Abstract
The development of the musculoskeletal system is a complex process that involves very precise control of bone formation and growth as well as remodeling during postnatal life. Although the understanding of the transcriptional mechanisms of osteogenesis has increased considerably, the molecular regulatory basis, especially the gene regulatory network of osteogenic differentiation, is still poorly understood. This review provides the reader with an overview of the key transcription factors that govern bone formation, highlighting their function and regulation linked to Runt-related transcription factor 2 (Runx2). Runx2 as the master transcription factor of osteoblast differentiation, Twist, Msh homeobox 2 (Msx2), and promyelocytic leukemia zinc-finger protein (PLZF) acting upstream of Runx2, Osterix (Osx) acting downstream of Runx2, and activating transcription factor 4 (ATF4) and zinc-finger protein 521 (ZFP521) acting as cofactors of Runx2 are discussed, and their relevance for tissue engineering is presented. References are provided for more in-depth personal study.
Introduction
The development of the musculoskeletal system is a complex process that involves very precise control of bone formation, growth, and remodeling during postnatal life. Bone consists of a dense organic matrix and an inorganic, mineral component. During embryonic development, bone develops in two ways: by intramembranous ossification and endochondral ossification. Correspondingly, there are two types of bone: intramembranous bones or flat bones such as the skull and pelvis, and endochondral bones, which include the long, short, and irregular bones. Bone is formed and maintained through a tightly controlled balance between osteoblasts and osteoclasts, the former synthesizing the mineralized extracellular matrix, the latter absorbing the bone.
The process of intramembranous ossification begins with mesenchymal stem cells (MSCs), and the bone tissue is generated by the development of MSCs into osteoblasts and osteocytes. The osteocytes are the most numerous cells in mature bone, which reside in a mineralized extracellular matrix.1 So far, the process of osteocytogenesis is largely unknown. A cartilage intermediate is not involved during intramembranous ossification. Bone formation by endochondral ossification is a complex phenomenon whereby recruitment, replication, and condensation of mesenchymal precursors occur at sites of future skeletal elements. Within these condensations, cells differentiate into chondrocytes followed by replacement by bone.2 The process involves preosteoblasts, osteoblasts, mature osteoblasts, and ultimately the accumulation and mineralization of the extracellular matrix. A number of factors influence development, growth, and repair of bone. The mediators involved in osteogenesis include transcription factors, growth factors, cytokines, metabolites, hormones, mechanical loading, and aging. Hormones are involved in bone formation. Estrogen deficiency causes bone loss and osteoporosis by increasing the generation and activity of osteoclasts.3 Excessive glucocorticoid also results in osteoporosis, which is a major cause for bone fractures in the elderly, by decreasing bone formation4 and promoting bone resorption.5 These data show that glucocorticoid signaling is required for normal bone formation. A recent report showed that Runt-related transcription factor 2 (Runx2) played a role in glucocorticoid-mediated BIM induction and apoptosis of leukemia cells.6 Studies by Phillips et al.7 also showed that the coordinated action of dexamethasone and the osteogenic transcription factor Runx2/Cbfa1 synergistically increased osteogenic gene expression, including osteocalcin (OC), bone sialoprotein (BSP), and alkaline phosphatase (ALP), and biological mineral deposition in primary dermal fibroblasts.
Major advances in the genetics of skeletogenesis have been occurred over past 15 years, especially in the identification of genetic factors regulating osteogenesis. However, the gene regulatory network (GRN) of osteoblast differentiation still remains poorly understood. Understanding the transcription factors affecting osteogenesis and related GRN is crucial to harness the inherent regenerative potential of skeletal tissues for possible use for gene therapy in bone repair and regeneration. This review provides the reader with an overview of the transcription factors linked to Runx2, the master transcription factor of osteoblast differentiation, highlighting the transcriptional cascade of osteogenesis involved in bone formation and remodeling. The master transcription factor of osteoblast differentiation Runx2, its upstream factors Twist, Msh homeobox 2 (Msx2), and promyelocytic leukemia zinc-finger protein (PLZF), downstream factor osterix (Osx), and coactivators activating transcription factor 4 (ATF4) and zinc-finger protein 521 (ZFP521) are discussed in the review. References are provided for more in-depth personal study.
Runx2, the Master Transcription Factor for Osteogenesis
Runx2 is a transcription factor that belongs to the runt homology domain protein family, which contains a glutamine-/alanine-rich domain at its N-terminal end (a runt domain) and a proline/serine/threonine (PST)-rich region at the C-terminus. Runx2 is also called core-binding factor-alpha (CBFA1), Acute Myeloid Leukemia 3 (AML3), PEBP2alphaA, and osteoblast-specific factor 2 (OSF2). To unify the naming system for this exciting class of transcription factors and facilitate cross-referencing of articles, the Nomenclature Committee of the Human Genome Organization (HUGO) adopted the use of the term RUNX to refer to genes encoding the runt-related proteins in November 1999. Runx2 is expressed in cells prefiguring the skeleton as early as E10.5,8 at which stage cells still have the capacity to differentiate into osteoblasts or chondrocytes. Runx2 is expressed in the osteoblast lineage,9 hypertrophic chondrocytes,10 odontoblasts, and ameloblasts.11 Runx2 decreases and eventually vanishes at E16.5 in differentiating chondrocytes.10
