultured neurons and hippocampal slices. Among the many mRNAs identified, we observed a large fraction of known synaptic proteins including signaling molecules, scaffolds and receptors. These results reveal a previously unappreciated enormous potential for the local protein synthesis machinery to supply, maintain and modify the dendritic and synaptic proteome.
INTRODUCTION
In eukaryotic cells, the localization of mRNA is an important mechanism to establish or maintain cell polarity, regulate gene expression, and sequester the activity of proteins. Neurons, with their complex dendritic and axonal structure, represent a special class of polarized cells with 103–104 synapses that can be modified independently. The establishment, maintenance, and regulation of this specificity are mediated by differences in protein composition within synapses. In neurons, mRNAs as well as polyribosomes have been observed throughout the dendritic arbor, often hundreds of microns from the cell body (Steward and Levy, 1982). In the developing hippocampus, between 8% and 16% of dendritic spines possess a polyribosome under control conditions (Ostroff et al., 2002).
Although protein synthesis in neuronal cell bodies is undoubtedly important, emerging data indicate that local protein translation can play an important role in synaptic development and plasticity (Martin and Ephrussi, 2009; Richter and Klann, 2009; Sutton and Schuman, 2006). The synaptic potentiation induced by BDNF requires local translation (Kang and Schuman, 1996) as do other forms of plasticity including long-term facilitation in Aplysia (Martin et al., 1997), long-term depression elicited by metabotropic glutamate receptor activation (Huber et al., 2000), late-phase LTP (Bradshaw et al., 2003), dopamine-induced plasticity (Smith et al., 2005), and homeostatic plasticity induced by a blockade of spontaneous neurotransmitter release (Sutton et al., 2004, 2006, 2007). In most cases above, the specific proteins that are locally synthesized during plasticity have not been identified.
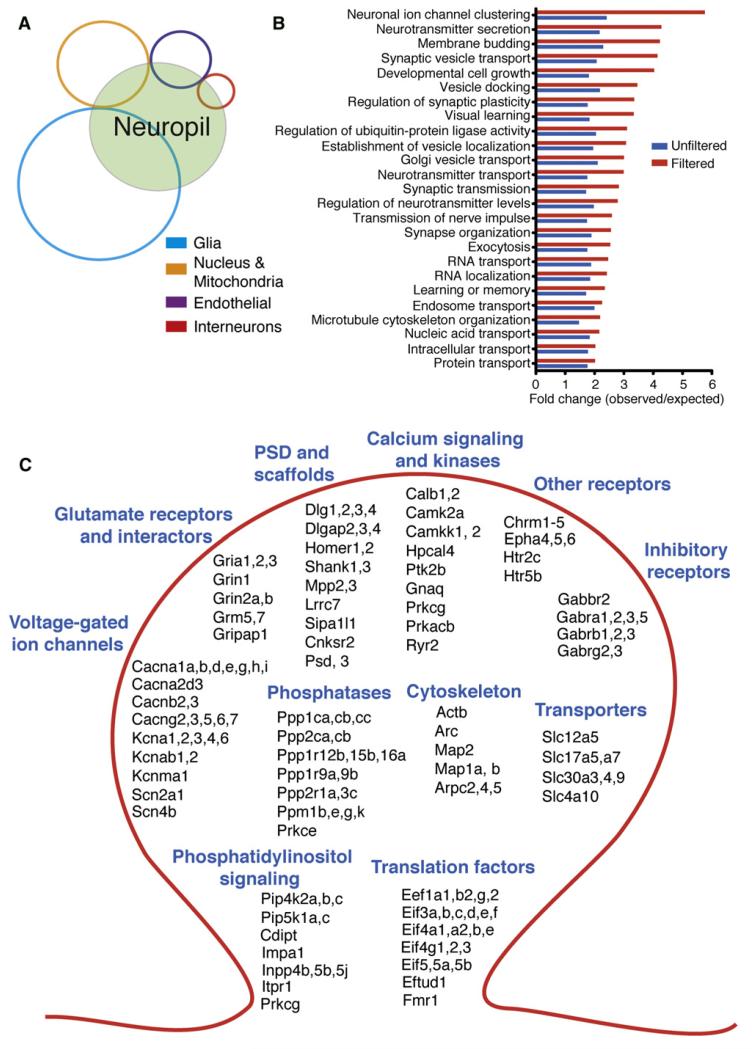
Several individual mRNAs have been visualized in dendrites using in situ hybridization, including the mRNA for the Ca2+-calmodulin-dependent protein kinase alpha subunit, CaMKIIα (Burgin et al., 1990; Mayford et al., 1996), MAP2 (Garner et al., 1988), Shank (Böckers et al., 2004), and β-actin (Tiruchinapalli et al., 2003). The mRNAs localized to growth cones of retinal ganglion cells have recently been elucidated (Zivraj et al., 2010). Recent microarray approaches using tissue enriched for dendrites expanded the local transcriptome to ~285 mRNAs (Poon et al., 2006; Zhong et al., 2006) and the high-throughput in situ hybridization screen performed by the Allen Brain Project identified 68 mRNAs in the synaptic neuropil (Lein et al., 2007). Analysis of the overlap between the various studies, however, yields a surprisingly small number of mRNAs discovered by two or more studies (Figure 1A), suggesting that the identification of the local mRNA population is not near saturation. Here, we used deep RNA sequencing to identify the full complement of mRNAs present in synaptic regions (Figure 2). We focused our attention on the CA1 area of the rat hippocampus because, as indicated above, synapses in this region express several forms of plasticity that require local translation. Following sequencing and bioinformatic analysis with other data sets, we identified 2,550 mRNAs that are associated with the dendrites and/or axons in the hippocampal neuropil. High-resolution imaging allowed us to validate, independently, a subset of these mRNAs and to localize them specifically to the dendrites of hippocampal neurons.
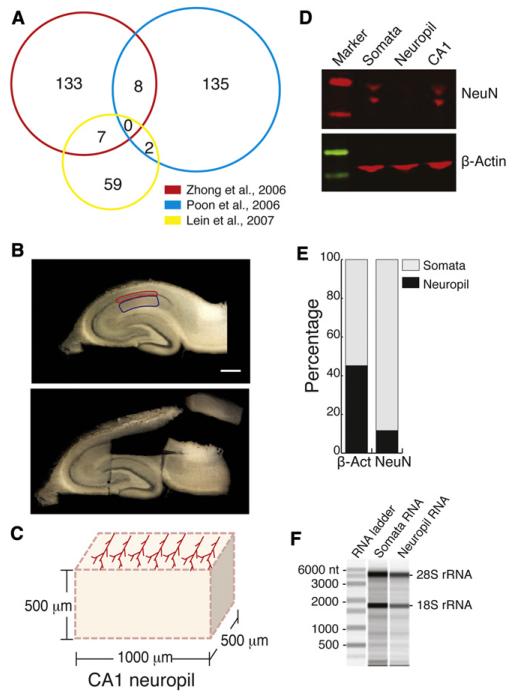
Figure 1. Isolation of Hippocampal RNA for Deep Sequencing.
(A) Venn diagram showing overlap between mRNAs identified in three recently published neuropil transcriptome data sets.
(B) Photograph of a slice before and after microdissection with somatic (red) and neuropil (blue) segments outlined. Scale bar = 1 mm.
(C) Scheme of an individual microdissected slice, showing the orientation of CA1 neuron dendrites (for slice preparation, see Experimental Procedures).
(D) Microdissected regions containing the somatic layer of CA1 are enriched with the neuron-specific transcription factor NeuN, whereas neuropil microdissected slices are de-enriched for NeuN. Protein lysates were prepared from neuropil, somata, and whole CA1 (CA1) tissues and analyzed by western blot using antibodies against NeuN and β-actin.
(E) Graph shows the enrichment of proteins in the different CA1 regions.
(F) Total RNA was isolated from the tissue corresponding to the microdissected neuropil and the microdissected somatic layer. The purity of the sample was analyzed using an Agilent 2100 Bioanalyzer. mRNAs are distributed throughout the lanes.
Figure 2. Flowchart of Experiments and Analysis.
Shown is a description of the experiments, beginning with sample preparation and concluding with the analysis and validation of the neuropil mRNA population.
