Abstract
Aim
This study compared the subgingival microbiota of subjects with refractory periodontitis (RP) to those in subjects with treatable periodontitis (GR) or periodontal health (PH) using the Human Oral Microbe Identification Microarray (HOMIM).
Methods
At baseline, subgingival plaque samples were taken from 47 periodontitis and 20 PH individuals, and analyzed for the presence of 300 species by HOMIM. The periodontitis subjects were classified as RP (n=17) based on mean attachment loss (AL) and/or >3 sites with AL ≥2.5 mm after SRP, surgery and systemically administered amoxicillin and metronidazole or as GR (n=30) based on mean attachment gain and no sites with AL ≥2.5 mm after treatment. Significant differences in taxa among groups were sought using the Kruskal Wallis and Chi-square tests.
Results
More species were detected in diseased patients (GR or RP) than those without disease (PH). RP subjects were distinguished from GR and PH by a significantly high frequency of putative periodontal pathogens such as, Parvimonas micra, Campylobacter gracilis, Eubacterium nodatum, Selenomonas noxia, Tannerella forsythia, Porphyromonas gingivalis, Prevotella spp., Treponema spp., Eikenella corrodens, as well as “unusual” species (Pseudoramibacter alactolyticus, TM7 spp. oral taxon (OT) 346/356, Bacteroidetes spp. OT 272/274, Solobacterium moorei, Desulfobulbus sp. OT 041, Brevundimonas diminuta, Sphaerocytophaga sp. OT 337, Shuttleworthia satelles, Filifactor alocis, Dialister invisus/pneumosintes, Granulicatella adiacens, Mogibacterium tidmidum, Veillonella atypica, Mycoplasma salivarium, Synergistes sp. cluster II, Acidaminococcaceae [G-1] sp. OT 132/150/155/148/135) [p<0.05]. Species that were more prevalent in PH than in periodontitis patients included Actinomyces sp. OT 170, Actinomyces spp. cluster I, Capnocytophaga sputigena, Cardiobacterium hominis, Haemophilus parainfluenzae, Lautropia mirabilis, Propionibacterium propionicum, Rothia dentocariosa/mucilagenosa, Streptococcus sanguinis (p<0.05).
Conclusion
RP patients present a distinct microbial profile compared to patients in the GR and PH groups as determined by HOMIM.
Keywords: Refractory periodontitis, subgingival microbiota, periodontal pathogen, HOMIM, periodontal therapy
Introduction
Over the years, microbiological studies based on cultural and molecular methods have identified over 700 bacterial species in the human oral cavity.1–6 More than 400 of these species have been detected in the periodontal pocket, and approximately half of the taxa have not yet been cultivated.4, 5 Furthermore, substantial microbial diversity among different sites in the oral cavity, and/or people has also been observed.4–8 Although most of these organisms are commensal, several bacterial species, including those that cannot be grown in vitro, have been associated with either periodontal health or disease.2–4, 8, 9 Consequently, knowledge of the bacterial diversity in the subgingival biofilm is important for the diagnosis and rational treatment of periodontal diseases.
The major goal of periodontal therapy is to reduce or eliminate the pathogenic species and maintain colonization by host-compatible species.10, 11 Most periodontal diseases are treated predictably by conventional periodontal therapy, including scaling, periodontal surgery, systemically administered antibiotics, and regular maintenance.12, 13 Certain subjects with destructive periodontal disease, however, respond poorly to conventional therapy and continue to show loss of periodontal attachment despite treatment.14, 16 These subjects have been referred to as exhibiting “refractory periodontal disease”.17 The biological basis for refractory periodontitis is poorly understood, and studies have indicated that these individuals exhibit clinical, microbiological, and immunological heterogeneity.18–28 Several hypotheses have been proposed to explain this condition, including the existence of an infection caused by specific pathogenic species (possibly newly recognized or “uncultivable” species) in a susceptible host.23–26 Therefore, identification of the microbiota in refractory periodontitis is warranted, in order to lead to a more effective therapy.29 In the current study, we used a new 16S rRNA-based microarray method, the Human Oral Microbe Identification Microarray (HOMIM),30 to test the hypothesis that subjects with refractory periodontal disease (RP) can be distinguished from subjects with treatable periodontitis (GR= Good Responders) and periodontal health (PH) based upon their baseline subgingival microbial profiles.
Material and methods
Subject Population
The subject population was comprised of 67 patients in good general health, recruited between April 2003 and February 2007, from the Clinical Center for Periodontal Research at The Forsyth Institute, and the Clinical Research Center at the Boston University Goldman School of Dental Medicine. Informed consent was obtained from all enrolled individuals. The study protocol was reviewed and approved by the Institutional Review Boards at The Forsyth Institute and Boston University Medical Center. All subjects had at least 20 natural teeth and were over 20 years of age. Subjects with periodontitis were selected that had ≥ 5 sites with pocket depth and attachment loss ≥ 6 mm at baseline. Subjects in the PH group had no pockets > 3 mm and no attachment loss > 2 mm at any site. Exclusion criteria included pregnancy, lactation, systemic conditions that could affect the progression or treatment of periodontal diseases, and any known allergy to amoxicillin and/or metronidazole. In addition, subjects who had received systemic antibiotics or periodontal therapy in the previous 6 months were excluded.
Clinical Monitoring
Periodontal clinical measurements were performed at 6 sites per tooth at all teeth excluding third molars,31 and included: probing depth (PD) and clinical attachment level (CAL) measured using a North Carolina periodontal probe#, presence or absence of gingival redness, suppuration, bleeding on probing, and supragingival plaque accumulation. Pocket depth and attachment level measurements were repeated within a subject at each visit. Duplicate measurements were compared to ensure a standard deviation of <1.0 mm, and the means of pairs of attachment level measurements at different visits used to determine disease progression. The same examiner performed all measurements for each subject at every visit. Examiners at both centers were trained and calibrated on the periodontal measurements. Clinical measurements were taken at baseline and at 3, 6, 9, 12 and 15 months post therapy.
Treatment Protocol
After completion of baseline monitoring, subjects with evidence of destructive periodontal disease received full mouth scaling and root planing (SRP) under local anesthetic,32 and instruction in proper home care procedures within a 4-week period. Approximately 2 months after the completion of SRP, the subjects received modified Widman flap surgery in quadrants with residual pockets > 4 mm. At the first surgical visit, subjects were asked to take 500 mg amoxicillin and 250 mg metronidazole 3 times daily for 14 days. The surgical/antibiotic phase was completed within 4 weeks. Subjects then entered a maintenance and clinical monitoring phase with visits at 3, 6, 9, 12 and 15 months where clinical measurements, maintenance scaling and reinforcement of home care procedures were performed. GR had full mouth mean attachment level gain and no sites with attachment loss ≥ 2.5 mm during one year after either SRP or surgery with antibiotics. Any subject showing mean attachment loss, and/or > 3 sites with attachment loss ≥ 2.5 mm from the baseline visit to any monitoring visit within 1 year post-therapy was defined as having refractory periodontal disease (RP).
Subgingival Sample Taking and Isolation of Bacterial DNA
After removal of supragingival biofilm with sterile gauze, individual subgingival biofilm samples were taken from the mesio-buccal aspect of up to 14 teeth in different quadrants (1 and 3 or 2 and 4) per subject using sterile periodontal curettes#. Each sample was placed in separate 1.5 ml tubes containing 50 µl of TE (50 mM Tris-HCl, 1 mM EDTA, pH 7.6). For bacterial DNA extraction, 44 µl of each sample was taken, and 0.5% Tween 20 ** and 1 µl of Proteinase K (10 mg/ml) †† were added. The samples were heated at 55°C for 2 h, and at 95°C for 5 min for inactivation of Proteinase K. All samples were stored in a freezer at −80 °C prior to PCR amplification.