In vitro and vivo studies show that Runx2 is an essential transcription factor for osteoblast differentiation, matrix production, and mineralization during bone formation. Runx2 controls the expression of major bone matrix protein genes through a direct binding site called osteoblast-specific cis-acting element (OSE2), which is present in the promoter of several osteoblast-specific genes such as OC,9 osteopontin (OPN),9 BSP,9 and collagen, type I, alpha 1 (Col1A1).9,12 Runx2 also directly regulates cranisynostosis-associated gene NEL-like 1 (NELL1),13 skeletal tissue-enriched gene Pannexin 3 (PANX3),14 and zinc-dependent endopeptidases matrix metallopeptidase 9 (MMP9)15 and MMP13.16
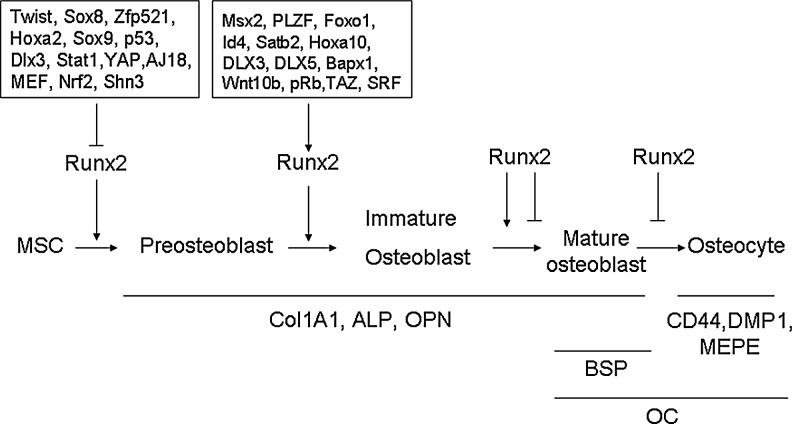
Runx2 binds OSE2 in the promoter to initiate mesenchymal condensations of the developing skeleton and regulate osteogenesis. In immature stage, Runx2 enhances osteogenesis. However, Runx2 is not essential to maintain the expression of the major bone matrix protein genes in mature osteoblasts and needs to be suppressed to form mature bone17 (Figs. 1 and 2). Overexpression of Runx2 in adipose tissue-derived MSCs inhibits adipogenesis as demonstrated by decreased lipoprotein lipase and peroxisome proliferator-activated receptor (PPARγ) expression and reduced lipid droplet formation. In osteogenesis, Runx2-overexpressing adipose tissue-derived MSCs undergo rapid and marked osteoblast differentiation as determined by osteoblastic gene expression, ALP activity, and mineral deposition.18 Even forced expression of Runx2 in nonosteoblastic cells induces the expression of the major osteoblast-specific genes.9 Runx2 is also a critical transcription factor that promotes chondrocyte maturation (Fig. 1). The formation of hypertrophic chondrocytes is severely impaired in some skeletal elements in Runx2-knockout mice.19 Expression of Runx2 in nonhypertrophic Col2a1-expressing chondrocytes accelerates chondrocyte differentiation and partially rescues the chondrocyte phenotype in Runx2 knockout mice.20
FIG. 1.
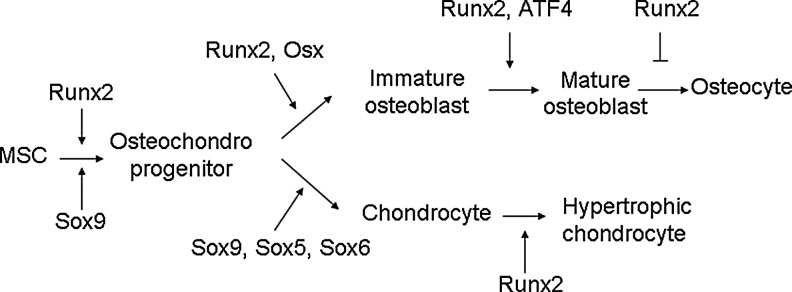
Role of transcription factors in cell differentiation along the osteoblast and chondrocyte lineage. Runt-related transcription factor 2 (Runx2) is an essential transcription factor of early osteoblast differentiation as the master gene of bone formation. However, Runx2 is not essential for late stage of osteoblast differentiation and needs to be suppressed for terminal differentiation into osteocytes. Runx2 is also required for chondrocyte maturation.
FIG. 2.
Role and delicate regulation of Runx2 during osteoblast differentiation. Runx2 is essential for skeletal development. In osteoblast differentiation, Runx2 expression is detected in preosteoblasts expressing type I collagen weakly and is upregulated in immature osteoblasts expressing osteopontin. However, Runx2 expression is downregulated in mature osteoblasts expressing osteocalcin (OC). As the osteoblast transitions to an osteocyte expressing secreted proteins CD44, DMP1, and MEPE, alkaline phosphatase is reduced, whereas OC is elevated. The expression and transcriptional activity of Runx2 are tightly regulated by multiple proteins during osteoblast differentiation.
Runx2 expression level is important for normal bone development. Decreased Runx2 expression results in abnormal bone development. Mice with a homozygous mutation in Runx2 died just after birth without breathing, and analysis of their skeletons revealed lack of ossification of bone due to the maturational arrest of osteoblasts characterized by lack of expression of a later-stage marker OC, demonstrating that Runx2 plays an essential role in osteogenesis.21,22 Runx2 mutations cause cleidocranial dysplasia (CCD) in humans, an autosomal-dominant condition exhibiting defective endochondral and intramembranous bone formation, which is characterized by hypoplasia/aplasia of clavicles, patent fontanelles, supernumerary teeth, short stature, and other changes in skeletal patterning and growth.23 Heterozygous loss of Runx2 function in mice is sufficient to produce CCD disorders similar to humans.21,23 Further data demonstrate that truncated Runx2 protein severely impaired Runx2 transactivation activity and failed to interact with and respond to Smads to induce osteogenesis and cause CCD.24
Transgenic data show that increased Runx2 expression also cause bone dysplasia. By overexpression of Runx2 under the type I collagen promoter in transgenic mice, Runx2 transgenic mice end up with osteopenia with multiple fractures. Thin, porous, and OPN-enriched cortical bone is invaded by osteoclasts, despite the absence of acceleration of osteoclastogenesis. The number of neonatal osteoblasts increases, but matrix production and mineralization are impaired. Terminally differentiated osteoblasts and osteocytes decrease greatly, whereas less-mature osteoblasts accumulate in adult bone, demonstrating that Runx2 inhibits osteoblast differentiation at late stage by blocking maturation of osteoblasts.17 Runx2-overexpressing mice by the MMP13 promoter show increased mRNA expression of bone-forming genes and decreased MMP13, which plays a role in recruiting osteoclasts to the bone surface in the tibiae of transgenic mice. Further histological analyses of the proximal tibiae show increased bone mineralization surface, mineral apposition rate, and bone formation rate, but decreased osteoblast number.25 These data suggest that the Runx2 expression level is very important to maintain the balance between the bone formation–bone resorption process. Overall, Runx2 is the master transcription factor of bone formation.
Regulators of Runx2: Delicate Regulation of Osteogenesis
Bone development is a complex process, and three types of cells, including osteoblasts, osteoclasts, and chondrocytes, need be elaborately orchestrated to result in normal bone development. The expression and transcriptional activity of Runx2 are tightly regulated by upstream factors and cofactors to control downstream factors and function in skeletogenesis as the master transcription factor of osteogenesis.