RESULTS
Next Generation Sequencing of Neuropil RNA Samples Reveals a Large Number of Previously Undetected Neuropil mRNAs
To discover the full local transcriptome, we first microdissected individual synaptic neuropil (stratum radiatum and lacunosum moleculare) segments from area CA1 of the adult rat hippocampus (Figures 1B and 1C). This synaptic neuropil comprises dendrites, axons, glia, and a sparse population of interneurons, but lacks principal neuron cell bodies (Figures 1D and 1E). Microdissection of CA1 synaptic neuropil from 120 individual slices yielded sufficient RNA for a single deep sequencing run (Figure 1F; 454 Technology, Roche). To maximize coverage of the local mRNA population, poly(A) RNA was isolated and then normalized cDNA libraries were prepared (Patanjali et al., 1991) to enhance sensitivity to lower abundance transcripts. Two different neuropil sequencing runs (using starting material from two different dissections) yielded 550,442 and 571,554 reads for a total of 1,121,196 reads with a mean read length of ~400 nucleotides (Figure S1 available online). Reads were annotated to identify the genes represented (see Experimental Procedures; Figure S1). We chose 50% coverage of the coding sequence (Table S1, Column F) as a threshold value for inclusion in our subsequent analysis of the neuropil data sets yielding 8,379 unique mRNAs (Table S1). We compared this data set with the three most recently published neuropil transcriptome data sets obtained from microarrays (Poon et al., 2006; Zhong et al., 2006) and high-throughput in situ hybridization analysis (Lein et al., 2007). Using the above data set of 8,379 unique mRNAs we found substantial overlap between our data and the other three data sets (86%, 86%, and 91%, respectively, for Zhong et al., 2006; Poon et al., 2006; and Lein et al., 2007) (Figure S1; Table S4).
Gene Ontology Analysis of Neuropil Transcriptome and Validation and Quantification of Target Transcripts using nCounter Nanostring
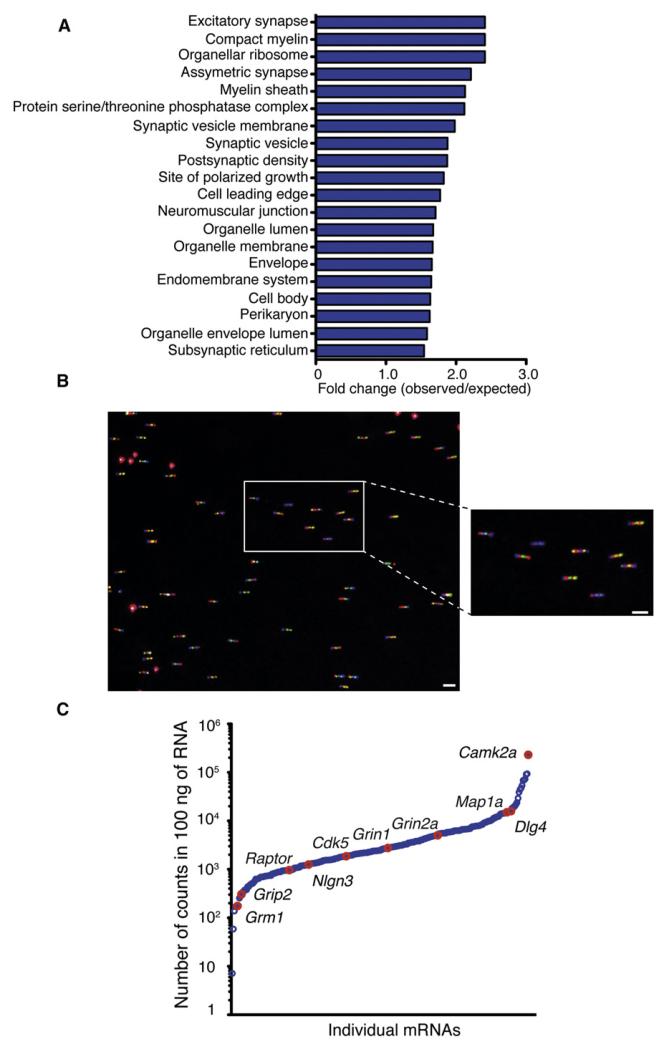
We used Gene Ontology (GO) to identify the gene families and protein functions significantly represented by our neuropil data set. As shown in Figure 3A and Table S2, many transcripts fall into categories associated with aspects of neuronal function including genes associated with dendrites, spines, and axons. To independently validate a subset of the above genes, we used a new technique (Nanostring nCounter; (Geiss et al., 2008) that permits high-resolution visualization of single mRNA molecules and allows one to obtain quantitative estimates of the abundance of a given mRNA species. For each mRNA of interest, two specific nucleotide probes were designed, one that contains a six molecule fluorescent barcode and the other that contains a biotin group to enable binding of a hybridized mRNA to a substrate. Following hybridization with both probes, individual mRNAs were imaged (Figure 3B) and counted based on their identifying barcode. We detected 290 of the 292 target mRNAs in our sample (Figure 3C), as well as several positive controls. None of the negative control probes were detected. To quantify the abundance of our target mRNAs, we spiked our sample with several control mRNAs at known quantities (see Experimental Procedures). This allowed us to obtain concentration estimates for our target mRNAs and to observe their relative abundance (Figure 3C; Table S5). As shown in Figure 3C, Camk2a (CAMKIIα) is the most abundant mRNA detected in the neuropil, consistent with its role as an organizer and regulator of synaptic function, and its detection as a localized mRNA in earlier studies (Miller et al., 2002; Ouyang et al., 1999). Other relatively abundant mRNAs included Shank1, Dlg4 (PSD-95), Ddn (Dendrin), and Map1a, all previously identified in published studies (Böckers et al., 2004; Herb et al., 1997; Muddashetty et al., 2007; Tucker et al., 1989). The power of deep sequencing, however, is its ability to detect transcripts of lesser abundance. Indeed, we identified in the neuropil many previously undetected mRNAs such as synGAP, Snap25, Cyfip2, and Rptor. The abundance of different mRNAs varied over 3 orders of magnitude. We also performed additional validation of 15 synaptic targets by real-time PCR (Table S6).
Figure 3. Gene Ontology Analysis of Neuropil Transcriptome and Validation and Quantification of Target Transcripts Using Nanostring.
(A) Bar graph illustrating some transcript families from Gene Ontology (GO) that are significantly overrepresented in our samples.
(B) Example of Nanostring image showing unique fluorescent bar codes for individual mRNA transcripts. Bright red spots are fiducial markers used to register high magnification images into a montage. Scale bar = 2 μm.
(C) Plot of individual mRNA species and their abundance from Nanostring experiments (n = 3 experiments). A selection of mRNAs representing synaptically relevant transcripts is indicated by red circles. For full data set with numbers, including many more synaptically relevant transcripts see Table S5.
See also Figure S1.
Relative Enrichment of Transcripts in the Neuropil, Glia, and Somata
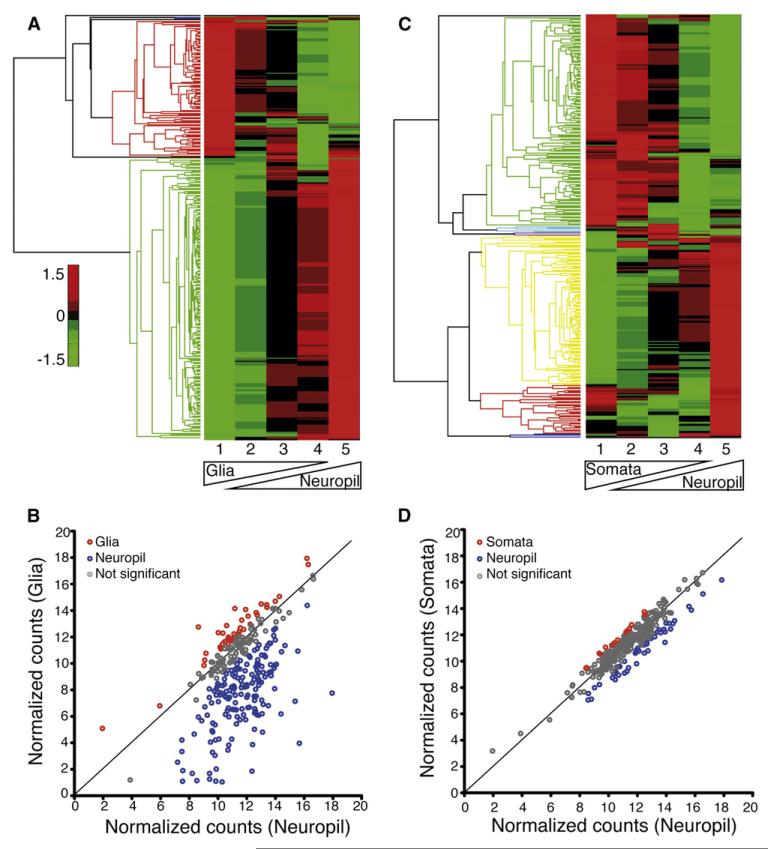
In addition to axons and dendrites, the synaptic neuropil also contains glial cells. We initially determined the contribution of putative glial transcripts to our data set by conducting Nanostring analysis of a preparation of glial cells grown in culture (see Experimental Procedures; Figure S2). In a series of “ramp” experiments, we tested whether the glial cells were a significant source of the identified neuropil transcripts by varying the relative amounts of glial-derived sample from 100% to 0% and, in the opposite manner, varying the relative amount of neuropil-derived sample (Figure 4A). If the glial cells were a significant source of a given transcript, then we would expect this transcript to exhibit high (red) levels in the 100% glial sample, and progressively lower levels as the relative glial contribution was reduced. For a majority (n = 167) of the neuropil transcripts we examined, we observed the opposite pattern: most were relatively de-enriched in the 100% glial sample (green in Figure 4A) and showed progressive enrichment as the glial contribution was reduced, relative to the neuropil sample (red in Figure 4A). For a relatively small group of targets, we observed a significant enrichment in the glial sample, these transcripts include some well-established glial genes such as Gfap (Figure 4B; Table S7).