Amplification of 16S rRNA Genes
The 16S rRNA genes of each sample were amplified in two separate PCR reactions using different primer sets. In the first PCR reaction, a mix of two forward primers at 1:1 ratio (4F: 5`-CCAGAGTTTGATYMTGGC-`3 and 6F:5`-GACTAGAGTTTGATYMTGGC–`3), and the reverse primer 1541R (5`-GAAGGAGGTGWTCCADCC-`3) ‡‡ were used. One microliter of the DNA template was added to a reaction mixture (25 µL, final volume) containing 5 pmol of the 4F and 6F primer mix, 15 pmol of the 1541R primer, 2.5 µL 10·X high fidelity PCR buffer, 2 mM MgSO4, 0.5 µL deoxynucleotide triphosphate mixture (10 mM), and 1 U Taq polymerase §§. In the second PCR amplification, the forward primer mix described above and the 1492R reverse primer (5`-GYTACCTTGTTACGACTT-`3) ‡‡ were used. One microliter of the DNA template was added to a reaction mixture (25 µL final volume) containing 15 pmol of the forward primer mix, 5 pmol of the 1492R primer, 2.5 µL 10·X high fidelity PCR buffer, 2 mM MgSO4, 0.5 µL deoxynucleotide triphosphate mixture (10 mM, 0.2 mM final concentration of each deoxynucleotide), and 1 U Taq polymerase §§. The PCR program was carried out in thin-walled tubes with a thermocycler ║║ and included an initial denaturation step at 94°C for 2 minutes, followed by 32 cycles of denaturation at 94°C for 30s, annealing at 55°C for 30s, elongation at 68°C for 1.5 minute with an additional 1 s for each cycle, and a final elongation step at 68°C for 10 minutes. The PCR products were analyzed on a 1.5% agarose gel electrophoresis in Tris-borate-EDTA buffer, stained ¶¶ and visualized under short-wavelength UV light. The PCR reactions were combined for each sample providing a 50 µL PCR product. The PCR products were purified using a DNA purification kit ## according to the manufacturer’s instructions for further labeling.
Labeling of Samples
Purified PCR products were labeled via the incorporation of Cy3-dCTP *** during another amplification reaction. Briefly, 7 µL of each product were added into a 25 µL reaction mixture containing 15 pmol of the forward primer 9F (5`-GAGTTTGATYMTGGCTCAG-3`) ‡‡, 5 pmol of the 1492R primer ‡‡, 2.5 µL 10·X high fidelity PCR buffer, 2 mM MgSO4, 2.5 µL of deoxynucleotide triphosphate mixture without deoxycytidine triphosphate (10 mM, 0.2 mM final concentration of each deoxynucleotide), 2.5 µL of deoxycytidine triphosphate mixture (1 µL of 1 mM Cy3-dCTP + 4 µL of 2 mM dCTP, 0.2 mM final concentration), 0.25 µL 10% Triton X, and 1.25 U Taq polymerase §§. The PCR program included an initial denaturation step at 94°C for 2 minutes, followed by 40 cycles of denaturation at 94°C for 30s, annealing at 55°C for 30s, elongation at 68°C for 1.5 minute with an additional 1 s for each cycle, and a final elongation step at 68°C for 10 minutes. The PCR products were purified using a purification kit ## according to manufacturer instructions.
Capture Probes and Slide Printing
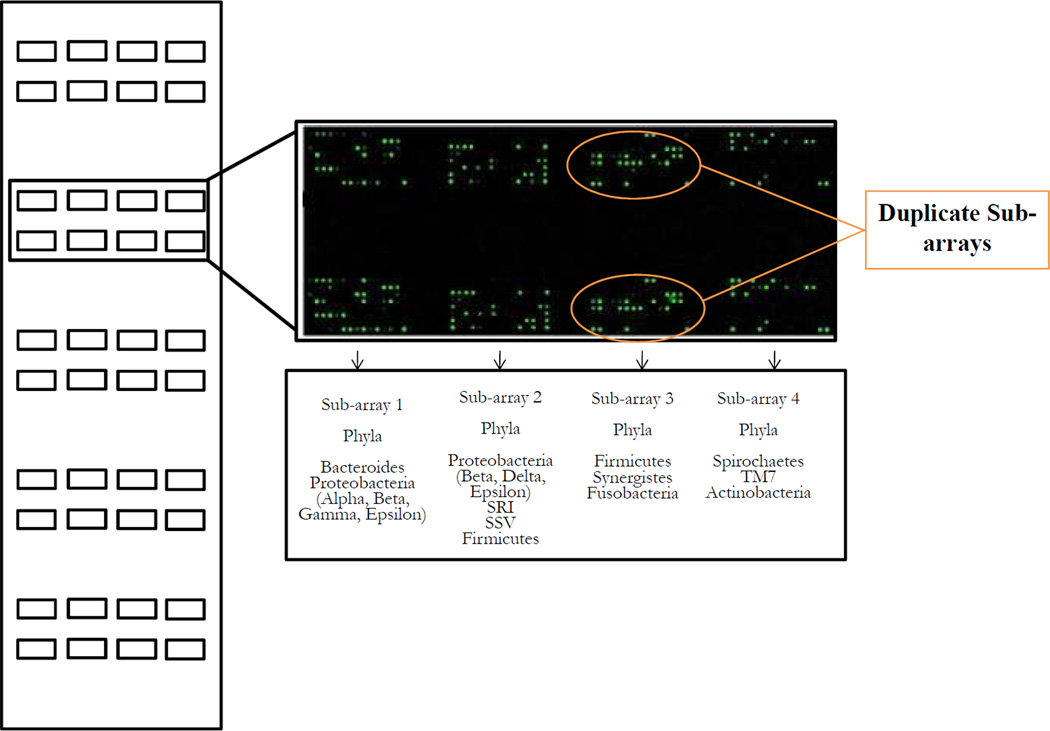
16S rRNA-based oligonucleotide reverse capture probes were custom synthesized ††† with a 5’-(C6)-amine modified base, eight spacer thymidines and 18 to 20-nucleotides of target sequence, and printed (Michigan State University Research Technology Support Facility) on 25 × 76 mm aldehyde-coated glass slides ‡‡‡. A total of 400 oligonucleotide probes targeting over 300 bacterial taxa 33 (Table S1 in the supplemental material) were printed on each microarray. Probes targeting more than two closely related species appeared as clusters (37 altogether). Oral taxon designations for each species are provided as defined in the Human Oral Microbiome Database.34 Included on each array were positive controls, i.e. “universal” probes that hybridize with all or most bacterial species, and provide both array orientation and labeling efficiency, as well as negative controls to determine array background levels. A universal 16S rRNA probe was also printed in a series of concentrations in order to monitor signal linearity. Moreover, probes were designed to have the same melting temperature (51–53°C), and G+C content. Regarding sensitivity, the lower limit of detection for the HOMIM array is about ≥ 104 bacterial cells. Five copies of the array were printed per slide, each one printed as four 8 × 15 duplicate sub-arrays. Probes were arranged phylogenetically on each sub-array (Figure 1).
Figure 1.
Schematic layout of a HOMIM slide. Five identical arrays containing four 8 × 15 duplicate sub-arrays are printed on an aldehyde-coated glass slide. Probes are organized phylogenetically on each sub-array (Sub-arrays 1 to 4). Up to five samples can be hybridized simultaneously against probes to 300 different species per slide.