Upstream factors of Runx2
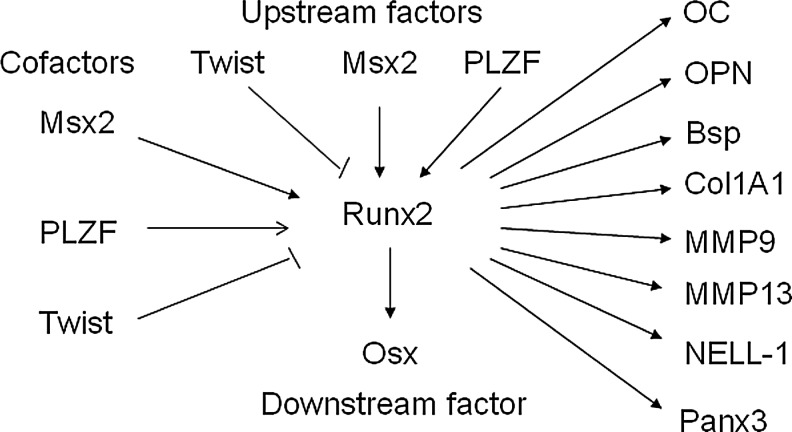
Many factors such as Twist, Msx2, and PLZF have been identified to be involved in the regulation of Runx2 expression or transcription activity as upstream factors and therefore regulate osteogenesis (Fig. 3). It has also been shown that SRY (sex-determining region Y)-box 8 (Sox8)26 and homeobox A2 (Hoxa2)27 reduce expression of Runx2 to regulate bone formation. Sox9 directly interacts with Runx2 and represses its activity via their evolutionarily conserved high-mobility-group and runt domains.28 p53-null osteoprogenitor cells show increased expression of Runx2.29 Conversely, some transcriptional factors increase the expression of Runx2 to regulate osteogenesis. Forkhead box protein O1 (Foxo1) directly interacts with the promoter of Runx2 and regulates its expression.30 SATB homeobox 2 (Satb2),27 Hoxa10,31 distal-less homeobox 3 (Dlx3),32 and Dlx533 activate the expression of Runx2 and osteogenic genes. Very interestingly, Dlx3 exerts both positive and negative regulation of gene transcription by a different molecular mechanism. Dlx3 protein–DNA interactions increase OC promoter activity, whereas Dlx3–Runx2 protein–protein interactions decrease Runx2-mediated transcription.34 Mice lacking Bapx1 die at birth showing dysplasia of the axial skeleton and strongly decreased Runx2, indicating that Bapx1 is required for Runx2 expression.35 Wnt10b directs cell fate toward the osteoblast lineage by inducing osteoblastogenic transcription factors Runx2, Dlx5, and Osx.36 Serum response factor (SRF) deficiency decreases the transcriptional activity of Runx2, whereas overexpression of SRF induces Runx2 transactivity, suggesting that SRF regulates bone formation through Runx2 (Fig. 2).37
FIG. 3.
Runx2 as the master regulator of osteoblast differentiation is modulated by multiple transcription factors to regulate downstream factors and osteoblast differentiation. The expression and transcriptional activities of Runx2 are tightly modulated by upstream transcription factors and cofactors to regulate downstream transcription factors. As the master regulator of osteoblast differentiation, Runx2 directly binds and regulates the expression of multiple osteogenic genes that determine the osteoblast phenotype.
Twist, antagonist of Runx2
Runx2 directly binds and regulates the expression of OC. However, OC is expressed 4–5 days later than Runx2 expression during mouse development. It has been proposed that Runx2 function is transiently inhibited by other genes. Twist is a basic helix-loop-helix transcription factor. Twist1 is highly expressed in freshly purified bone marrow-derived MSCs and decreased during ex vivo expansion.38 During early development, Twist-1 is coexpressed with Runx2 in cells destined to become osteoblasts and disappear when osteogenesis is initiated. Twist-1 inactivation alters Runx2 expression and Runx2-binding ability to the OC promoter.39 Highly expressing Twist1 increases the expression of MSC markers, but decreases osteogenesis, implicating that Twist mediates self-renewal and lineage commitment of MSCs.38 Haploinsufficiency of Twist1 causes Saethre-Chotzen syndrome, a form of craniosynostosis.40,41 Removing one allele of Twist1 is sufficient to correct the skull defects from heterozygous mutation of Runx2, and Twist-1 or 2 deficiency results in premature osteoblast differentiation. Conversely, Twist-1 overexpression inhibits osteogenesis without affecting Runx2 expression,42 whereas Twist-1 silencing enhances osteogenesis of MSCs.43 Further molecular analysis shows that Twist directly binds to the Runt DNA-binding domain of Runx2 to decrease binding of Runx2 to DNA.38 These data show that Twist determines the onset of osteogenesis as an antagonist of Runx2.42
Msx2, a regulator of bone development
Msx2 is a homeodomain transcription factor first identified in craniofacial bone and human femoral osteoblasts.44–46 Mice deficient in Msx2 manifest defects in skull ossification and a marked reduction in bone formation associated with decrease in osteoblast numbers, thus suggesting that Msx2 is involved in bone formation.45,46 Tracing the origin of the calvarial foramen defects shows that mesenchymal cell populations decreased due to defects in differentiation and proliferation, and not because of apoptosis or deficient migration of neural crest-derived precursor cells.47 Msx2 also affects tooth development. Msx2−/− homozygous mice develop compound amelogenesis imperfecta, dentinogenesis imperfecta, and periodontal osteopetrosis.48 The expression of Runx2 is strongly reduced in Msx2-deficient mice, suggesting that Msx2 acts as upstream factor of Runx2 to regulate osteoblast differentiation (Fig. 3). Overexpression of Msx2 affects bone formation. In vitro, Msx2 promotes osteogenesis of MSCs.49 However, a controversial result was reported in chick calvarial osteoblasts, showing that ectopic Msx2 overexpression prevents osteoblastic differentiation and mineralization, whereas Msx2 knockdown by antisense decreases proliferation and accelerates differentiation.50 Overexpression of Msx2 in the Runx2-deficient MSCs induces Osx expression, whereas knockdown of Msx2 inhibits Osx by BMP2.51 Msx2 transgenic mice increase bone formation by increasing osteoblast numbers. Further molecular studies reveal that the Msx2 transgene promotes osteogenic differentiation in part by reducing Dkk1 (an antagonist of Wnt receptors LRP5 and LRP6) expression and enhancing Wnt signaling.52 These data show that Msx2 plays an important role in regulating bone development.