Figure 4. Relative Enrichment of Transcripts in the Neuropil, Glia, and Somata.
(A) Heat map of a glia versus neuropil ramp experiment. Transcript enrichment is encoded in the heat map from low (green) to high (red). Transcripts that show similar expression patterns are clustered together, as indicated by the colored groups to the left of the heat map.
(B) Summary graph indicating the significant enrichment of transcripts in either the neuropil (blue) or glial samples (red), (n = 3).
(C) Heat map of a somatic layer versus neuropil ramp experiment. Transcript enrichment is encoded in the heat map from low (green) to high (red). Transcripts that show similar expression patterns are clustered together, as indicated by the colored groups to the left of the heat map. In the heat map it is evident that a clusters of ~105 and ~150 transcripts are enriched in the neuropil and somata, respectively.
(D) Graph indicating those transcripts which exhibited a significant enrichment in either the neuropil (blue; n = 123) or somata (red; n = 14) (n = 3 experiments). See also Figure S2.
How does the neuropil transcriptome compare to that found in the somatic compartment? As transcription occurs in the nucleus followed by export of the mRNA to the cytoplasm, all neuronal transcripts, regardless of their ultimate destination, reside in the cell body for some period of time. Thus, it is expected that, assuming perfect detection, all dendritic transcripts should also be discovered in the cell body. We compared our neuropil transcriptome to a somata data set, obtained from the microdissection of sister segments comprising the stratum pyramidale (cell body layer) of hippocampal area CA1. Deep sequencing (same protocol as above) of two different somatic tissue samples resulted in 1,099,501 reads that correspond to 8,044 unique mRNAs (Table S3). We used Nanostring to estimate the relative enrichment of a subset of mRNAs in somata versus neuropil. We varied the relative amount of somatic tissue to neuropil tissue and identified a subset of mRNAs that is indeed enriched in the neuropil (Figures 4C and 4D). A unique cluster of mRNAs is also apparently enriched in the cell body layer (Figures 4C and 4D). We note here that enrichment in somata is influenced by many variables including transcript abundance, decay rates, and transport rates that have not yet been carefully measured or quantified. Furthermore, relative enrichment in somata does not rule out a dendritic function. For example, the most abundant dendritic mRNA, Camk2a (Figure 3C), was not detected as a dendritically enriched transcript in two previous studies (Poon et al., 2006; Zhong et al., 2006).
A Conservative Estimate of mRNAs Present in Synaptic Regions
The neuropil is a composite tissue-comprising dendrites, axons, glial cells, interneurons, and some blood vessels. To refine our list of transcripts to those of dendritic and/or axonal origin we made use of recently published data sets to subtract transcripts enriched in other neuropilar cellular or subcellular compartments (Figure 2; Figure S3). First, we expanded our own list of glial-enriched transcripts with published data on transcripts enriched in astrocytes and oligodendrocytes obtained via cell-type-specific expression of a fluorescent protein (Cahoy et al., 2008; Okaty et al., 2011) and subtracted them from the neuropil transcriptome (Figure 5A; Table S8). We next developed a list of interneuron-enriched transcripts based on published studies (Klausberger and Somogyi, 2008; Okaty et al., 2011; Sugino et al., 2006) as well as in situ hybridization data (http://mouse.brain-map.org/) and subtracted these from our neuropil data set (Figure 5A; Table S8). We also subtracted mRNAs enriched in blood vessels (Daneman et al., 2010) and mitochondrion (http://mitominer.mrc-mbu.cam.ac.uk/release-1.1) as well as transcripts that code for nuclear proteins (Figure 5A). Following subtraction of all potential candidates, we obtained a list of 2,550 transcripts that are of dendritic or axonal origin (Figure S3A; Table S10). These 2,550 mRNAs code for proteins that are involved in most of the cell biological functions known to occur in dendrites and axons (Figure 5B); note that the subtraction of transcripts from other compartments significantly enhanced the enrichment in these functions. The analysis of the individual mRNAs that are nested in the above groups reveals a huge representation of previously undetected synaptic proteins mRNAs (Figure 5C).
Figure 5. A Conservative Estimate of mRNAs Present in Synaptic Regions.
(A) Venn diagram of the neuropil data set and its overlap with transcripts enriched in glia (astrocytes and oligodendroctyes) interneurons, mitochondria and nucleus as well as the endothelium (blood vessels). The remaining transcripts number is 2,550.
(B) Analysis of the filtered neuropil list, showing a significant enrichment of transcripts in categories related to synaptic function and cell biological processes.
(C) Scheme of a postsynaptic compartment highlighting some of the transcript families for synaptically relevant proteins represented in the filtered neuropil list. See also Figures S3 and S7.
High-Resolution Fluorescent In Situ Hybridization Detection of mRNA Transcripts in Hippocampal Dendrites
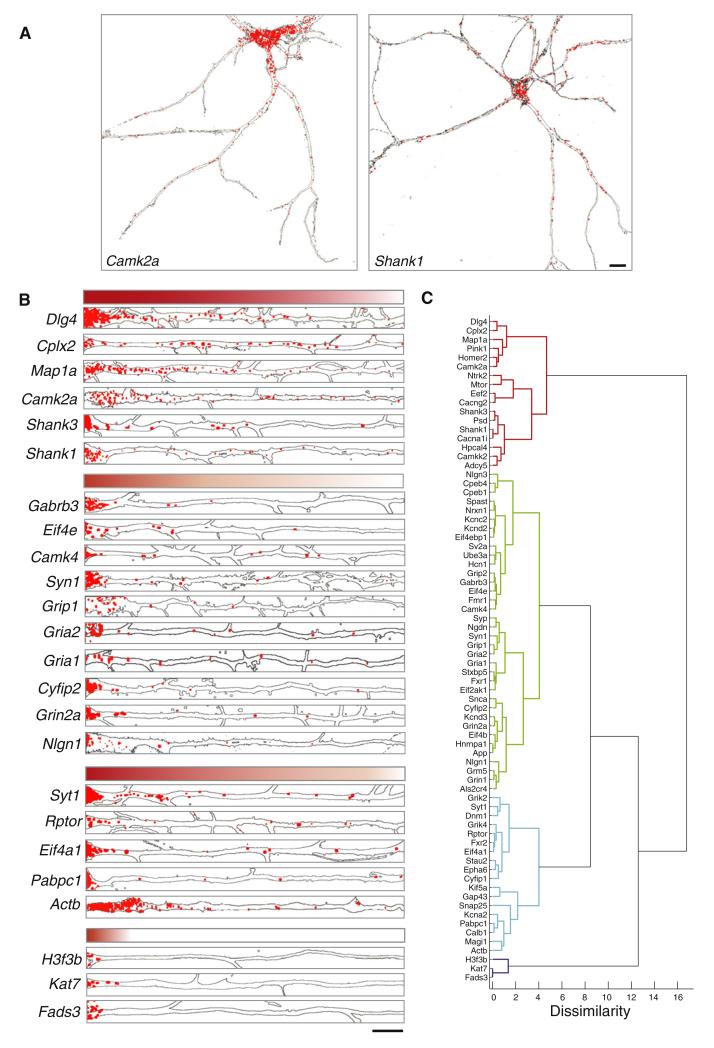
To visualize neuropil transcripts in dendrites we used high-resolution fluorescent in situ hybridization (Taylor et al., 2010). Using 71 probe sets specific for individual synaptic mRNAs we examined the subcellular localization in dissociated cultured hippocampal neurons. As previously observed and predicted by our Nanostring data, Camk2a and Shank1 mRNAs were abundant in the dendrites (Figure 6A). Indeed, all of the mRNAs for which we developed probes were detected in the dendrites (Figure 6B; Figures S4, S5, and S6). Control experiments, either lacking the initial probe or using a sense probe, showed no significant detectable signal (Figures S7A–S7D). Some mRNAs with high copy numbers within the dendrites included Cplx2, Map1a, and Cyfip2 (Figures 6A and 6B). The mRNAs for obligate subunits for ionotropic glutamate transmission (GluR1/a and 2/b, gene names: Gria1 and Gria2, respectively) were detectable at low copy number in the proximal dendrites, but not always present in the distal dendrites (Figure 6B). In contrast, transcripts predicted to reside in the cell body such as H3f3b, Kat7, and Fads3 did not show any appreciable dendritic in situ signal that extended beyond the proximal (approximately the first 25 μm) dendrite (Figure 6B). The abundance of different mRNA types varied both as a function of the initial concentration in the proximal dendrite and the rate of decline in the number of particles along dendritic length (Figure 6B; Figure S6). We performed an unbiased cluster analysis (see Experimental Procedures) to group the dendritic mRNAs that exhibit similar distribution patterns (Figures 6B and 6C); this clustering revealed three large groups that differ in the way in which they distribute their mRNA particles along the proximal-distal dendritic axis. Transcripts that are associated with astrocytes (Cahoy et al., 2008; Doyle et al., 2008), oligodendrocytes (Doyle et al., 2008), interneurons (Doyle et al., 2008; Sugino et al., 2006), and endothelial cells (Daneman et al., 2010) also did not exhibit any in situ signal in neuronal dendrites (Figures S7E–S7I). Analysis of the different mRNA distribution patterns indicates that the dendrite to soma ratio for distinct mRNAs is not a constant value and is not solely dependent on the apparent somatic concentration of an mRNA (see Figure 6B). In addition, we quantified the ratio of the Dlg4 mRNA between the dendrites and the cell body in single neurons and found a dendrite: soma ratio of ~30:70 (Figure S5).