Microarray Hybridization and Reading
Prior to hybridization, slides were blocked with sodium borohydride (NaBH4, 1× PBS, 99% ethanol) in order to reduce unreactive aldehyde groups, thus minimizing the fluorescent background. One cover slip was used to cover the pair of duplicate arrays, allowing for 5 separate experiments per slide. Fourteen microliter of each labeled and purified PCR sample were mixed with 6 µl of hybridization buffer containing 2× SSC, 0.2 µg/µl yeast tRNA, 0.1% SDS, and heated for 5 minutes at 100°C. After that, 10 µl of each solution were loaded slowly at one corner and under the cover slip, allowing capillary action to pull the solution over the array. Slides were then placed on incubation racks in a humidified chamber, and hybridization was then performed at 55°C for 16 h in a microarray incubator§§§. Slides were removed from the hybridization chamber and sunk into a water bath containing a washing solution (2× SSC, 10%SDS, at 55°C) to remove the cover slips. The slides were washed using an automated microarray robotic instrument║║║. A washing program was set up as follow: one washing in buffer (2× SSC, 10%SDS) at 55°C for 2 minutes, a second washing in 2× SSC buffer at room temperature for 2 minutes, and a final washing in 0.1× SSC buffer at room temperature for 2 minutes. Once the slides are finished spinning in the centrifuge, they were removed and placed in a light-proof container until scanning. The microarray slides were scanned using a scanner¶¶¶ and crude data was extracted using software for microarray image analysis.
Statistical Analysis
Statistical analyses were performed using statistical package software ****. Full-mouth clinical measurements were computed for each subject and then averaged across subjects within the 3 clinical groups. Differences in clinical parameters among groups were sought using Kruskal-Wallis, Mann-Whitney and Chi-square tests. The microbial data were generated from image files of scanned arrays using a HOMIM online analysis tool.35 The detection of a particular species in a sample was determined by the presence of a fluorescent spot for that unique probe. Qualified and adjusted spots of the same probe were then summed and a mean intensity calculated to represent the signal intensity for each specific probe. Signals were normalized by comparing individual signal intensities to the average of signals for the universal probes. The normalized signal intensities were raised to the power of 0.3 and the maximal transformed intensity was used to determine the range of the signal levels. In HOMIM, any original signal < 2 times the background value was reset to 1 and was assigned to the signal level 0 (no signal for the corresponding probe). All the values > 1 were categorized into scores 1 to 5, corresponding to different signal levels. The prevalence of each species (frequency of scores 0 to 5) was computed for each subject, and averaged within groups. Significant differences among groups were sought using the Kruskal-Wallis and Mann-Whitney tests, whereas differences between healthy sites or sites with attachment gain, and sites that lost attachment were analysed by Chi-square test. Any difference of p < 0.05 was considered significant.
Results
The demographic and baseline periodontal clinical features of the subject groups are shown in Table 1, and the baseline clinical data of the sites sampled for microbial analysis are presented in Table 2. The PH group presented lower proportions of males and had younger individuals than the periodontitis groups (p < 0.01, Chi-square test). A higher frequency of smokers was observed in the RP group compared to GR (p < 0.01, Chi-square test). No significant difference among groups was observed for race (Table 1). All clinical parameters of periodontal tissue destruction and inflammation, except SUP were significantly greater in the periodontitis groups compared to controls (p < 0.01; Mann-Whitney test). Moreover, RP patients showed higher mean CAL than GR (p < 0.01, Mann-Whitney test) (Tables 1 and 2).
Table 1.
Demographic and full-mouth baseline clinical parameters (mean ± SD) of the study population.
| Clinical Parameters |
Periodontally Healthy (N = 20) |
Good Responders (N = 30) |
Refractory Periodontitis (N = 17) |
|
|---|---|---|---|---|
| Age (years) * | 34 ± 12 | 48 ± 10 | 51 ± 11 | |
| % Males † | 20 | 60 | 82 | |
| % Smokers † § | 0 | 23 | 59 | |
| % Race | White | 75 | 40 | 53 |
| Black | 10 | 43 | 24 | |
| Hispanic | 15 | 3.5 | 12 | |
| Asians | 0 | 6.5 | 6 | |
| Others | 0 | 7 | 5 | |
| Probing depth (mm) * | 2.0 ± 0.2 | 3.7± 1.3 | 3.9 ± 0.9 | |
| Clinical attachment level (mm) * ‡ | 1.3 ± 0.5 | 3.2 ± 1.5 | 4.2 ± 1.2 | |
| N of missing teeth * | 0.7 ± 1.5 | 2.1 ± 2.3 | 3.3 ± 2.8 | |
| % of sites with | ||||
| Bleeding on probing * | 11 ± 6.5 | 56 ± 25 | 53 ± 28 | |
| Supragingival biofilm * | 30 ± 22 | 46 ± 30 | 65 ± 29 | |
| Gingival Redness * | 26 ± 18 | 85 ± 20 | 88 ± 18 | |
| Suppuration | 0 | 0.22 ± 0.7 | 0.33 ± 0.6 | |
p < 0.01, Kruskal-Wallis test;
p < 0.01, Chi-square test;
< 0.01, Mann-Whitney test between refractory and good responders;
p < 0.01, Chi-square test between refractory and good responders
Table 2.
Baseline clinical parameters of the sites sampled for microbial analysis from the three subject groups.
| Clinical Parameters | Periodontally Healthy (N = 91) |
Good Responders (N = 143) |
Refractory Periodontitis (N = 111) |
|---|---|---|---|
| Mean (± SD) Probing depth (mm) * † | 2.2 ± 0.5 | 4.3± 2.2 | 4.9 ± 2 |
| Mean (± SD) Clinical attachment level (mm) * † ‡ | 1.4 ± 0.7 | 3.7 ± 2.4 | 4.7 ± 2.6 |
| % Bleeding on probing § ║ | 18.7 | 63.6 | 59.5 |
| % Supragingival biofilm § ║ | 27.5 | 47.6 | 67.4 |
| % Gingival Redness § ║ | 35.2 | 83.0 | 85.6 |
| % Suppuration | 0 | 2.8 | 0 |
p < 0.01, Kruskal-Wallis test;
p < 0.01, Mann-Whitney test between periodontally healthy and refractory or periodontally healthy and good responders;