Plzf, an upstream factor of Runx2
PLZF encodes a zinc-finger-type transcription factor, comprising 9 Kruppel-like C2H2 zinc fingers located in the C-terminus of the protein53 and a BTB (bric-a-brac, tram track, broad complex)/POZ (poxvirus, zinc finger) domain in the N-terminal end. PLZF is shown to be one of the highly upregulated genes during osteogenesis and is functionally involved in osteogenesis of MSCs. PLZF knockdown results in decreased expression of osteoblast-specific genes, whereas PLZF overexpression improves osteogenesis.54,55 Similar effects of PLZF overexpression on immortalized MSCs are also observed.55 Deletion of the BTB domain abrogates the effect of PLZF on osteogenesis, showing that BTB plays a crucial role in osteogenesis. Further analyses show that PLZF affects Runx2, whereas Runx2 does not affect the expression of PLZF, suggesting that PLZF regulates osteogenesis as an upstream factor of Runx2.54 PLZF is also shown to regulate chondrogenesis of MSCs as an upstream factor of Sox9, the master regulator of chondrocyte differentiation. PLZF-overexpressing MSCs repaired cartilage defects better and earlier than control MSCs.55 It is proposed that PLZF regulates the osteogenic master regulator Runx2, which directly regulates osteoblast-specific genes, including OC,9 OPN,9 BSP,9 and Col1A19,12 during osteogenesis (Fig. 3). Therefore, PLZF represents a very promising strategy for bone regeneration and repair.
Osx, downstream factors of Runx2
Runx2 regulates downstream genes that determine the osteoblast phenotype and function in skeletogenesis. Besides major osteoblast-specific genes such as OC, Col1A1, BSP, and OPN,9 Runx2 also regulates Osx, which is essential for osteoblast differentiation. Osx is a zinc-finger-containing transcription factor expressed in osteoblasts56 and chondrocytes.57 Inactivation of Osx in mice results in perinatal lethality owing to a complete absence of bone formation and completely arrested osteoblast differentiation displaying lack of early and late markers of osteoblast differentiation.56 A homozygous single-base-pair deletion in Osx resulted in recessive osteogenesis imperfecta in an Egyptian child.58 Osx-deficient mice lack a mineralized matrix in bones formed by intramembraneous ossification, which is different from Runx2-deficient mice, which show an entirely nonmineralized skeleton. Osx-deficient bones formed by endochondral ossification contain some mineralized matrix, which resembles calcified cartilage, not mineralized bone matrix, showing that Osx is not required for chondrocyte hypertrophy. Osx is not expressed in Runx2-deficient embryos, whereas Runx2 is normally expressed in Osx-deficient embryos, showing that Osx is a downstream factor of Runx2 during osteoblast differentiation. Further data show that Runx2 directly binds to Osx at a responsive element in the promoter of the Osx gene.59 These data demonstrate that Osx specifically induces osteoblast differentiation and bone formation in vivo. Further molecular data show that the nuclear factor of activated T cells (NFAT) and Osx form a complex that binds to the osteoblast-specific Col1a1 promoter and synergistically stimulates its activity.60 All these data suggest that Runx2 and Osx belong to two independent pathways; Osx is a Runx2-dependent transcription factor and is absolutely required for bone formation.
Cofactors of Runx2-regulating osteogenesis
Runx2 is a transcription factor essential for osteoblast commitment and early stages of osteoblast differentiation, whose activity is tightly controlled by transcriptional factors through protein–DNA or protein–protein interactions. It was shown that Stat1 interacts with Runx2 to inhibit its nuclear localization.61 Yes-associated protein (YAP) suppresses Runx2 transcriptional activity in a dose-dependent manner.62 AJ18 modulates Runx2 activity and osteogenic differentiation by suppressing Runx2-mediated transactivation of an OC promoter.63 Myeloid Elf-1 like factor (MEF) forms a complex with Runx2 to interfere with binding of Runx2 to the OC promoter at the OSE2 site.64 Nrf2 interacts with Runx2 to inhibit Runx2-dependent stimulation of OC promoter activity and recruitment of Runx2 on the OC promoter without affecting the expression of Runx2 mRNA.65 pRb serves as a linker connecting p204 and Runx2 by forming a ternary complex to stimulate Runx2 activity.66 Schnurri 3 (Shn3) controls the protein level of Runx2 by mediating recruitment of E3 ubiquitin ligase WW domain-containing protein 1 (WWP1) to alter the availability of Runx2 in the nucleus.67 Core-binding factor-beta (CBFbeta)68 and Smads24 interact with Runx2 to regulate skeletal development. Smad does not directly induce Runx2 expression, and Runx2 is induced by the receptor-activated Smad through Smad-induced junB functions.69 TAZ (transcriptional coactivator with the PDZ-binding motif) coactivates Runx2-dependent gene transcription while repressing PPAR-gamma-dependent gene.70 Id4 promotes osteoblast differentiation by releasing Hes1 from Hes1–Hey2 complexes to increase the stability and transcriptional activity of Runx2.71 GLI family zinc-finger 2 (GLI2) interacts with Runx2 to enhance osteogenesis.72 Chicken ovalbumin upstream promoter–transcription factor II (COUP-TFII) physically interacts with Runx2 to impair the Runx2-dependent activation of the OC promoter73 (Table 1 and Fig. 2).
Table 1.
Coactivators of Runx2 -Osteoblast Differentiation
| Genes | Features | Models of action | Refs |
|---|---|---|---|
| Stat1 | STAT family transcription factor | Inhibit nuclear localization of Runx2 | 61 |
| YAP | Homolog of TAZ | Suppresses Runx2 transcriptional activity | 62 |
| AJ18 | Zinc-finger transcription factor | Suppresses Runx2-mediated transactivation | 63 |
| MEF | ETS transcription factor | Forms a complex with Runx2 to interfere with Runx2 binding to OC promoter | 64 |
| Nrf2 | The cap-n-collar transcription factor | Inhibits binding of Runx2 to OC promoter | 65 |
| pRb | Tumor suppressor nuclear protein | Links p204 and Runx2 by forming a ternary complex to stimulate Runx2 activity | 66 |
| Shn3 | Zinc-finger adaptor protein | Controls protein level of Runx2 | 67 |
| CBFbeta | PEBP2/CBF transcription factor | Interacts with Runx2 | 68 |
| Smad | Downstream of TGF-β signaling | Interacts with Runx2 | 24 |
| TAZ | Transcriptional coactivator with PDZ-binding motif | Coactivates Runx2-dependent gene transcription | 70 |
| Id4 | bHLH transcription factor | Increases the stability and transcriptional activity of Runx2 | 71 |
| ATF4 | CREB transcription factor-containing basic zipper | Interacts with Runx2 through Satb2 to synergize their activities | 27 |
| Zfp521 | Zinc-finger protein | Represses Runx2-mediate gene activation | 80 |
| COUP-TFII | Orphan nuclear receptor | Interacts with Runx2 to repress the DNA binding of Runx2 to OC promoter | 73 |
| GLI2 | Zinc-finger protein | Interacts with Runx2 to enhance its activity | 72 |
ATF4, activating transcription factor 4; ZFP521, zinc-finger protein 521; OC, osteocalcin; Satb2, SATB homeobox 2; YAP, yes-associated protein; MEF, myeloid Elf-1-like factor; Rb, retinoblastoma; Shn3, schnurri 3; CBFbeta, core-binding factor-beta; COUP-TFII, chicken ovalbumin upstream promoter–transcription factor II.