Figure 6. High-Resolution Fluorescent In Situ Hybridization Detection of mRNA Transcripts in the Dendrites of Cultured Hippocampal Neurons.
(A) Fluorescent in situ hybridization (FISH) signal in cultured hippocampal neurons (DIV 21) showing the presence of Camk2a and Shank1 mRNA molecules (red particles) within the cell body and entire dendritic arbor. The neuron was immunostained with an antibody to MAP2 (green) to generate a mask that outlines the dendrites. Scale bar = 20 μm.
(B) FISH signal in individual dendrites from cultured hippocampal neurons linearized for analysis. MAP2 immunostaining was used to generate a mask that outlines the dendritic structure. The red particles show the transcripts for each indicated gene within the dendrites. The transcripts vary in their abundance as well as their distribution along the length of the dendrite. Dendrites are shown in groups that display similar distribution patterns. Scale bar = 20 μm.
(C) The transcripts are represented in groups identified by cluster analysis (see Experimental Procedures) based the distribution of mRNA particles along the dendritic axis. The cophenetic correlation coefficient for the dendrogram is 0.8058 and the generation of each of the four labeled clusters is significant: p < 0.001.
See also Figures S4, S5, S6, and S7.
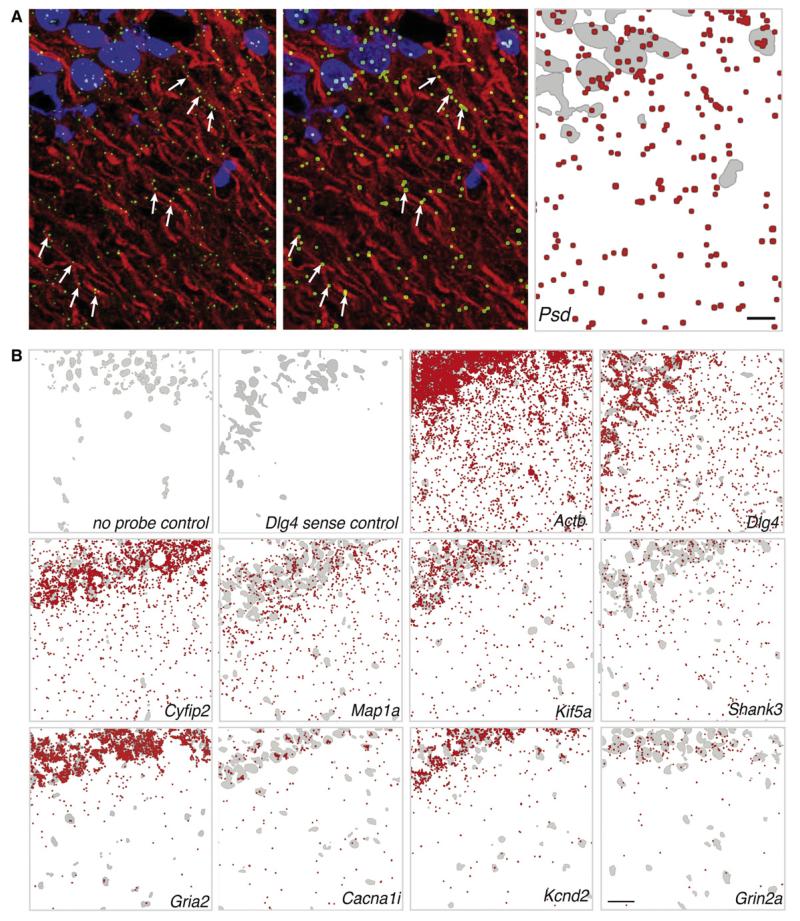
The above experiments validate the presence of mRNAs we identified via deep sequencing in the dendrites of cultured hippocampal neurons. To examine the localization of a subset of mRNAs in a more realistic context, we adapted the high resolution in situ hybridization technique for use in rat hippocampal slices and combined it with immunohistochemical labeling of dendrites using an antibody to MAP2 (Figure 7A). We focused our analysis on area CA1, the region from which we microdissected tissue for deep sequencing and the site of several forms of plasticity that require local translation. We examined the localization of 19 (Dlg4, Map1a, Cacng2, Shank3, Psd, Shank1, Cacna1i, Hpcal4, Nlgn3, Kcnd2, Camk4, Gria2, Cyfip2, Grin2a, Grik2, Kif5a, Kcna2, Actb, and Pclo; 11 are shown in Figure 7) different transcripts and found positive evidence for their presence within the synaptic neuropil (Figures 7A and 7B). In some fortuitous cases, the MAP2-labeled dendrites were sufficiently well-resolved to allow us to visualize labeled mRNAs associated with dendrites (Figure 7A). Taken together, these data indicate that the mRNAs identified by deep sequencing can be observed by high-resolution imaging to reside in the synaptic neuropil of hippocampal area CA1.
Figure 7. High-Resolution Fluorescent In Situ Hybridization Detection of mRNA Transcripts in the Synaptic Neuropil of Area CA1 of the Rat Hippocampus.
(A) Raw image from an in situ hybridization experiment using antisense probes against the Psd (pleckstrin and Sec7 domain protein) mRNA (green) in area CA1 of rat hippocampal slices (left). Dendrites were immunostained using an anti-Map2 antibody (red). The nuclei were stained with DAPI (blue). There are several examples where mRNA puncta are clearly present within the dendrites (arrows). In order to increase visibility when displaying larger areas of area CA1 we processed the images by dilating the mRNA puncta in ImageJ after setting a threshold (middle). For printing purposes, images were converted for display on white background (right). The DAPI channel was converted to a binary image, inverted, and displayed in gray to provide orientation in the CA1 region, the mRNA signal was converted to red, and the two channels were merged using Adobe Photoshop. Scale bar = 4 μm.
(B) Distribution of mRNA puncta in CA1 from in situ hybridization experiments in hippocampal slices displayed as indicated in (A) (right panel). In some control experiments, slices were not exposed to the probe (no probe) but to all subsequent amplification steps; in other controls, a sense probe followed by all subsequent amplification steps was used. Experiments were performed with the indicated probes. Scale bar = 25 μm.
DISCUSSION
A Dramatically Different Landscape for Local Translation
The proteome of an individual synapse is the physical entity that determines the response of a given synapse to an input, and it is clear that, like other proteomes (e.g., Ingolia et al., 2009), the synaptic proteome is subject to ongoing and dynamic modification by regulated protein synthesis and degradation. The prior identification of mRNAs resident in dendrites and axons have yielded a largely heterogeneous mix of a small number (100 or so) transcripts that did not suggest an ongoing role for local protein synthesis in synaptic function, but rather suggested that local synthesis might be used in special cases during some forms of synaptic plasticity. Here, using next generation sequencing of hippocampal neuropil RNA samples we reveal a surprisingly large number of previously undetected neuropil mRNAs, suggesting that mRNA localization may be more of a rule, than the exception. In addition, many of the proteins that populate the synapse may originate from a local source. Based on current reference databases (NCBI, Rattus norvegicus transcriptome version rn4.2 2010), we estimate that the number of unique transcripts associated with the various cells and compartments (axons, dendrites, glia, and interneurons) of the entire hippocampal neuropil is about 8,379 mRNAs. Using published data sets and experimental data to subtract genes that are overrepresented in other cell types (glia, interneurons) or compartments (mitochondria and nucleus), we arrive at a dendritic-axonal data set of ~2,550 mRNAs (Table S10). Considered together, these data sets suggest an enormous potential for protein translation that is independent of the principal cell somata, resident locally within the neuropil.
A Large Heterogeneity in mRNA Species Abundance and Distribution
We used high-resolution imaging techniques to validate, quantify, and localize a subset of the transcripts identified through deep sequencing. Using Nanostring, we detected neuropil mRNAs that vary in their abundance over three orders of magnitude, highlighting the sensitivity of our approaches. Indeed, previous studies failed to identify most of the lesser abundant mRNAs, presumably owing to the lower sensitivity of microarray-based approaches (Figure S1). (The dynamic range to quantify gene expression levels is up to a few hundred fold for microarrays and >8,000-fold for RNA-Seq, Wang et al., 2009). It is possible that some the of low-abundance transcripts we identified are concentrated in subsets of pyramidal neurons, rather than equally distributed across the population, as would be expected if pyramidal cells are molecularly heterogeneous (Doyle et al., 2008; Sugino et al., 2006). Our high-resolution in situ hybridization data indicate that the distribution pattern of transcripts within dendrites is also heterogeneous. We identified three main groups that differ in their spatial allocation of mRNA particles along the proximal to distal dendrite axis. Gradients of localized mRNAs might be used to establish or maintain gradients of protein distribution or to create local computationally relevant subdomains within dendritic branches (Govindarajan et al., 2011).