p < 0.01, Mann-Whitney test between refractory and good responders;
p < 0.01, Chi-square test;
p < 0.01, Chi-square test between periodontally healthy and refractory or periodontally healthy and good responders;
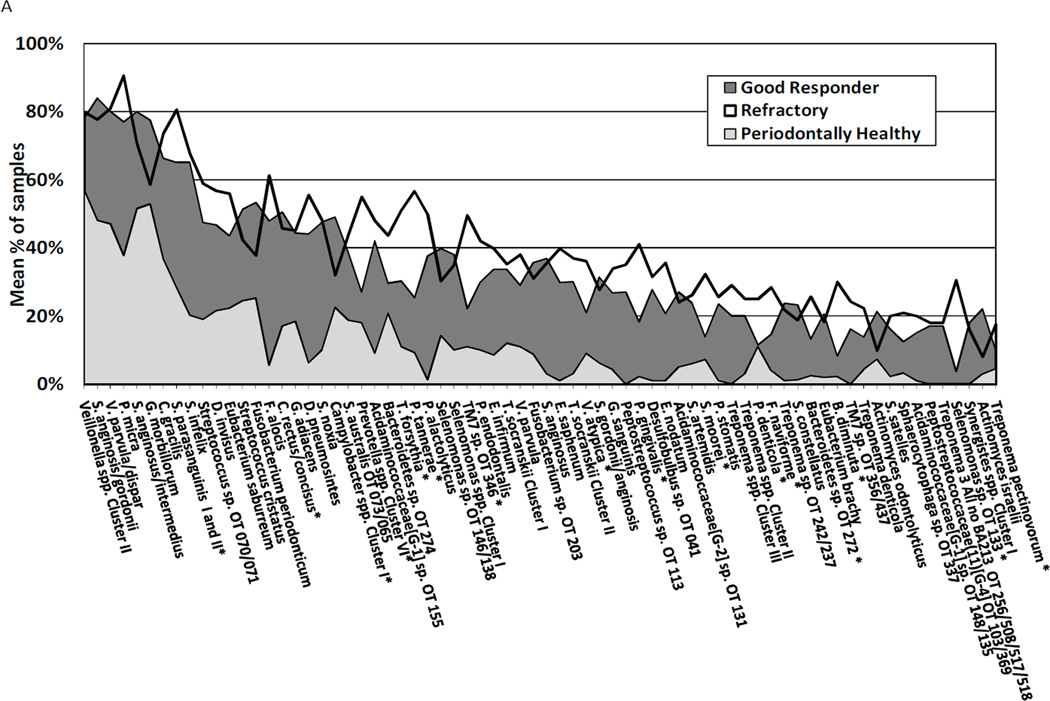
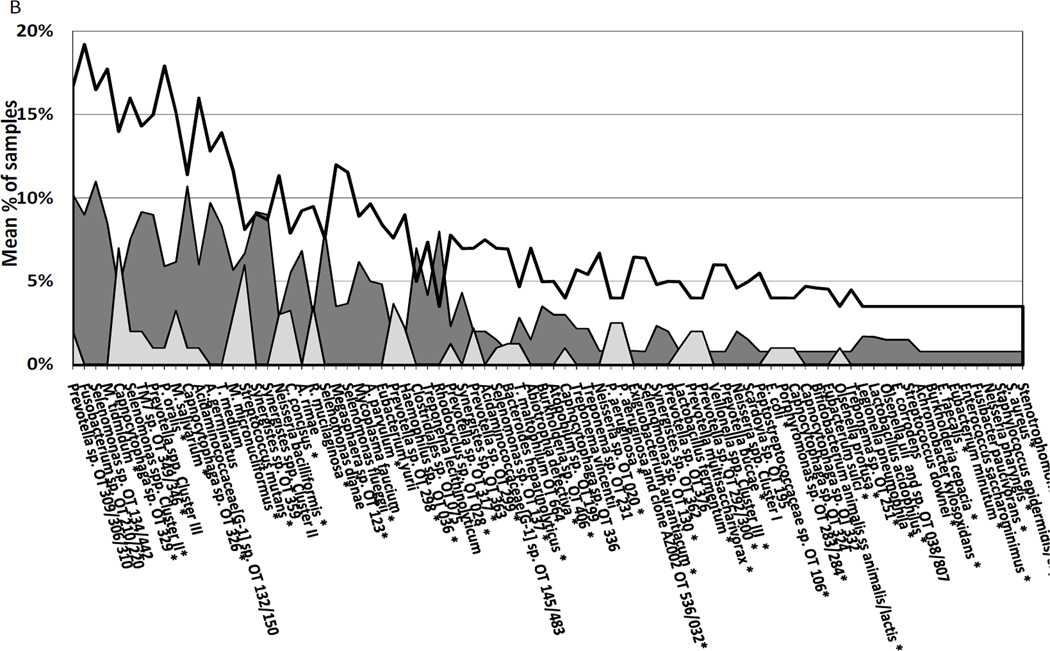
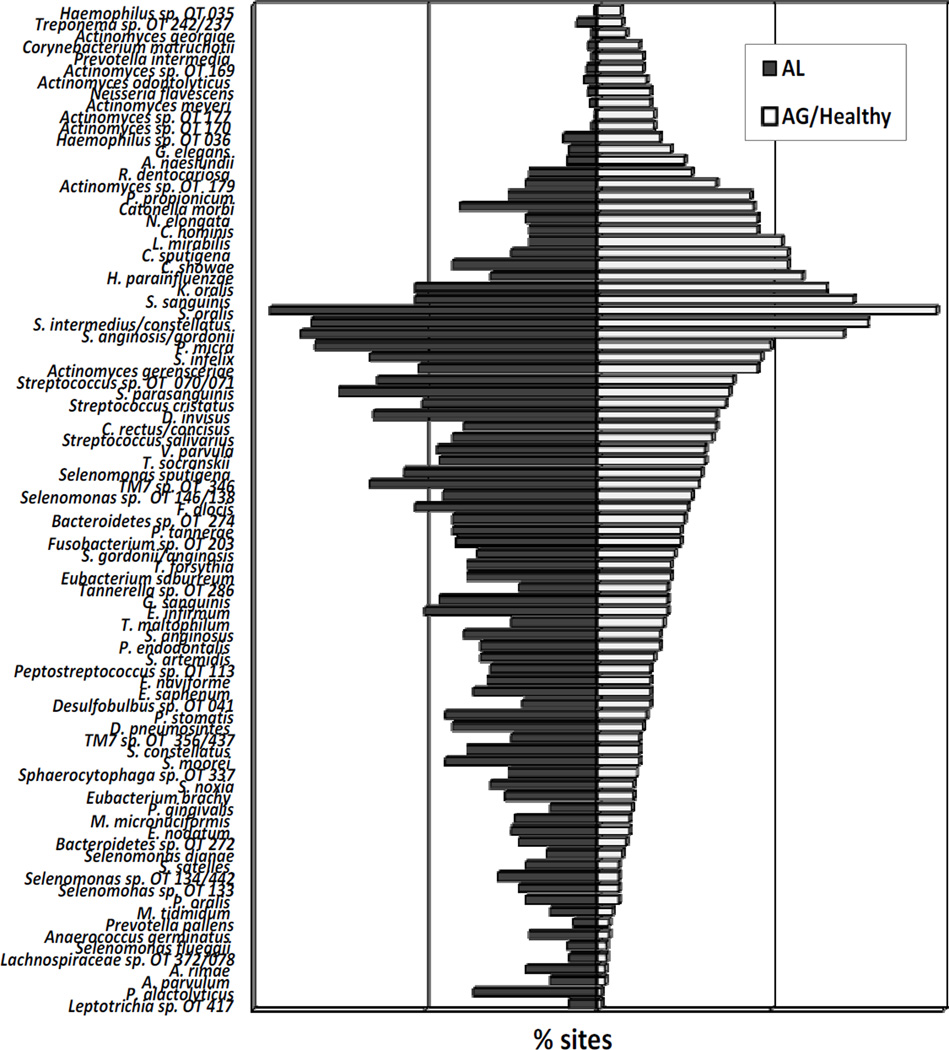
The microbiological profiles of the three subject groups at baseline are presented in Figures 2A and 2B. Overall, the most prevalent species/clusters (>50% of all samples) were Streptococcus spp. clusters II/III (oral taxa [OT] 758/755/071/768/767/745/734/728/721/707), Streptococcus intermedius/anginosis, Streptococcus intermedius/constellatus, Streptococcus anginosis/gordonii, Streptococcus anginosus/intermedius, Streptococcus sanguinis, Streptococcus parasanguinis, Streptococcus oralis, Gemella morbillorum, Fusobacterium nucleatum ss polymorphum, Leptotrichia spp. cluster I (OT 563/498/463/462), Veillonella parvula/dispar, Veillonella spp. cluster II (OT 524/161/160), Capnocytophaga granulosa/sputigena, Parvimonas micra, Actinomyces spp. cluster I (OT 708/701/688/671), Campylobacter rectus/concisus, Campylobacter gracilis, Campylobacter showae, Dialister invisus, Rothia dentocariosa/mucilaginosa, Kingella oralis, Selenomonas infelix (data not shown). The majority of the species evaluated were more frequently detected in samples from diseased than from PH subjects. Approximately 28% of all species/genera/clones were not detected in any sample from PH individuals. RP subjects harbored significantly higher frequencies of putative periodontal pathogens, as well as microorganisms not usually associated with periodontitis in comparison to GR and/or PH individuals. Periodontal pathogens significantly prevalent in RP subjects included P. micra, C. gracilis, Tannerella forsythia, Prevotella spp., Eubacterium nodatum, Selenomonas noxia, Porphyromonas gingivalis, Treponema spp., Eikenella corrodens, and Campylobacter concisus. Among the “unusual” species, the most detected ones in RP subjects were S. parasanguinis I and II, S. infelix, Selenomonas artemidis, Selenomonas spp. OT 133/134/442/478/149/136, Streptotoccus sp. OT 070/071, Filifactor alocis, Dialister invisus, Dialister pneumosintes, Granulicatella adiacens, Streptococcus australis OT 073/065, Acidaminococcaceae [G-1] spp. OT 155/148/135, Bacteroidetes sp. OT 272/274, Pseudoramibacter alactolyticus, TM7 spp. OT 356/437, Eubacterium infirmum, Eubacterium saphenum, Veillonella atypica, Gemella sanguinis, Peptostreptococcus stomatis, Solobacterium moorei, Desulfobulbus sp. OT 041, Fusobacterium naviforme, Sphaerocytophaga sp. OT 337, Brevundimonas diminuta, Shuttleworthia satelles, (Figure 2A). Other bacterial species less frequently detected (<10% of samples) are depicted in figure 2B. The only species significantly more prevalent in PH than periodontitis patients were Actinomyces sp. oral taxon 170, oral clone AP064, Actinomyces Cluster I (oral taxa 708, 701, 688, 671), Capnocytophaga sputigena, Cardiobacterium hominis, Haemophilus parainfluenzae, Lautropia mirabilis, Propionibacterium propionicum, Rothia dentocariosa/mucilagenosa and Streptococcus sanguinis (p < 0.05, Mann-Whitney test) (Table 3). Comparisons between the subgingival microbiota of sites that lost attachment and healthy sites or sites that gained attachment were also performed (Figure 3). In this analysis, baseline samples from PH and post-therapy samples from periodontitis patients were included. Likewise, a high prevalence of periodontal pathogens and novel species, particularly S. intermedius/constellatus, Streptococcus anginosus, P. micra, Selenomonas spp., S. parasanguinis, Streptococcus sp. OT 070/071, F. alocis, D. invisus, D. pneumosintes, C. rectus/concisus, TM7 spp. OT 346/356/437, T. socranskii, T. maltophilum, Bacteroidetes sp. OT 274/272, Prevotella tannerae, T. forsythia, Eubacterium spp., G. sanguinis, Porphyromonas endodontalis, Peptostreptococcus sp. OT 113, Desulfobulbus sp. OT 041, P. stomatis, S. moorei, Sphaerocytophaga sp. OT 337, P. gingivalis, M. micronuciformis, S. satelles, Prevotella oralis, M. tidmidum, A. germinatus, A. rimae, A. parvulum, P. alactolyticus was observed significantly more often in sites losing attachment than sites gaining attachment or with no disease (p < 0.05, Chi-square test). In contrast, S. sanguinis, H. parainfluenza, C. sputigena, C. hominis, L. mirabilis, Neisseria elongata, P. propionicum, R. dentocariosa, Actinomyces sp. OT 179, Actinomyces naeslundii, K. oralis, Granulicatella elegans were highly prevalent in healthy sites or sites with attachment gain (Figure 3).
Figure 2.
A and B. Mean frequency of bacterial species significantly different (p<0.05; Kruskal-Wallis test) among refractory, good responders and periodontally healthy subjects at baseline. (A) Represents the species that were detected in >10% of all samples. (B) Represents the species that were detected in <10% of all samples. OT means oral taxon designation. *Refers to significant differences between good responders and refractory subjects (p<0.05; Mann-Whitney test).
Table 3.
Mean frequency (±SD) of bacterial species detected significantly more often in subgingival plaque samples from periodontally healthy than periodontitis subjects.
| Species | Periodontally Healthy |
Good Responders |
Refractory Periodontitis |
p |
|---|---|---|---|---|
| Actinomyces sp oral taxon 170 | 33±29% | 9±18% | 6±13% | 0.001 |
| Actinomyces Cluster I (oral taxa 708, 701, 688, 671) | 75±33% | 71±30% | 45±29% | 0.049 |
| Capnocytophaga sputigena | 70±36% | 40±35% | 38±32% | 0.012 |
| Cardiobacterium hominis | 63±28% | 32±31% | 23±28% | 0.0001 |
| Haemophilus parainfluenzae | 73±26% | 33±35% | 36±39% | 0.001 |
| Lautropia mirabilis | 59±38% | 36±35% | 30±35% | 0.026 |