Activating transcription factor 4
ATF4 encodes a transcription factor that plays a crucial role in osteoblast differentiation and function. In vitro kinase assay shows that AFT4 is strongly phosphorylated by ribosomal S6 serine/threonine kinase 2 (RSK2) as a critical substrate, and this phosphorylation is undetectable in osteoblasts from mice lacking RSK2, encoding a growth factor-regulated kinase, in which the mutation results in DuCoffin-Lowry syndrome (CLS). CLS is an X-linked mental retardation condition associated with skeletal abnormalities. In addition, ATF4 knockout mice displayed delayed skeletal development, decreased bone formation, and thereafter develop a severe low-bone-mass phenotype.74 Molecular studies reveal that ATF4 binds to an osteoblast-specific element in the OC promoter and directly activates its transcription.74 ATF4 is also required for proper synthesis of Col1A1. Col1A1 synthesis is decreased in ATF4-decifient osteoblasts compared with wild-type osteoblasts. This decrease in Col1A1 synthesis is not due to decreased gene expression of Col1A1, Runx2, and Osx, as their expression is normal in mutant osteoblasts compared with wild type osteoblasts, suggesting that ATF4 affects synthesis of Col1A1 through a post-transcriptional mechanism. Addition of nonessential amino acids in a culture medium rescues the defect in collagen synthesis in osteoblasts lacking ATF4, suggesting that ATF4 is required for efficient amino acid import into osteoblasts.74 Forced expression of a constitutively active form of RSK2 (RSK2-T707A)75 enhances the activity of the pOG2-luc reporter in COS cells, whereas RSK2-T707A is not able to increase the activity of the ATF4 mutant that cannot be phosphorylated by RSK2,74 suggesting that phosphorylation of ATF4 by RSK2 enhances its transactivation ability. Further studies showed that ATF4 interacts with Runx2 through Satb2 to synergize their activities by binding proximal binding sites at the OC promoter.27
ATF4 also regulates osteoclast differentiation and ultimately bone resorption through its expression in osteoblasts.76,77 This can be explained by the binding of ATF4 to the promoter of the receptor activator of nuclear factor-KB ligand (RANKL) gene,78 which encodes a factor secreted by osteoblasts and binds to its receptor RANK on osteoclasts to trigger intricate and distinct signaling cascades that control lineage commitment and activation of osteoclasts. RANKL binding to RANK is negatively regulated by OPG to inhibit bone turnover by osteoclasts.79 ATF4-deficient mice have decreased osteoclast numbers owing to reduced RANKL expression, demonstrating that ATF4 is involved in the control of bone resorption.72 These data demonstrate that ATF4 is a major regulator of osteoblast differentiation.
Zfp521, a binding partner of Runx2
Zfp521, a 30-zinc-finger protein, is a new player in bone formation. Zfp521 is a nuclear protein highly expressed at the periphery of mesenchymal condensations, and plays an important role in developing bones, including the perichondrium and periosteum, osteoblast precursors, osteoblasts, ostocytes, chondroblasts, and prehypertrophic chondrocytes. Zfp521 is also highly expressed at the periphery of mesenchymal condensations and calvarial osteoblasts.80,81 Zfp521 strongly represses activation of the Runx2-mediated reporter gene. Zfp521 calvarial cells from Zfp521-transgenic mice show decreased osteogenesis as determined by less nodule formation and mineralization, and decreased early osteoblast marker genes,82 suggesting that Zfp521 antagonizes early stages of osteoblast differentiation. Conversely, depletion of Zfp521 increases expression of Runx2 and Runx2 target genes.83 Forced expression of Zfp521 in osteoblasts in vivo leads to an increase in bone mass due to a marked increase in the osteoblast number and bone-forming activity.81 Removing one copy of Zfp521 can rescue the CCD phenotype due to lack of one copy of Runx2 in mice, whereas overexpressing Zfp521 exacerbates the bone defects.83 Zfp521 is an important PTHrP target gene.82,84 Further molecular data have shown that Zfp521 blocks Runx2 in the presence of HDAC3 by recruiting the histone deacetylase HDAC3 to repress its transcriptional activity.83 Zfp521 coimmunoprecipitates with Runx2 in the same complex to repress its transcriptional activity, demonstrating that Zfp521 is a binding partner of Runx2. These data show that Zfp521 restricts early stages of osteoblast differentiation, but promotes late stages of osteoblast maturation by antagonizing Runx2 transcriptional activity.
HDACs, cofactors of Runx2
Runx2 is the master regulator of osteogenesis and is crucial for regulating the expression of bone-specific genes. Runx2 either activates or represses transcription of tissue-specific genes by binding specific DNA sequences or interacting with transcriptional coactivators and corepressors to regulate bone formation. It has been shown that transcriptional activity of Runx2 is inhibited by HDACs. HDACs are a class of enzymes that remove acetyl groups from an ɛ-N-acetyl lysine amino acid on a histone, allowing the histones to wrap the DNA more tightly to regulate DNA expression by acetylation and deacetylation. HDAC activity plays important roles in the development of bone formation by post-translational modification. HDAC4 and HDAC5 inhibit transcriptional activity of Runx2 by deacetylating.85 HDAC4 also inhibits Runx2 transcriptional activity by binding the Runt domain and interfering with DNA binding.86 In a different manner, HDAC3 and HDAC6 repress Runx2-mediated transcription by binding the aminoterminal domain87 and the carboxy terminus88 of Runx2 to deacetylate lysines in the Runx2 protein, respectively. In a deacetylase-independent manner, HDAC7 represses Runx2 transcriptional activity by indirectly binding Runx2 through the aminoterminus of HDAC.89
Identification of Genetic Factors Regulating Osteogenesis
Great progress has been made over the past 15 years in identification of genetic factors involved in skeletogenesis and the understanding of the molecular events of bone development. To take these studies further, it is important to make use of human disease models, animal models, and well-designed in vitro studies.