Our data, combined with previously published data sets (Table S14) validates with in situ hybridization 140 mRNAs (Table S14) within the dendrites of hippocampal slices or dissociated hippocampal neurons that were also identified by our deep sequencing. Taking into account our data set and internal (in situ hybridization) and external (previously published studies) sources for validation, we assign a 95% confidence level of dendritic localization for 90% (2,295/2,550) of our transcripts.
mRNAs for Most Protein Families Are Present in the Local Transcriptome
The transcriptome identified here includes mRNAs that belong to diverse classes of synaptically relevant proteins, including ionotropic and metabotropic neurotransmitter receptors, adhesion molecules, synaptic scaffolding molecules, signaling molecules as well as components and regulators of the protein synthesis and degradation machinery (Figure 5C; Table S11). This expanded list indicates that many of the proteins that populate the synapse could arise from a local, rather than somatic, source.
We also detected many mRNAs for proteins associated with the presynaptic terminal, including mRNAs encoding synapsins, synaptotagmins, synaptophysins, and active-zone molecules (Table S10). Although axonal protein synthesis has been clearly documented during development and regeneration (Andreassi et al., 2010; Lin and Holt, 2008) and a large number of mRNAs have been detected in growth cones (Zivraj et al., 2010), it remains unclear whether mature axons of the CNS are capable of local protein synthesis. Here, we demonstrate mRNAs coding for proteins associated with presynaptic function are present in the mature rat neuropil, suggesting the possibility that healthy adult axons are the sites of protein synthesis.
We also detected the mRNAs for many membrane proteins, including a large number of voltage-gated ion channels: 5 distinct Na+, 15 Ca2+, and 33 K+ channel subunits (Table S10). It is known that many of these channels are expressed in gradients from the soma to the dendrites, resulting in local control of signaling as well as the excitability of the dendrites (Johnston and Narayanan, 2008; Makara et al., 2009). For example, synaptic excitation has been shown to suppress translation of Kv1.1 (Raab-Graham et al., 2006), resulting in enhanced excitability of pyramidal neurons. The presence of multiple K+, Ca2+, and Na+ channel subunits mRNAs in our dendritic/axonal data set suggests that local translation could establish, maintain, and regulate these protein gradients, resulting in local control of the dendritic integrative properties. If membrane protein mRNAs are translated locally then the machinery required for co- and posttranslational processing of these proteins should also be localized. While it is clear that there are some components of ER and Golgi present (Gardiol et al., 1999; Horton and Ehlers, 2003; Horton et al., 2005; Torre and Steward, 1996), it remains a matter of debate as to the nature and location of membrane protein processing. It is thus interesting that we identified mRNAs for components of the secretory pathway as well as many enzymes associated with the N-glycosylation pathway including key enzymes that influence ER export and complex type N-glycan biosynthesis. The glycosylation status of a membrane protein influences its folding, trafficking, as well as membrane residence time and function. The detection of mRNAs for membrane proteins as well as secretory pathway components and enzymes strengthen the view that membrane protein synthesis and processing might occur locally (Gardiol et al., 1999; Torre and Steward, 1996) (Table S11).
Local translation has been implicated in neurodevelopmental, psychiatric or degenerative diseases (Swanger and Bassell, 2011). In the local transcriptome, we discovered many mRNAs relevant for these and other diseases including, but not limited to, multiple Bbs transcripts (Bardet Biedl syndrome), Dgcr8 (diGeorge syndrome) Cyfip1, Fmr1 and Fxr1 and 2 (Fragile X syndrome), Nlgn1,3, and Shank3 (Autism-spectrum disorders), Snca (Alzheimer’s disease), and Ube3a (Angelman’s syndrome). The localization of these mRNAs within the processes suggests the possibility that dysregulation of mRNA localization or translation may give rise to some of the phenotypes associated with these diseases.
What fraction of a single cell’s transcriptome exhibits localization within the dendrites and/or axons? One previous study provided an estimate of the CA1 neuron transcriptome number to be ~4,500 genes (Kamme et al., 2003). Our own analysis, combining the unique mRNAs expressed in the somata (Tables S9 and S12) and axodendritic compartments provides an estimate of 3,508 genes (Table S13). We thus estimate that greater than one-half of the CA1 neuron transcriptome can be detected in the axons and dendrites. Once established within a network, most of a neuron’s important moment-to-moment function occurs in dendrites and axons. In addition, in an individual CA1 pyramidal neuron the volume of axons and dendrites is about 30–60 times greater than that of the soma, indicating that a huge majority of the total cellular proteome function in the neuropil, rather than the somata. Thus, viewed from either a functional or morphological perspective, it is perhaps not surprising that most transcripts are found in the dendrites and/or axons. A previous study demonstrated that deletion of Camk2a mRNA from the dendrites resulted in an 85% loss of the synaptic CaMKIIα protein (Miller et al., 2002). This observation, together with the expanded local transcriptome identified here, suggests that a substantial fraction of the dendritic and synaptic proteins may be translated at a local, rather than somatic, source.
EXPERIMENTAL PROCEDURES
Tissue Microdissection and RNA Isolation
Hippocampal slices were prepared as previously described (Aakalu et al., 2001). The CA1 neuropil and cell body layers were carefully microdissected by hand from each slice. One cut was made at the stratum pyramidale-stratum radiatum border. Another cut was made at the stratum lacunosum moleculare-hippocampal fissure border. Lateral cuts were made at the CA2-CA1 border and near the end of region inferior in area CA1. To prepare sufficient tissue for a single deep sequencing run, we dissected both hippocampi from 6 male rats, yielding 12 hippocampi, and 120 microdissected slices. From 120 microdissected slices, we obtained ~25 μg of RNA from which we estimate we obtained 3 × 109 to 8 × 109 molecules of mRNA (Sambrook and Russell, 2001).
After microdissection, the tissue was transferred to a tube containing RNAlater (Ambion) in order to stabilize and prevent degradation of RNA. Total RNA was extracted using Trizol (Invitrogen) following the manufacturer’s recommendations. Briefly, the microdissected slices were homogenized in 1 ml of Trizol using a Teflon homogenizer. The homogenate was incubated on ice for 5 min. Two hundred microliters of chloroform was added to the samples and mixed for 15 s. Then the samples were centrifuged for 15 min (13,000 rpm; 4°C). The aqueous (upper) phase was collected and transferred to a new microtube. Five hundred microliters of isopropanol was added and the RNA was precipitated at −20°C for 30 min. The samples were centrifuged at 10,000 rpm for 10 min. After centrifugation, the isopropanol was removed, the pellet was washed with 1 ml of 70% ethanol and samples were centrifuged for 5 min at 5,000 rpm. Finally, the ethanol was discarded and the RNA pellet was air-dried.
Preparation of Glial Cell-Enriched Cultures
The cortex and hippocampus from P2 rat pups were removed and collected in DMEM (+ Glucose, + Glutamine) containing 1× PenStrep on ice. They were subsequently transferred to a Petri dish containing cold DMEM (+ Glucose). The medium was aspirated and the tissue was cut into small pieces. The chopped tissue was strained (BD Falcon Cell Strainer 40 μm; Catalog #352340) into a Falcon tube containing cold DMEM. The solution was centrifuged for 10 min at 600 rpm. The pellet was resuspended in 20 ml ice-cold DMEM (+ glucose, + glutamine), 10% fetal calf serum, 1% pyruvate per brain and 20 ml were plated per 10 cm tissue culture dish. The media was exchanged every 2 days. The relatively late stage of the rat pups, the mechanical disruption of cells and the lack of neuronal growth factors in the media promotes glial, but not neuronal, growth. It is also possible that the above process may alter glial transcription resulting in differences with glial transcriptome observed in vivo.
Preparation of Normalized cDNA Libraries
Twenty-five micrograms of total RNA (per sequencing run) was used as starting material. For cDNA synthesis the RNA was treated with DNase and poly(A) mRNA was isolated. (Note that the isolation of poly(A) mRNA dramatically reduces the presence of noncoding RNAs in our sample, and, as such, we focus on the mRNA population in this study). First-strand cDNA synthesis was conducted with a N6 randomized primer. Normalization of the sample was carried out by one cycle of denaturation and reassociation of the cDNAs. Reassociated double stranded-cDNAs were separated from the single-stranded cDNAs (normalized cDNA) by passing the mixture over a hydroxylapatite column. The cDNAs in the size range of 600–800 bp were eluted from preparative agarose gels. Then 454 adapters were ligated to the 5′ and 3′ ends of the cDNAs and they were finally amplified with 16 PCR cycles using a proofreading enzyme.