| Propionibacterium propionicum | 55±35% | 32±31% | 22±20% | 0.011 |
| Rothia dentocariosa/mucilaginosa | 78±31% | 47±32% | 49±26% | 0.004 |
| Streptococcus sanguinis | 79±20% | 51±35% | 58±30% | 0.012 |
Figure 3.
Frequency of detection of bacterial species significantly different (p<0.05; Chi square test) among sites that lost attachment, healthy sites or sites that gained attachment. OT means oral taxon designation.
Discussion
The ultimate goal of characterizing the microbiota of different forms of periodontal diseases is to discriminate individuals at greater risk for treatment failure, and consequently, continuous periodontal attachment loss. In order to achieve that, it is important first to distinguish among various microbial profiles and determine which may be considered “normal” or “disease-related”. The purpose of the present study was to discriminate subjects with refractory periodontitis from individuals who were successfully treated or who were periodontally healthy based upon their baseline subgingival microbial profiles. The existence of refractory periodontal disease has been controversial for many years. One of the reasons proposed to explain the poor response of “refractory” subjects to conventional therapies is the predominance of an “unusual” virulent periodontal microbiota. Studies have shown that the microbiota of refractory patients is generally similar to that of chronic periodontitis subjects, although some differences have been described.18, 22, 23, 28 Most of these studies, however, have examined a particular segment of the subgingival periodontal microbiota, so that microbial data on these subjects have been limited to some bacterial species. In the current investigation, we expanded the battery of species to be evaluated by using the HOMIM method.30 This technique allows the detection of about 300 species, including both cultivable and not yet-cultivable species.4 Our data showed that periodontitis patients presented a greater diversity in the subgingival microbiota compared to PH individuals at baseline. All species or not yet cultivated species evaluated were detected in at least one sample from periodontitis patients, whereas approximately 28% of the species tested were not detected in any sample from healthy subjects. Microorganisms that were significantly more prevalent in PH compared to periodontitis patients included Actinomyces spp. oral taxa 708/701/688/671/170, C. sputigena, C. hominis, H. parainfluenzae, L. mirabilis, P. propionicum, R. dentocariosa/mucilagenosa, and S. sanguinis. Previous studies have correlated the high frequency and levels of at least some of these species with periodontal health and/or clinical improvement after periodontal treatment.2, 3, 8, 10, 11, 32, 36 Of interest, Socransky et al. 28 demonstrated that refractory individuals exhibited a lower prevalence of Actinomyces species and S. sanguinis compared to PH, well-maintained elderly and periodontitis subjects. Furthermore, the species S. sanguinis and C. sputigena were shown to be in significantly lower counts in refractory than untreated periodontitis subjects.27 Information regarding the association between L. mirabilis and C. hominis and periodontal status is scarce. These traditional gram-negative species are considered normal human oropharyngeal microbiota and are rarely the cause of human infection.37, 38 L. mirabilis has been isolated from the oral cavity of HIV-infected children 39 and patients with respiratory infections.37 C. hominis is notorious for causing apparently culture-negative endocarditis, as are the other members of the HACEK group (i.e., Haemophilus spp., Aggregatibacter actinomycetemcomitans, Eikenella corrodens, and Kingella kingae).40
Among periodontitis patients, putative periodontal pathogens were found more commonly in RP than in GR (Fig. 2A, 2B). Likewise, sites that lost attachment had a greater prevalence of these pathogens than healthy sites or sites that gained attachment (Fig. 3). In particular, species such as P. micra, T. forsythia, Prevotella spp., P. gingivalis, E. nodatum, Eubacterium spp., Selenomonas spp. and Treponema spp. were remarkably more prevalent in RP than the other two groups, corroborating the data reported by other investigators.18–21, 41 Classical periodontal pathogens were less frequently observed in the refractory patients of Magnusson et al.,22 Colombo et al. 23 and Haffajee et al. 27 studies. These authors suggested that this observation may have resulted from repeated treatments that these refractory subjects had received prior to inclusion in the studies. In the present investigation, referred refractory periodontitis individuals were not included. In addition, subjects who had received systemic antibiotics or periodontal therapy in the previous 6 months were excluded. The high prevalence of putative periodontal pathogens in our refractory patients may also reflect the significantly greater mean attachment level and proportion of smokers in this group compared to GR at baseline (Table 2). There is accumulating evidence that smokers have a higher level of periodontal destruction, less reduction of periodontal pathogens and a poorer response to periodontal treatment than non-smokers.42, 43 Refractory periodontitis has also been associated with smoking.44 Regarding the influence of smoking in the subgingival microbiota, some investigators found no differences in the prevalence or counts of periodontal pathogens between smokers and non-smokers,45, 46 while other studies clearly did.47–49 The reasons for these discrepancies in colonization are not clear; however, some of these authors suggest that cigarette smoke could directly affect the pathogens or their microenvironment, as well as the host’s ability to control the infection. As a consequence, elimination or control of species would be more difficult, increasing the risk for treatment failure.