Methods to identify genetic factors regulating osteogenesis
Human disease cases with skeletal dysplasia
Naturally occurring human disease cases with bone dysplasia offer an excellent platform to explore the underlying genes causing the various bone abnormalities and their phenotypes. However, the incidence is rare. Studies of extreme phenotypes will yield very useful data that can be used for studies that may result in therapeutic intervention in the form of gene therapy.
Animal models
Animal models, especially mouse genetics models, allow researchers to investigate the role of genetic factors involved in skeletogenesis and their phenotypes, which would otherwise not be possible in humans. Producing genetically engineered animals by modulating in vivo gene expression is widely used to study the underlying molecular alterations and screen genes related to bone development, including mutated animals with homozygous or heterozygous mutations and transgenic animals. Conditional gene knockout by the Cre/loxP or FLP/FRT recombination system is widely used to study gene function in adult mice and selected cell types based on a tissue-specific inactivation of the gene of interest. This approach is advantageous over the conventional type in that conditional mice survive longer, especially when conventional knockout mice exhibit embryonic or early postnatal lethality, or when a gene alteration exerts its effects in multiple different cell and tissue types, which make it difficult to distinguish direct function in a particular tissue or secondary effects resulting from altered gene function in other tissues. The technique is also much cleaner and scientifically more precise for elimination of a specific target gene from a single organ in the body. Animal models can provide insights into the biological role in human disease and permit the identification of the genetic defect in humans. It was shown that mice with mutations in GMAP-210 were similar to patients with Achondrogenesis type 1A in humans, a neonatal lethal form of skeletal dysplasia, suggesting that Achondrogenesis type 1A may be caused by GMAP-210 deficiency. Further sequence analysis revealed mutations in the unrelated patients with Achondrogenesis type 1A.90
In vitro study
Microarrays and high-throughput sequencing technology are being employed to screen and identify novel molecular signature related to osteogenesis. Screened candidate genes will then be validated by gain or loss of function in cells in vitro. This will be very useful in studying human genes regulating osteogenesis without human disease cases.
Challenges of study in genetic factors regulating osteogenesis
Although extraordinary progress has been made in the identification of transcription factors involved in osteogenesis, detailed GRN of osteogenesis remains poorly understood. So far, the repair of massive bone defects still represents a major clinical orthopedic challenge. Bone is a highly vascularized tissue dependent on the close spatial and temporal connection between endothelial cells and bone cells to maintain skeletal integrity. Bone tissue engineering is a complex undertaking that involves the combination of growth factors (such as osteogenic and angiogenic factors), cells, and scaffolds. To repair the massive bone defects by tissue engineering, only study of the genetic factors in osteogenesis is not sufficient. To improve bone regeneration and repair, it is necessary to identify novel genetic factors regulating osteogenesis and decipher GRNs of osteogenesis. At the same time, it is also crucial to understand how osteogenic and angiogenic factors interact with each other and with cells during the multistem processes of bone development and repair. Combined strategies of osteogenic and angiogenic factors will enhance the regenerative capacity of bone. It is important to establish how osteogenic and angiogenic factors can be delivered in a spatial and temporal manner in biomimetic scaffolds so that bone regeneration can occur.
Transcription factor-targeted gene therapy for bone regeneration
Gene therapy represents one the most promising approaches for orthopedic regenerative medicine. The use of transcription factors for bone tissue engineering has been examined. It has been shown that Runx2 overexpression significantly upregulates osteoblastic differentiation and enhances mineralization of bone marrow stromal cells (BMSCs) in vitro and in vivo.91 Similar results are obtained in rat BMSCs.92 Further data show that Runx2-modified BMSCs are able to accelerate healing of critical-sized bone defects.93 In fibroblasts, effects of Runx2 overexpression on mineralization depend on the scaffold used; collagen foams exhibit 10-fold higher mineral volume compared to PCL and PLGA matrices in vitro,94 suggesting that the choice of scaffold will affect the reparative effects. Runx2-modified dermal fibroblasts form mineralized templates in vivo after subcutaneous implantation.95 Graded tissue interface is engineered by gradients of immobilized Runx2 retrovirus via deposition of controlled poly(l-lysine) densities.96 In addition, PLZF-modified MSCs are shown to be able to repair cartilage defects better and earlier than control MSCs.55 These data suggest that the use of transcription factors may be a promising gene strategy for bone and cartilage regeneration, especially in terms of obtaining the correct bone or cartilage phenotype. However, there are still challenges that have to be overcome to deliver them to defect sites in a safe and efficient manner. Safe and efficient delivery and controlled and sustained expression of the therapeutic genes should be considered with other strategies such as the choice of appropriate scaffolds, a conducive extracellular environment, and other bioactive factors to enhance bone regeneration in tissue engineering applications.
Conclusions
We have provided a detailed overview of the identity of the important mediators and the molecular mechanisms of the genetic factors linked to Runx2 as the master transcription factor of osteoblast differentiation. The information presented can form the basis for many researchers in the stem cell arena to embark on further studies to enhance bone healing and bone repair. Bone development and repair is a very complex process, and many related issues remain to be elucidated. Further scientific endeavors need be undertaken to deepen our understanding of osteogenesis, so that it can lead to better solutions for bone repair and regeneration.
Acknowledgment
This work is supported by a BMRC grant (R-175-000-085-305).
Disclosure Statement
The authors have declared that no competing interests exist.