454 Next-Generation Sequencing and Annotation
cDNAs were sequenced by GATC Biotech (Konstanz, Germany). The resulting reads were mapped to the Rattus norvegicus Transcriptome (version rn4.2 September 2010, ftp://ftp.ncbi.nih.gov/genomes/R_norvegicus/RNA).
Two sets of reference files were used. The fasta file (rna.fa) contains the sequences used in the alignment process, while the GenBank file (rna.gbk) provides information about start and stop positions of the coding regions in each transcript. Reads were aligned and genes were annotated using GMAP (Wu and Watanabe, 2005). Due to the unpredictable nature of predicted and hypothetical records, we intentionally excluded them from the analysis. In order to include a gene in our subsequent analysis, we selected for coding sequences in which reads spanned 50% or more. To give readers access to the full data set, we provide in Table S1 the identity of all genes identified with one or more reads in coding or untranslated regions.
Data Mining of Filter Lists
As the filtering process is designed to remove transcripts that are contributed from sources other than dendrites and axons in the neuropil region of area CA1 of the hippocampus, the first step was to identify these sources and data mine studies that correspond to the sources of contamination. We screened results from microarray studies performed on the same microarray chip in a different rodent species (Mus musculus) for glia, interneurons, and endothelilal cell-enriched candidates. The annotation of the chip probes was downloaded from the NCBI GEO DataSets Repository (http://www.ncbi.nlm.nih.gov/gds). In order to convert mouse gene ids to rat gene ids, we referred to homologous genes described by three different sources, namely, NCBI Homolog Genes, http://www.ncbi.nlm.nih.gov/homologene; MGI (mouse genome informatics database), ftp://ftp.informatics.jax.org/pub/reports/index.html#nomen; and RGD (rat genome database): ftp://rgd.mcw.edu/pub/data_release/.
The up-to-date homolog records between the three databases were unified and a reference local database was built, associating mouse gene records with rat gene records. This procedure was used to convert the lists of overenriched genes from the above studies into usable measures for our analysis. On average the lists showed more than 95% conversion rate. The remaining genes can be explained by gene family member isoforms not present in rat.
Mitochondria related genes were downloaded from MitoMiner Database (http://mitominer.mrc-mbu.cam.ac.uk/release-2.0/begin.do). The nucleus related genes were extracted from Gene Ontology annotation files (http://www.geneontology.org/GO.downloads.annotations.shtml). Candidate genes were selected from the Gene Ontology database based on a search for gene products within the GO term “nuclear.” Since it has been shown that transcription factors can have functions in neuronal processes, we included in the filter only proteins that are part of the core RNA Polymerase and the DNA replication machinery. The above lists were specific for Rattus norvegicus. Two further data sets were provided by personal communication from their authors including a microglia list from Ben Barres (personal communication) and a hippocampal interneuron list from Ed Lein (Allen Mouse Brain Atlas, 2011 and personal communication).
All incorporated references are as follows
Blood Brain Barier Enriched (Daneman et al., 2010)
Developmentally CNS Up Regulated Vascular (Daneman et al., 2010)
Endothelial Specific (Daneman et al., 2010)
Pericyte (Daneman et al., 2010)
Cortical Astrocytes (Doyle et al., 2008)
Cortical Oligodendrocytes Cmtm5 (Doyle et al., 2008)
Cortical Oligodendrocytes Olig2 (Doyle et al., 2008)
Glia Culture Enriched (Nanostring Ramp Experiment)
Forbrain Astrocytes (Cahoy et al., 2008)
Forebrain Microglia (B. Barres, personal communication)
Forebrain Oligodendrocytes (Cahoy et al., 2008)
Hippocampus Interneurons (E. Lein, personal communication)
CA1 Underrepresented (Allen Mouse Brain Atlas, 2011)
Hippocampus Interneurons (Sugino et al., 2006)
Cortical Interneurons (Doyle et al., 2008)
Mitochondrion Related (mitoMiner Database 2011)
Nucleus Related (Gene Ontology Annotation 2011)
In order to incorporate these lists in our filter data set, we first selected only those genes that were present in the initial rat transcriptome set for alignment. In that step we equalized all lists to the initial gene pool, thus allowing us to conduct statistics for list comparison by treating the data sets as random samples with common source. For calculating overlapping probabilities we used a cumulative hypergeometric distribution as described by (Fury et al., 2006). To ensure that we compared the same labels by genes, we compiled a table where each record contains the following fields:
Official Gene Symbol by Nomenclature
Entrez Gene ID from Entrez Reference Sequence Database (NCBI)
Offical Gene Name by Nomenclature
Reference Source
The Entrez Gene ID was used as a key field for comparison. The gene information was extracted from weekly updated gene information files on NCBI repository (ftp://ftp.ncbi.nih.gov/gene/DATA/GENE_INFO/Mammalia/).
Filter Process
The filter process is conducted in two main steps. First, comparison is done between our query data set and the filter data. We split the query list in two: potential filtered transcripts and potential dendritic/axonal transcripts. The second step is the assessment of false candidate after filtering.
False-negative candidates arise in the filtered list due coexpression of those candidates in different cell types. Such records are identified and rescued by comparing the filtered list to transcripts that are present in hippocampus pyramidal neurons (Sugino et al., 2006) or are identified by in situ methods either conducted by us (71 in situ probes) or by previous studies (Table S14).
False-positive candidates arise in the cleaned list due to genes that were detected by 454 but were not present on the microarray chip from the reference studies. Those genes were checked in the Allen Brain Atlas for pyramidal neuron expression in area CAI of the hippocampus. The genes that were de-enriched in the investigated area were pulled out of the result list and a false-positive rate was determined.
Gene Ontology
The Gene Ontology analysis was conducted using the Bingo Plug-In (v 2.44) for Cytoscape (Maere et al., 2005). The Cytoscape output is a text file with the following parameters: term id, term name, p value, x (number of genes from the query list annotated to a certain term), X (number of genes from the query list that are annotated to a specific ontology), n (total number of genes annotated to a certain term by the rat genome database), N (total number of genes annotated to an ontology by the rat genome database). One file was generated per ontology (biological process, molecular function, and cellular component).
We calculated cluster frequency, total frequency, fold change, for each term graph level, where:
Three biographs with the ancestors of all overrepresented terms in the corresponding ontology were built. An application was developed in order to search for the shortest path from each overrepresented term to the root of its graph and assign the distance as the depth level for the term (Dijkstra, 1959). We used an additional custom application to combine the results from the three ontologies in one file. The file was imported to Microsoft Excel in order to obtain one table per query list. The table was sorted by depth level and fold change. All terms in the table are overrepresented with p value less than 0.05, after correction with the Benjamini & Hochberg false discovery rate method.
Digital Analysis of Gene Expression Using Nanostring
Custom codesets were manufactured based on the accession numbers (NCBI) of the transcripts. Each mRNA was detected by two probes of 50 nucleotide length: a target-specific capture and a reporter probe. The reporter probe was linked to a six-color fluorescent barcode and the capture probe had biotin attached in order to bind the surface of the Nanostring cartridge. All probes were designed against coding sequences (Geiss et al., 2008). One hundred nanograms of total RNA treated with DNaseI and cleaned with the RNeasy MinElute cleanup kit (QIAGEN) was hybridized to capture and reporter probes for at least 16 hr at 65°C. Samples were prepared for data collection using an automated fluidic handling system (nCounter Prep Station). After posthybridization processing, the nCounter Digital Analyzer collected the data by taking images of the immobilized fluorescent reporters with a CCD camera through a microscope objective lens. All cartridges were imaged using a 60× objective with 1,150 fields of view.
Each reaction contained positive and negative controls for hybridization. The positive controls were from the External RNA Control Consortium (ERCC) sequences with a synthetic template spiked in at different concentrations. The ERCC sequences were developed by a consortium looking for nonbiological sequences to be used as controls for gene expression experiments. The negative controls were also obtained from the ERCC set, but are spiked without any RNA, to provide an estimate of the background signal. Genes with stable mRNA levels throughout the different conditions were identified by the geNorm method in order to normalize the data for concentration variation; these mRNAs included Map1lc3b, Htt, Clcn3, Pten, Gsk3b, Cript, and Mtor. Hierarchical clustering was applied to the normalized counts to identify clusters in the ramp experiments. To address overrepresentation in the different compartments, a t test was conducted. Cutoff parameters for biological significance were *p < 0.05 and fold change >2. To correct for false p values each test was bootstrapped 1,000 times.
Real-Time PCR
One microgram of RNA was treated with DNase I and subsequently reverse-transcribed using the QuantiTect Reverse Transcription Kit (QIAGEN). Dilutions (1:100) were used as template in the PCR. Each reaction contained 5 μl of template, 1× primers (QuantiTect primer assays from QIAGEN), and 1× SYBR Green PCR master mix (Applied Biosystems). The cycling parameters used were those recommended by the QuantiTect primer assays.