Although differences among studies do exist, there is a consensus that refractory subjects are heterogeneous in terms of their subgingival microbiota.18, 23, 28 For instance, Socransky et al. 28 reported 4 cluster groups with distinct microbial profiles in a group of refractory subjects. The predominant cluster exhibited high counts and proportions of members of the orange and red complexes (particularly T. forsythia), and low proportions of Actinomyces spp. and Veillonella parvula. A second cluster was comprised of high proportions of orange complex species and very low total bacterial counts. A third cluster was marked by high proportions of Actinomyces spp. and V. parvula and moderate levels of red and orange complex species. The fourth refractory cluster was dominated by Streptococcus species. Members of the “milleri” streptococci group (S. anginosus, S. constellatus and S. intermedius) have been implicated in refractory forms of periodontitis by other investigators.18, 22, 23 In the present study, the baseline distribution of S. anginosus, S. constellatus and S. anginosus/intermedius was similar between GR and RP individuals (Fig. 2A). On the other hand, these species were detected significantly more often in sites that lost attachment compared to healthy sites or sites that gained attachment after therapy (Fig. 3). Therefore, it is possible that periodontal treatment, particularly SRP plus systemically administered amoxicillin and metronidazole, selected a microbiota dominated by Streptococcus species, as may be seen in some RP subjects.10, 11, 22, 28, 32
Data from various investigations indicate that classical periodontal pathogens such as the red and orange complex members may play a role in the pathogenesis of RP.18–21 Thus, lowering the levels and proportions of these pathogenic species may control disease progression and provide long term stability of the periodontium in most of these individuals.27 Nevertheless, there are still some patients who present a modest response to therapy, even after reduction of these putative pathogens. It has been hypothesized that these RP subjects either have a poor host response to infection, high burden of other virulent species, or both. Indeed, we were able to show, by the HOMIM technique, that RP individuals do harbor high frequency of several species not commonly associated with periodontitis at baseline. Among them are TM7 spp., Bacteroidetes sp., V. atypica, Desulfobulbus sp., B. diminuta, Sphaerocytophaga sp., S. satelles, S. australis, G. sanguinis, M. tidmidum, M. salivarium, A. geminatus, M. micronuciformis, S. parasanguinis, S. infelix, S. artemidis, G. adiacens, Acidaminococcaceae [G-1] spp., E. infirmum, E. saphenum, P. stomatis, F. naviforme, T. medium (Fig. 2A). Few studies have reported on the relationship of these microorganisms and periodontal diseases.2, 4, 9, 50 Many of these species cannot yet be grown or are difficult to grow in culture. Moreover, some of them act as opportunist pathogens in immunocompromised patients.51, 52 Although in low frequency, known bacterial pathogens including E. coli, S. aureus and P. aeruginosa were also more predominant in RP than GR and PH subjects. Likewise, other investigators have found an association of these species with periodontitis.21, 23, 53–56 Other microorganisms frequently detected in RP and/or sites loosing attachment were P. alactolyticus, a species associated with endodontic infections,57 S. moorei, a species associated with halitosis,58 and F. alocis, D. pneumosintes and D. invisus, slow growing anaerobic microorganisms commonly associated with primary and persistent endodontic infections,59 as well as different forms of periodontitis.4, 9, 50, 60
The role that these species play in the initiation and/or progression of RP; the effect of periodontal therapy on this “unusual” microbiota; and the interactions between these species and oral microorganisms are unknown. One could speculate that SRP combined with systemic antibiotics may be very effective against the pathogenic oral microbiota, but it may not be effective against “unusual” species, resulting in the overgrowth of these microorganisms and persistence of periodontal destruction. Further studies evaluating different microbial profiles of periodontitis patients, as well as the effect of periodontal treatments on these profiles will be required to answer these questions.
Supplementary Material
Acknowledgments
This work was supported by NIDCR grants DE11443 and RR025771; and by National Council for Scientific and Technological Development (CAPES), Brazil.
Sources of support: Supported by NIDCR grants DE11443 and RR025771; CAPES, Brazil. The authors report no conflicts of interest related to this study.
Summary of findings: The subgingival microbiota of refractory periodontitis (RP), successfully treated and periodontally healthy individuals were compared by HOMIM. RP subjects presented a highly diverse microbiota, including high frequencies of putative periodontal pathogens and novel species. The role of these “unusual” species in the pathogenesis of refractory disease needs further investigation.
Footnotes
The authors report no conflicts of interest related to this study.