References
- 1.Van Der Plas A. Aarden E.M. Feijen J.H. de Boer A.H. Wiltink A. Alblas M.J., et al. Characteristics and properties of osteocytes in culture. J Bone Miner Res. 1994;9:1697. doi: 10.1002/jbmr.5650091105. [DOI] [PubMed] [Google Scholar]
- 2.DeLise A.M. Stringa E. Woodward W.A. Mello M.A. Tuan R.S. Embryonic limb mesenchyme micromass culture as an in vitro model for chondrogenesis and cartilage maturation. Methods Mol Biol. 2000;137:359. doi: 10.1385/1-59259-066-7:359. [DOI] [PubMed] [Google Scholar]
- 3.Hughes D.E. Dai A. Tiffee J.C. Li H.H. Mundy G.R. Boyce B.F. Estrogen promotes apoptosis of murine osteoclasts mediated by TGF-beta. Nat Med. 1996;2:1132. doi: 10.1038/nm1096-1132. [DOI] [PubMed] [Google Scholar]
- 4.Weinstein R.S. Jilka R.L. Parfitt A.M. Manolagas S.C. Inhibition of osteoblastogenesis and promotion of apoptosis of osteoblasts and osteocytes by glucocorticoids. Potential mechanisms of their deleterious effects on bone. J Clin Invest. 1998;102:274. doi: 10.1172/JCI2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weinstein R.S. Chen J.R. Powers C.C. Stewart S.A. Landes R.D. Bellido T., et al. Promotion of osteoclast survival and antagonism of bisphosphonate-induced osteoclast apoptosis by glucocorticoids. J Clin Invest. 2002;109:1041. doi: 10.1172/JCI14538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heidari N. Miller A.V. Hicks M.A. Marking C.B. Harada H. Glucocorticoid-mediated BIM induction and apoptosis are regulated by Runx2 and c-Jun in leukemia cells. Cell Death Dis. 2012;3:e349. doi: 10.1038/cddis.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phillips J.E. Gersbach C.A. Wojtowicz A.M. García A.J. Glucocorticoid-induced osteogenesis is negatively regulated by Runx2/Cbfa1 serine phosphorylation. J Cell Sci. 2006;119(Pt 3):581. doi: 10.1242/jcs.02758. [DOI] [PubMed] [Google Scholar]
- 8.Ducy P. Cbfa1: a molecular switch in osteoblast biology. Dev Dyn. 2000;219:461. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1074>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 9.Ducy P. Zhang R. Geoffroy V. Ridall A.L. Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 10.Villavicencio-Lorini P. Kuss P. Friedrich J. Haupt J. Farooq M. Türkmen S., et al. Homeobox genes d11–d13 and a13 control mouse autopod cortical bone and joint formation. J Clin Invest. 2010;120:1994. doi: 10.1172/JCI41554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D'Souza R.N. Aberg T. Gaikwad J. Cavender A. Owen M. Karsenty G., et al. Cbfa1 is required for epithelial-mesenchymal interactions regulating tooth development in mice. Development. 1999;126:2911. doi: 10.1242/dev.126.13.2911. [DOI] [PubMed] [Google Scholar]
- 12.Kern B. Shen J. Starbuck M. Karsenty G. RUNX2 contributes to the osteoblast-specific expression of type I collagen genes. J Biol Chem. 2001;276:7101. doi: 10.1074/jbc.M006215200. [DOI] [PubMed] [Google Scholar]
- 13.Truong T. Zhang X. Pathmanathan D. Soo C. Ting K. Craniosynostosisassociated gene nell-1 is regulated by runx2. J Bone Miner Res. 2007;22:7. doi: 10.1359/jbmr.061012. [DOI] [PubMed] [Google Scholar]
- 14.Bond S.R. Lau A. Penuela S. Sampaio A.V. Underhill T.M. Laird D.W., et al. Pannexin 3 is a novel target for Runx2, expressed by osteoblasts and mature growth plate chondrocytes. J Bone Miner Res. 2011;26:2911. doi: 10.1002/jbmr.509. [DOI] [PubMed] [Google Scholar]
- 15.Pratap J. Javed A. Languino L.R. van Wijnen A.J. Stein J.L. Stein G.S., et al. The Runx2 osteogenic transcription factor regulates matrix metalloproteinase 9 in bone metastatic cancer cells and controls cell invasion. Mol Cell Biol. 2005;25:8581. doi: 10.1128/MCB.25.19.8581-8591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boumah C.E. Lee M. Selvamurugan N. Shimizu E. Partridge N.C. Runx2 recruits p300 to mediate parathyroid hormone's effects on histone acetylation and transcriptional activation of the matrix metalloproteinase-13 gene. Mol Endocrinol. 2009;23:1255. doi: 10.1210/me.2008-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu W. Toyosawa S. Furuichi T. Kanatani N. Yoshida C. Liu Y., et al. Overexpression of RUNX2 in osteoblasts inhibits osteoblast maturation and causes osteopenia with multiple fractures. J Cell Biol. 2001;155:157. doi: 10.1083/jcb.200105052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X. Yang M. Lin L. Chen P. Ma K.T. Zhou C.Y., et al. Runx2 overexpression enhances osteoblastic differentiation and mineralization in adipose-derived stem cells in vitro and in vivo. Calcif Tissue Int. 2006;79:169. doi: 10.1007/s00223-006-0083-6. [DOI] [PubMed] [Google Scholar]
- 19.Inada M. Yasui T. Nomura S. Miyake S. Deguchi K. Himeno M., et al. Maturational disturbance of chondrocytes in Cbfa1-deficient mice. Dev Dyn. 1999;214:279. doi: 10.1002/(SICI)1097-0177(199904)214:4<279::AID-AJA1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 20.Takeda S. Bonnamy J.P. Owen M.J. Ducy P. Karsenty G. Continuous expression of Cbfa1 in nonhypertrophic chondrocytes uncovers its ability to induce hypertrophic chondrocyte differentiation and partially rescues Cbfa1-deficient mice. Genes Dev. 2001;15:467. doi: 10.1101/gad.845101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otto F. Thornell A.P. Crompton T. Denzel A. Gilmour K.C. Rosewell I.R., et al. RUNX2, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 22.Komori T. Yagi H. Nomura S. Yamaguchi A. Sasaki K. Deguchi K., et al. Targeted disruption of RUNX2 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 23.Mundlos S. Otto F. Mundlos C. Mulliken J.B. Aylsworth A.S. Albright S., et al. Mutations involving the transcription factor RUNX2 cause cleidocranial dysplasia. Cell. 1997;89:773. doi: 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y.W. Yasui N. Ito K. Huang G. Fujii M. Hanai J., et al. A RUNX2/PEBP2alpha A/CBFA1 mutation displaying impaired transactivation and Smad interaction in cleidocranial dysplasia. Proc Natl Acad Sci U S A. 2000;97:10549. doi: 10.1073/pnas.180309597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Selvamurugan N. Jefcoat S.C. Kwok S. Kowalewski R. Tamasi J.A. Partridge N.C. Overexpression of Runx2 directed by the matrix metalloproteinase-13 promoter containing the AP-1 and Runx/RD/Cbfa sites alters bone remodeling in vivo. J Cell Biochem. 