High-Resolution RNA In Situ Hybridization and Immunostaining
We prepared and maintained dissociated hippocampal neurons as previously described (Aakalu et al., 2001). In situ hybridization was performed using the QuantiGene (QG) ViewRNA kit from Panomics as previously described (Taylor et al., 2010) with the following modifications. Cells (DIV 18-24) were fixed for 30 min at room temperature using a 4% paraformaldehyde solution (4% paraformaldehyde, 5.4% Glucose, 0.01 M sodium metaperiodate, in lysine-phosphate buffer). The Proteinase K treatment was omitted in order to preserve the integrity of the dendrites. After completion of in situ hybridizations, cells were washed with PBS 1× and incubated in blocking buffer (4% goat serum in PBS 1×) for 1 hr. Neurons were subsequently processed for immunofluorescence using standard methods (Aakalu et al., 2001). Images were obtained from 10 micron z-stacks (~20 images) that were acquired with 2,048 × 2,048 pixel resolution. They were analyzed using custom applications created in MATLAB. Briefly, dendrites were straightened and maximum intensity projections were generated. Areas of local maxima were detected and binary masks were created. The areas were distance-transformed and a watershed algorithm was applied in order to detect single puncta. For representation purposes, the channels corresponding to the detected mRNA and the MAP2 staining were converted to binary images. The mRNA puncta were dilated two times. An outline of the dendrite or soma was generated using the MAP2 immunostaining as a mask (see Figure S5). Both processed channels were merged using Adobe Photoshop. In order to quantify the number of puncta for the Dlg4 (PSD-95) mRNA in the neuronal compartments (Figures S5D and S5E), we acquired two sets of images from each cell. Puncta in the entire dendritic field were obtained from maximum intensity projections of 10-micron z-stacks (~20 images) acquired using a 40× oil objective (0.7× digital zoom) with 2,048 × 2,048 pixel resolution. Puncta in the cell body were counted from maximum intensity projections of images taken using the same parameters described above but using a 2× digital zoom in order to increase the resolution of the particles. Individual punctae were counted manually from both sets of images. Great care was taken to resolve and differentiate individual punctae.
For in situ hybridization in tissue, 500 μm hippocampal slices were cut with a tissue chopper (Stölting), collected in ACSF, immersion fixed for 30 min at room temperature using a 4% paraformaldehyde solution (4% paraformaldehyde, 5.4% glucose, 0.01 M sodium metaperiodate, in lysine-phosphate buffer) and cryoprotected by sequential incubation in 10% (1 hr, 4°C), 20% (1 hr, 4°C), and 30% sucrose (over night, 4°C) in PBS. Hippocampal slices were embedded in tissue-tek (Sakura) and cryostat sectioned at a thickness of 4 μm. Sections were collected on superfrost+ slides and stored at −80°C. On the day of the experiment, the slices were air-dried for 10 min at room temperature before they were covered by secure seal gaskets (Invitrogen). To remove embedding medium, sections were washed three times in PBS and postfixed for 10 min at room temperature in the above-mentioned fixative. Permeabilization and in situ hybridization were carried out essentially as described for hippocampal neurons but with additional washing steps. After in situ hybridization, sections were washed in PBS, blocked for 1 hr with blocking buffer (4% goat serum in PBS) and incubated for 3 hr at room temperature with rabbit anti-MAP2 (Millipore, 1:1000) and mouse Smi312 (Covance, 1:4000) in blocking buffer. Secondary antibodies were applied in blocking buffer for 30 min at room temperature, nuclei were stained for 3 min with DAPI and samples were mounted in Aqua Polymount (Polysciences). The CA1 region of hippocampal slices was imaged using a Zeiss LSM780 confocal microscope and a 40× oil objective (plan achromate, NA 1.4). Z-stacks spanning the entire thickness of the slice were obtained and channels were separated and collapsed to a maximum intensity projection in ImageJ. For representation purposes, the channels corresponding to the detected mRNA and the DAPI staining were converted to binary images with fixed thresholds within an experiment for control and experimental sections. The mRNA puncta were dilated three times for better visualization. Both processed channels were merged using Adobe Photoshop.
Clustering of mRNA Localization Patterns
Using the in situ hybridization data, each investigated dendrite was divided in bins of 25 μm and signal puncta were counted per bin. A master dendrite was made for every transcript with the average number of puncta per bin assigned to the bin. We used sum norm to normalize the row expression vector for each candidate to make transcripts comparable, normalizing for differences in total expression levels. A hierarchical clustering algorithm was used to group the normalized expression vectors of all transcripts. As a dissimilarity measure, we used 1 minus the standardized covariance of the signal and the linkage option was the average of the dissimilarities. We visualized the resulting dendrogram in MATLAB. Four main clusters were identified by the above procedure. In order to measure how faithfully the dendrogram preserves the pairwise distances between the original unmodeled data points, we calculated the cophenetic correlation coefficient. We also addressed the significance of the generation of the four main clusters as previously described (Varshavsky et al., 2008).
Supplementary Material
ACKNOWLEDGMENTS
Financial support was provided in part by the DFG-funded Collaborative Research Center 902: “Molecular Principles of RNA-based Regulation.” We are extremely grateful to Mona Khan, Christian Lozanoski, and Peter Mombaerts for assistance with the Nanostring technology. We thank Ben Barres for discussions on glial transcriptomes. We thank Ed Lein for assistance in compiling interneuron-enriched transcripts. We thank Ina Bartnik for the preparation of cultured hippocampal neurons. We thank Gilles Laurent, Mona Khan, and Schuman lab members for comments on the manuscript.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and fourteen tables and can be found with this article online at doi:10.1016/j.neuron.2012.02.036.
REFERENCES
- Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron. 2001;30:489–502. doi: 10.1016/s0896-6273(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Allen Mouse Brain Atlas. Allen Institute for Brain Science; Seattle (WA): 2011. http://mouse.brain-map.org/ [Google Scholar]
- Andreassi C, Zimmermann C, Mitter R, Fusco S, De Vita S, Saiardi A, Riccio A. An NGF-responsive element targets myo-inositol monophosphatase-1 mRNA to sympathetic neuron axons. Nat. Neurosci. 2010;13:291–301. doi: 10.1038/nn.2486. [DOI] [PubMed] [Google Scholar]
- Böckers TM, Segger-Junius M, Iglauer P, Bockmann J, Gundelfinger ED, Kreutz MR, Richter D, Kindler S, Kreienkamp HJ. Differential expression and dendritic transcript localization of Shank family members: identification of a dendritic targeting element in the 3′ untranslated region of Shank1 mRNA. Mol. Cell. Neurosci. 2004;26:182–190. doi: 10.1016/j.mcn.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Bradshaw KD, Emptage NJ, Bliss TV. A role for dendritic protein synthesis in hippocampal late LTP. Eur. J. Neurosci. 2003;18:3150–3152. doi: 10.1111/j.1460-9568.2003.03054.x. [DOI] [PubMed] [Google Scholar]
- Burgin KE, Waxham MN, Rickling S, Westgate SA, Mobley WC, Kelly PT. In situ hybridization histochemistry of Ca2+/calmodulin-dependent protein kinase in developing rat brain. J. Neurosci. 1990;10:1788–1798. doi: 10.1523/JNEUROSCI.10-06-01788.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneman R, Zhou L, Agalliu D, Cahoy JD, Kaushal A, Barres BA. The mouse blood-brain barrier transcriptome: a new resource for understanding the development and function of brain endothelial cells. PLoS ONE. 2010;5:e13741. doi: 10.1371/journal.pone.0013741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkstra EW. A note on two problems in connexion with graphs. Numerische mathematik. 1959;1:269–271. [Google Scholar]
- Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, Doughty ML, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fury W, Batliwalla F, Gregersen PK, Li W. Overlapping probabilities of top ranking gene lists, hypergeometric distribution, and stringency of gene selection criterion. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2006;1:5531–5534. doi: 10.1109/IEMBS.2006.260828. [DOI] [PubMed] [Google Scholar]
- Gardiol A, Racca C, Triller A. Dendritic and postsynaptic protein synthetic machinery. J. Neurosci. 1999;19:168–179. doi: 10.1523/JNEUROSCI.19-01-00168.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner CC, Tucker RP, Matus A. Selective localization of messenger RNA for cytoskeletal protein MAP2 in dendrites. Nature. 1988;336:674–677. doi: 10.1038/336674a0. [DOI] [PubMed] [Google Scholar]
- Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- Govindarajan A, Israely I, Huang SY, Tonegawa S. The dendritic branch is the preferred integrative unit for protein synthesis-dependent LTP. Neuron. 2011;69:132–146. doi: 10.1016/j.neuron.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herb A, Wisden W, Catania MV, Maréchal D, Dresse A, Seeburg PH. Prominent dendritic localization in forebrain neurons of a novel mRNA and its product, dendrin. Mol. Cell. Neurosci. 1997;8:367–374. doi: 10.1006/mcne.1996.0594. [DOI] [PubMed] [Google Scholar]
- Horton AC, Ehlers MD. Dual modes of endoplasmic reticulum-to-Golgi transport in dendrites revealed by live-cell imaging. J. Neurosci. 2003;23:6188–6199. doi: 10.1523/JNEUROSCI.23-15-06188.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton AC, Rácz B, Monson EE, Lin AL, Weinberg RJ, Ehlers MD. Polarized secretory trafficking directs cargo for asymmetric dendrite growth and morphogenesis. Neuron. 2005;48:757–771. doi: 10.1016/j.neuron.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Huber KM, Kayser MS, Bear MF. Role for rapid dendritic protein synthesis in hippocampal mGluR-dependent long-term depression. Science. 2000;288:1254–1257. doi: 10.1126/science.288.5469.1254. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston D, Narayanan R. Active dendrites: colorful wings of the mysterious butterflies. Trends Neurosci. 2008;31:309–316. doi: 10.1016/j.tins.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Kamme F, Salunga R, Yu J, Tran DT, Zhu J, Luo L, Bittner A, Guo HQ, Miller N, Wan J, Erlander M. Single-cell microarray analysis in hippocampus CA1: demonstration and validation of cellular heterogeneity. J. Neurosci. 2003;23:3607–3615. doi: 10.1523/JNEUROSCI.23-09-03607.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Schuman EM. A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science. 1996;273:1402–1406. doi: 10.1126/science.273.5280.1402. [DOI] [PubMed] [Google Scholar]
- Klausberger T, Somogyi P. Neuronal diversity and temporal dynamics: the unity of hippocampal circuit operations. Science. 2008;321:53–57. doi: 10.1126/science.1149381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Lin AC, Holt CE. Function and regulation of local axonal translation. Curr. Opin. Neurobiol. 2008;18:60–68. doi: 10.1016/j.conb.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- Makara JK, Losonczy A, Wen Q, Magee JC. Experience-dependent compartmentalized dendritic plasticity in rat hippocampal CA1 pyramidal neurons. Nat. Neurosci. 2009;12:1485–1487. doi: 10.1038/nn.2428. [DOI] [PubMed] [Google Scholar]
- Martin KC, Casadio A, Zhu H, Yaping E, Rose JC, Chen M, Bailey CH, Kandel ER. Synapse-specific, long-term facilitation of aplysia sensory to motor synapses: a function for local protein synthesis in memory storage. Cell. 1997;91:927–938. doi: 10.1016/s0092-8674(00)80484-5. [DOI] [PubMed] [Google Scholar]
- Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayford M, Baranes D, Podsypanina K, Kandel ER. The 3′-untranslated region of CaMKII alpha is a cis-acting signal for the localization and translation of mRNA in dendrites. Proc. Natl. Acad. Sci. USA. 1996;93:13250–13255. doi: 10.1073/pnas.93.23.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S, Yasuda M, Coats JK, Jones Y, Martone ME, Mayford M. Disruption of dendritic translation of CaMKIIalpha impairs stabilization of synaptic plasticity and memory consolidation. Neuron. 2002;36:507–519. doi: 10.1016/s0896-6273(02)00978-9. [DOI] [PubMed] [Google Scholar]
- Muddashetty RS, Kelić S, Gross C, Xu M, Bassell GJ. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J. Neurosci. 2007;27:5338–5348. doi: 10.1523/JNEUROSCI.0937-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okaty BW, Sugino K, Nelson SB. A quantitative comparison of cell-type-specific microarray gene expression profiling methods in the mouse brain. PLoS ONE. 2011;6:e16493. doi: 10.1371/journal.pone.0016493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostroff LE, Fiala JC, Allwardt B, Harris KM. Polyribosomes redistribute from dendritic shafts into spines with enlarged synapses during LTP in developing rat hippocampal slices. Neuron. 2002;35:535–545. doi: 10.1016/s0896-6273(02)00785-7. [DOI] [PubMed] [Google Scholar]
- Ouyang Y, Rosenstein A, Kreiman G, Schuman EM, Kennedy MB. Tetanic stimulation leads to increased accumulation of Ca(2+)/calmodulin-dependent protein kinase II via dendritic protein synthesis in hippocampal neurons. J. Neurosci. 1999;19:7823–7833. doi: 10.1523/JNEUROSCI.19-18-07823.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patanjali SR, Parimoo S, Weissman SM. Construction of a uniform-abundance (normalized) cDNA library. Proc. Natl. Acad. Sci. USA. 1991;88:1943–1947. doi: 10.1073/pnas.88.5.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon MM, Choi SH, Jamieson CA, Geschwind DH, Martin KC. Identification of process-localized mRNAs from cultured rodent hippocampal neurons. J. Neurosci. 2006;26:13390–13399. doi: 10.1523/JNEUROSCI.3432-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab-Graham KF, Haddick PC, Jan YN, Jan LY. Activity- and mTOR-dependent suppression of Kv1.1 channel mRNA translation in dendrites. Science. 2006;314:144–148. doi: 10.1126/science.1131693. [DOI] [PubMed] [Google Scholar]
- Richter JD, Klann E. Making synaptic plasticity and memory last: mechanisms of translational regulation. Genes Dev. 2009;23:1–11. doi: 10.1101/gad.1735809. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- Smith WB, Starck SR, Roberts RW, Schuman EM. Dopaminergic stimulation of local protein synthesis enhances surface expression of GluR1 and synaptic transmission in hippocampal neurons. Neuron. 2005;45:765–779. doi: 10.1016/j.neuron.2005.01.015. [DOI] [PubMed] [Google Scholar]
- Steward O, Levy WB. Preferential localization of polyribosomes under the base of dendritic spines in granule cells of the dentate gyrus. J. Neurosci. 1982;2:284–291. doi: 10.1523/JNEUROSCI.02-03-00284.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino K, Hempel CM, Miller MN, Hattox AM, Shapiro P, Wu C, Huang ZJ, Nelson SB. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat. Neurosci. 2006;9:99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- Sutton MA, Ito HT, Cressy P, Kempf C, Woo JC, Schuman EM. Miniature neurotransmission stabilizes synaptic function via tonic suppression of local dendritic protein synthesis. Cell. 2006;125:785–799. doi: 10.1016/j.cell.2006.03.040. [DOI] [PubMed] [Google Scholar]
- Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Sutton MA, Taylor AM, Ito HT, Pham A, Schuman EM. Postsynaptic decoding of neural activity: eEF2 as a biochemical sensor coupling miniature synaptic transmission to local protein synthesis. Neuron. 2007;55:648–661. doi: 10.1016/j.neuron.2007.07.030. [DOI] [PubMed] [Google Scholar]
- Sutton MA, Wall NR, Aakalu GN, Schuman EM. Regulation of dendritic protein synthesis by miniature synaptic events. Science. 2004;304:1979–1983. doi: 10.1126/science.1096202. [DOI] [PubMed] [Google Scholar]
- Swanger SA, Bassell GJ. Making and breaking synapses through local mRNA regulation. Curr. Opin. Genet. Dev. 2011;21:414–421. doi: 10.1016/j.gde.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM, Dieterich DC, Ito HT, Kim SA, Schuman EM. Microfluidic local perfusion chambers for the visualization and manipulation of synapses. Neuron. 2010;66:57–68. doi: 10.1016/j.neuron.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiruchinapalli DM, Oleynikov Y, Kelic S, Shenoy SM, Hartley A, Stanton PK, Singer RH, Bassell GJ. Activity-dependent trafficking and dynamic localization of zipcode binding protein 1 and beta-actin mRNA in dendrites and spines of hippocampal neurons. J. Neurosci. 2003;23:3251–3261. doi: 10.1523/JNEUROSCI.23-08-03251.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre ER, Steward O. Protein synthesis within dendrites: glycosylation of newly synthesized proteins in dendrites of hippocampal neurons in culture. J. Neurosci. 1996;16:5967–5978. doi: 10.1523/JNEUROSCI.16-19-05967.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker RP, Garner CC, Matus A. In situ localization of microtubule-associated protein mRNA in the developing and adult rat brain. Neuron. 1989;2:1245–1256. doi: 10.1016/0896-6273(89)90309-7. [DOI] [PubMed] [Google Scholar]
- Varshavsky R, Horn D, Linial M. Global considerations in hierarchical clustering reveal meaningful patterns in data. PLoS ONE. 2008;3:e2247. doi: 10.1371/journal.pone.0002247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Watanabe CK. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21:1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- Zhong J, Zhang T, Bloch LM. Dendritic mRNAs encode diversified functionalities in hippocampal pyramidal neurons. BMC Neurosci. 2006;7:17. doi: 10.1186/1471-2202-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivraj KH, Tung YC, Piper M, Gumy L, Fawcett JW, Yeo GS, Holt CE. Subcellular profiling reveals distinct and developmentally regulated repertoire of growth cone mRNAs. J. Neurosci. 2010;30:15464–15478. doi: 10.1523/JNEUROSCI.1800-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.