Hu-Friedy, Chicago, IL
Sigma-Aldrich, Inc., St. Louis, MO
Promega, Madison, WI
Invitrogen, Carlsbad, CA
Platinum® Taq DNA Polymerase High Fidelity, Invitrogen, Carlsbad, CA
Perkin-Elmer 9700, Applied Biosystems, Foster City, CA
SYBR ® Safe, Molecular Probes, Inc., Eugene, OR
QIAquick® PCR Purification Kit, Qiagen Inc., Valencia, CA
GE Healthcare Bio-Sciences Corp., Piscataway, NJ
Sigma Genosys, The Woodlands, TX
Schott-Nexterion® Slide AL, Louisville, KY
Hybex® Microarray Incubation System, SciGene Corporation, Sunnyvale, CA
Little Dipper™ Microarray Processor, SciGene Corporation, Sunnyvale, CA
Axon GenePix® 4000B, MDS Analytical Technologies, Sunnyvale, CA
Microarray Image Analysis GenePix Pro 6.0, MDS Analytical Technologies, Sunnyvale, CA
Statistical Package for the Social Sciences - SPSS Inc®.v.16 Chicago, IL, USA
References
- 1.Choi BK, Paster BJ, Dewhirst FE, Gobel UB. Diversity of cultivable and uncultivable oral spirochetes from a patient with severe destructive periodontitis. Infect Immun. 1994;62:1889–1895. doi: 10.1128/iai.62.5.1889-1895.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore WEC, Moore LH. The bacteria of periodontal diseases. Periodontol 2000. 1994;5:66–77. doi: 10.1111/j.1600-0757.1994.tb00019.x. [DOI] [PubMed] [Google Scholar]
- 3.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 4.Paster BJ, Boches SK, Galvin JL, et al. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paster BJ, Olsen I, Aas JA, Dewhirst FE. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol 2000. 2006;42:80–87. doi: 10.1111/j.1600-0757.2006.00174.x. [DOI] [PubMed] [Google Scholar]
- 6.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mager DL, Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Distribution of selected bacterial species on intraoral surfaces. J Clin Periodontol. 2003;30:644–654. doi: 10.1034/j.1600-051x.2003.00376.x. [DOI] [PubMed] [Google Scholar]
- 8.Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontol 2000. 2005;38:135–187. doi: 10.1111/j.1600-0757.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- 9.Kumar PS, Griffen AL, Barton JA, Paster BJ, Moeschberger ML, Leys EJ. New bacterial species associated with chronic periodontitis. J Dent Res. 2003;82:338–344. doi: 10.1177/154405910308200503. [DOI] [PubMed] [Google Scholar]
- 10.Haffajee AD, Teles RP, Socransky SS. The effect of periodontal therapy on the composition of the subgingival microbiota. Periodontol 2000. 2006;42:219–258. doi: 10.1111/j.1600-0757.2006.00191.x. [DOI] [PubMed] [Google Scholar]
- 11.Teles RP, Haffajee AD, Socransky SS. Microbiological goals of periodontal therapy. Periodontol 2000. 2006;42:180–218. doi: 10.1111/j.1600-0757.2006.00192.x. [DOI] [PubMed] [Google Scholar]
- 12.Lindhe J, Westfelt E, Nyman S, Socransky SS, Haffajee AD. Long-term effect of surgical/non-surgical treatment of periodontal disease. J Clin. Periodontol. 1984;11:448–458. doi: 10.1111/j.1600-051x.1984.tb01344.x. [DOI] [PubMed] [Google Scholar]
- 13.Caffesse RG, Sweenwy PL, Smith BA. Scaling and root planning with and without periodontal flap surgery. J Clin. Periodontol. 1986;13:205–210. doi: 10.1111/j.1600-051x.1986.tb01461.x. [DOI] [PubMed] [Google Scholar]
- 14.Hirschfeld L, Wasserman B. A long-term survey of tooth loss in 600 treated periodontal patients. J Periodontol. 1978;49:225–237. doi: 10.1902/jop.1978.49.5.225. [DOI] [PubMed] [Google Scholar]
- 15.MacFall WT., Jr Tooth loss in 100 treated patients with periodontal disease. A long-term study. J Periodontol. 1982;53:539–549. doi: 10.1902/jop.1982.53.9.539. [DOI] [PubMed] [Google Scholar]
- 16.Adams D. Diagnosis and treatment of refractory periodontitis. Curr Opin Dent. 1992;2:33–38. [PubMed] [Google Scholar]
- 17.Flemmig TF. Periodontitis. Ann Periodontol. 1999;4:32–8. doi: 10.1902/annals.1999.4.1.32. [DOI] [PubMed] [Google Scholar]
- 18.Haffajee AD, Socransky SS, Dzink JL, et al. Clinical, microbiological and immunological features of subjects with refractory periodontal diseases. J Clin Periodontol. 1988;15:390–398. doi: 10.1111/j.1600-051x.1988.tb01017.x. [DOI] [PubMed] [Google Scholar]
- 19.Choi JI, Nakagawa T, Yamada S, Takazoe I, Okuda K. Clinical, microbiological and immunological studies in recurrent periodontal disease. J Clin Periodontol. 1990;17:426–434. [PubMed] [Google Scholar]
- 20.Collins JG, Offenbacher S, Arnold RR. Effects of a combination therapy to eliminate Porphyromonas gingivalis in refractory periodontitis. J Periodontol. 1993;64:998–1007. doi: 10.1902/jop.1993.64.10.998. [DOI] [PubMed] [Google Scholar]
- 21.Listgarten MA, Lai CH, Young V. Microbial composition and pattern of antibiotic resistance in subgingival microbial samples from patients with refractory periodontitis. J Periodontol. 1993;64:155–161. doi: 10.1902/jop.1993.64.3.155. [DOI] [PubMed] [Google Scholar]
- 22.Magnusson I, Marks RG, Clark WB, Walker CB, Low SB, MacArthur WP. Clinical, microbiological and immunological characteristics of subjects with"refractory" periodontal disease. J Clin Periodontol. 1991;18:291–299. doi: 10.1111/j.1600-051x.1991.tb00431.x. [DOI] [PubMed] [Google Scholar]
- 23.Colombo AP, Haffajee AD, Dewhirst FE, et al. Clinical and microbiological features of refractory periodontitis subjects. J Clin Periodontol. 1998;25:169–180. doi: 10.1111/j.1600-051x.1998.tb02424.x. [DOI] [PubMed] [Google Scholar]
- 24.Colombo AP, Eftimiadi C, Haffajee AD, Cugini MA, Socransky SS. Serum IgG2 level, Gm(23) allotype and FcgammaRIIa and FcgammaRIIIb receptors in refractory periodontal disease. J Clin Periodontol. 1998;25:465–474. doi: 10.1111/j.1600-051x.1998.tb02475.x. [DOI] [PubMed] [Google Scholar]
- 25.Colombo AP, Sakellari D, Haffajee AD, Tanner A, Cugini MA, Socransky SS. Serum antibodies reacting with subgingival species in refractory periodontitis subjects. J Clin Periodontol. 1998;25:596–604. doi: 10.1111/j.1600-051x.1998.tb02493.x. [DOI] [PubMed] [Google Scholar]
- 26.Colombo AP, Haffajee AD, Smith CM, Cugini MA, Socransky SS. Discrimination of refractory periodontitis subjects using clinical and laboratory parameters alone and in combination. J Clin Periodontol. 1999;26:569–576. doi: 10.1034/j.1600-051x.1999.260902.x. [DOI] [PubMed] [Google Scholar]
- 27.Haffajee AD, Uzel NG, Arguello EI, Torresyap G, Guerrero DM, Socransky SS. Clinical and microbiological changes associated with the use of combined antimicrobial therapies to treat refractory periodontitis. J Clin Periodontol. 2004;31:869–877. doi: 10.1111/j.1600-051X.2004.00573.x. [DOI] [PubMed] [Google Scholar]
- 28.Socransky SS, Smith C, Haffajee AD. Subgingival microbial profiles in refractory periodontal disease. J Clin Periodontol. 2002;29:206–268. doi: 10.1034/j.1600-051x.2002.290313.x. [DOI] [PubMed] [Google Scholar]
- 29.Fine DH. Microbial identification and antibiotic sensitivity testing, an aid for patients refractory to periodontal disease. J Clin Periodontol. 1994;21:98–106. doi: 10.1111/j.1600-051x.1994.tb00286.x. [DOI] [PubMed] [Google Scholar]
- 30.Preza D, Olsen I, Willumsen T, et al. Microarray analysis of the microflora of root caries in elderly. Eur J Clin Microbiol Infect Dis. 2009 doi: 10.1007/s10096-008-0662-8. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haffajee AD, Socransky SS, Goodson JM. Comparison of different data analyses for detecting changes in attachment level. J Clin Periodontol. 1983;10:298–310. doi: 10.1111/j.1600-051x.1983.tb01278.x. [DOI] [PubMed] [Google Scholar]
- 32.Haffajee AD, Cugini MA, Dibart S, Smith C, Kent RL, Jr, Socransky SS. The effect of SRP on the clinical and microbiological parameters of periodontal diseases. J Clin Periodontol. 1997;24:324–334. doi: 10.1111/j.1600-051x.1997.tb00765.x. [DOI] [PubMed] [Google Scholar]
- 33.Human Oral Microbe Identification Microarray (HOMIM) [Accessed January 2009]. Available at: http://mim.forsyth.org/homim.html. [Google Scholar]
- 34.Human Oral Microbiome Database (HOMD) at The Forsyth Institute. [Accessed January 2009]. Available at: http://www.homd.org/ [Google Scholar]
- 35.Human Oral Microbe Identification Microarray (HOMIM) [Accessed March-December 2008]. Available at: http://bioinformatics.forsyth.org/homim/ [Google Scholar]
- 36.Darveau RP, Tanner A, Page RC. The microbial challenge in periodontitis. Periodontol 2000. 1997;14:12–32. doi: 10.1111/j.1600-0757.1997.tb00190.x. [DOI] [PubMed] [Google Scholar]
- 37.Gerner-Smidt P, Keiser-Nielsen H, Dorsch M, et al. Lautropia mirabilis gen. nov., sp. nov. a Gram-negative motile coccus with unusual morphology isolated from the human mouth. Microbiology. 1994;140:1787–1797. doi: 10.1099/13500872-140-7-1787. [DOI] [PubMed] [Google Scholar]
- 38.Wormser GP, Bottone EJ. Cardiobacterium hominis: review of microbiologic and clinical features. Rev. Infect. Dis. 1993;5:680–691. doi: 10.1093/clinids/5.4.680. [DOI] [PubMed] [Google Scholar]
- 39.Rossmann SN, Wilson PH, Hicks J, et al. Isolation of Lautropia mirabilis from Oral Cavities of Human Immunodeficiency Virus-Infected Children. J. Clin. Microbiol. 1998;36:1756–1760. doi: 10.1128/jcm.36.6.1756-1760.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Das M, Badley AD, Cockerill FR, Steckelberg JM, Wilson WR. Infective endocarditis caused by HACEK microorganisms. Annu. Rev. Med. 1997;48:25–33. doi: 10.1146/annurev.med.48.1.25. [DOI] [PubMed] [Google Scholar]
- 41.Winkel EG, vanWinkelhoff AJ, Timmerman MF, Vansted T, van der Velden U. Effects of metronidazole in patients with refractory periodontitis associated with Bacteroides forsythus. J Clin Periodontol. 1997;24:573–579. doi: 10.1111/j.1600-051x.1997.tb00231.x. [DOI] [PubMed] [Google Scholar]
- 42.Bergstrom J, Preber H. Tobacco use as a risk factor. J Periodontol. 1994;65:545–550. doi: 10.1902/jop.1994.65.5s.545. [DOI] [PubMed] [Google Scholar]
- 43.Darby IB, Hodge PJ, Riggio MP, Kinane DF. Clinical and microbiological effect of scaling and root planing in smoker and non-smoker chronic and aggressive periodontitis patients. J Clin Periodontol. 2005;32:200–206. doi: 10.1111/j.1600-051X.2005.00644.x. [DOI] [PubMed] [Google Scholar]
- 44.Macfarlane GD, Herzberg M, Wolff L, Hardie N. Refractory periodontitis associated with abnormal polymorphonuclear leukocyte phagocytosis and cigarette smoking. J Periodontol. 1992;63:908–913. doi: 10.1902/jop.1992.63.11.908. [DOI] [PubMed] [Google Scholar]
- 45.Stoltenberg JL, Osborn JB, Pihlstrom BL, et al. Association between cigarette smoking, bacterial pathogens, and periodontal status. J Periodontol. 1993;64:1225–1230. doi: 10.1902/jop.1993.64.12.1225. [DOI] [PubMed] [Google Scholar]
- 46.Boström L, Bergstrom J, Dahle´n G, Linder LE. Smoking and subgingival microflora in periodontal disease. J Clin Periodontol. 2001;28:212–219. doi: 10.1034/j.1600-051x.2001.028003212.x. [DOI] [PubMed] [Google Scholar]
- 47.Zambon JJ, Grossi SG, Machtei EE, Ho AW, Dunford R, Genco RJ. Cigarette smoking increases the risk for subgingival infection with periodontal pathogens. J Periodontol. 1996;67:1050–1054. doi: 10.1902/jop.1996.67.10s.1050. [DOI] [PubMed] [Google Scholar]
- 48.Kamma JJ, Nakou M, Baehni PC. Clinical and microbiological characteristics of smokers with early onset periodontitis. J Periodont Res. 1999;34:25–33. doi: 10.1111/j.1600-0765.1999.tb02218.x. [DOI] [PubMed] [Google Scholar]
- 49.Haffajee AD, Socransky SS. Relation of cigarette smoking to the subgingival microbiota. J Clin Periodontol. 2001;28:377–388. doi: 10.1034/j.1600-051x.2001.028005377.x. [DOI] [PubMed] [Google Scholar]
- 50.Hutter G, Schlagenhauf U, Valenza G, et al. Molecular analysis of bacteria in periodontitis: evaluation of clone libraries, novel phylotypes and putative pathogens. Microbiology. 2003;149:67–75. doi: 10.1099/mic.0.25791-0. [DOI] [PubMed] [Google Scholar]
- 51.Arora U, Kaur S, Devi P. Ochrobactrum anthropi septicaemia. Indian J Med Microbiol. 2008;26:81–83. doi: 10.4103/0255-0857.38868. [DOI] [PubMed] [Google Scholar]
- 52.Han XY, Andrade RA. Brevundimonas diminuta infections and its resistance to fluoroquinolones. J Ant Chemoth. 2005;55:853–859. doi: 10.1093/jac/dki139. [DOI] [PubMed] [Google Scholar]
- 53.Colombo AP, Teles R, Torres MC, Souto RM, Mendes MCS, Rosalem JRW, Uzeda M. Subgingival microbiota of Brazilian Subjects with untreated chronic periodontitis. J Periodontol. 2002;73:360–369. doi: 10.1902/jop.2002.73.4.360. 24. [DOI] [PubMed] [Google Scholar]
- 54.Gonçalves LS, Ferreira SMS, Souza CO, Souto RM, Colombo APV. Clinical and microbiological profiles of HIV-seropositive undergoing HAART, and HIV-seronegative Brazilians with chronic periodontitis. J Periodontol. 2007;78:87–96. doi: 10.1902/jop.2007.060040. [DOI] [PubMed] [Google Scholar]
- 55.Slots J, Rams TF, Listgarten MA. Yeasts, enteric rods and pseudomonads in the subgingival flora of severe adult periodontitis. Oral Microbiol Immunol. 1988;3:47–52. doi: 10.1111/j.1399-302x.1988.tb00080.x. [DOI] [PubMed] [Google Scholar]
- 56.Souto R, Colombo APV. Prevalence of Enterococcus faecalis in subgingival biofilm and saliva of subjects with chronic periodontal infection. Arch Oral Biol. 2008;53:155–160. doi: 10.1016/j.archoralbio.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 57.Siqueira JF, Jr, Rôças IN. Pseudoramibacter alactolyticus in primary endodontic infections. J Endodon. 2003;29:735–738. doi: 10.1097/00004770-200311000-00012. [DOI] [PubMed] [Google Scholar]
- 58.Haraszthy VI, Gerber D, Clark B, et al. Characterization and prevalence of Solobacterium moorei associated with oral halitosis. [Accessed January 16, 2009];J Breath Res. 2008 Mar;2(1) doi: 10.1088/1752-7155/2/1/017002. [serial online]. doc. 017002. [DOI] [PubMed] [Google Scholar]
- 59.Siqueira JF, Jr, Rôças IN. Polymerase chain reaction-based analysis of microorganisms associated with failed endodontic treatment. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2004;97:85–94. doi: 10.1016/s1079-2104(03)00353-6. [DOI] [PubMed] [Google Scholar]
- 60.Ferraro CT, Gornic C, Barbosa AS, et al. Detection of Dialister pneumosintes in the subgingival biofilm of subjects with periodontal disease. Anaerobe. 2007;13:244–248. doi: 10.1016/j.anaerobe.2007.09.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.