2006;99:545. doi: 10.1002/jcb.20878. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt K. Schinke T. Haberland M. Priemel M. Schilling A.F. Mueldner C., et al. The high mobility group transcription factor Sox8 is a negative regulator of osteoblast differentiation. J Cell Biol. 2005;168:899. doi: 10.1083/jcb.200408013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dobreva G. Chahrour M. Dautzenberg M. Chirivella L. Kanzler B. Fariñas I., et al. SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell. 2006;125:971. doi: 10.1016/j.cell.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 28.Zhou G. Zheng Q. Engin F. Munivez E. Chen Y. Sebald E., et al. Dominance of SOX9 function over RUNX2 during skeletogenesis. Proc Natl Acad Sci U S A. 2006;103:19004. doi: 10.1073/pnas.0605170103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lengner C.J. Steinman H.A. Gagnon J. Smith T.W. Henderson J.E. Kream B.E., et al. Osteoblast differentiation and skeletal development are regulated by Mdm2-p53 signaling. J Cell Biol. 2006;172:909. doi: 10.1083/jcb.200508130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teixeira C.C. Liu Y. Thant L.M. Pang J. Palmer G. Alikhani M. Foxo1, a novel regulator of osteoblast differentiation and skeletogenesis. J Biol Chem. 2010;285:31055. doi: 10.1074/jbc.M109.079962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hassan M.Q. Tare R. Lee S.H. Mandeville M. Weiner B. Montecino M., et al. HOXA10 controls osteoblastogenesis by directly activating bone regulatory and phenotypic genes. Mol Cell Biol. 2007;27:3337. doi: 10.1128/MCB.01544-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hassan M.Q. Tare R.S. Lee S.H. Mandeville M. Morasso M.I. Javed A., et al. BMP2 commitment to the osteogenic lineage involves activation of Runx2 by DLX3 and a homeodomain transcriptional network. J Biol Chem. 2006;281:40515. doi: 10.1074/jbc.M604508200. [DOI] [PubMed] [Google Scholar]
- 33.Lee M.H. Kim Y.J. Yoon W.J. Kim J.I. Kim B.G. Hwang Y.S., et al. Dlx5 specifically regulates Runx2 type II expression by binding to homeodomain-response elements in the Runx2 distal promoter. J Biol Chem. 2005;280:35579. doi: 10.1074/jbc.M502267200. [DOI] [PubMed] [Google Scholar]
- 34.Hassan M.Q. Javed A. Morasso M.I. Karlin J. Montecino M. van Wijnen A.J., et al. Dlx3 transcriptional regulation of osteoblast differentiation: temporal recruitment of Msx2, Dlx3, and Dlx5 homeodomain proteins to chromatin of the osteocalcin gene. Mol Cell Biol. 2004;24:9248. doi: 10.1128/MCB.24.20.9248-9261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tribioli C. Lufkin T. The murine Bapx1 homeobox gene plays a critical role in embryonic development of the axial skeleton and spleen. Development. 1999;126:5699. doi: 10.1242/dev.126.24.5699. [DOI] [PubMed] [Google Scholar]
- 36.Bennett C.N. Longo K.A. Wright W.S. Suva L.J. Lane T.F. Hankenson K.D., et al. Regulation of osteoblastogenesis and bone mass by Wnt10b. Proc Natl Acad Sci U S A. 2005;102:3324. doi: 10.1073/pnas.0408742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen J. Yuan K. Mao X. Miano J.M. Wu H. Chen Y. Serum response factor regulates bone formation via IGF-1 and Runx2 signals. J Bone Miner Res. 2012;27:1659. doi: 10.1002/jbmr.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Isenmann S. Arthur A. Zannettino A.C. Turner J.L. Shi S. Glackin C.A., et al. TWIST family of basic helix-loop-helix transcription factors mediate human mesenchymal stem cell growth and commitment. Stem Cells. 2009;27:2457. doi: 10.1002/stem.181. [DOI] [PubMed] [Google Scholar]
- 39.Yousfi M. Lasmoles F. Marie P.J. TWIST inactivation reduces RUNX2/RUNX2 expression and DNA binding to the osteocalcin promoter in osteoblasts. Biochem Biophys Res Commun. 2002;297:641. doi: 10.1016/s0006-291x(02)02260-x. [DOI] [PubMed] [Google Scholar]
- 40.El Ghouzzi V. Le Merrer M. Perrin-Schmitt F. Lajeunie E. Benit P. Renier D., et al. Mutations of the TWIST gene in the Saethre-Chotzen syndrome. Nat Genet. 1997;15:42. doi: 10.1038/ng0197-42. [DOI] [PubMed] [Google Scholar]
- 41.Howard T.D. Paznekas W.A. Green E.D. Chiang L.C. Ma N. Ortiz de Luna R.I., et al. Mutations in TWIST, a basic helix-loop-helix transcription factor, in Sathre-Chotzen syndrome. Nat Genet. 1997;15:36. doi: 10.1038/ng0197-36. [DOI] [PubMed] [Google Scholar]
- 42.Bialek P. Kern B. Yang X. Schrock M. Sosic D. Hong N., et al. A twist code determines the onset of osteoblast differentiation. Dev Cell. 2004;6:423. doi: 10.1016/s1534-5807(04)00058-9. [DOI] [PubMed] [Google Scholar]
- 43.Miraoui H. Severe N. Vaudin P. Pagès J.C. Marie P.J. Molecular silencing of Twist1 enhances osteogenic differentiation of murine mesenchymal stem cells: implication of FGFR2 signaling. J Cell Biochem. 2010;110:1147. doi: 10.1002/jcb.22628. [DOI] [PubMed] [Google Scholar]
- 44.Jabs E.W. Müller U. Li X. Ma L. Luo W. Haworth I.S, et al. A mutation in the homeodomain of the human MSX2 gene in a family affected with autosomal dominant craniosynostosis. Cell. 1993;75:443. doi: 10.1016/0092-8674(93)90379-5. [DOI] [PubMed] [Google Scholar]
- 45.Wilkie A.O. Tang Z. Elanko N. Walsh S. Twigg S.R. Hurst J.A., et al. Functional haploinsufficiency of the human homeobox gene MSX2 causes defects in skull ossification. Nat Genet. 2000;24:387. doi: 10.1038/74224. [DOI] [PubMed] [Google Scholar]
- 46.Satokata I. Ma L. Ohshima H. Bei M. Woo I. Nishizawa K., et al. Msx2 deficiency in mice causes pleiotropic defects in bone growth and ectodermal organ formation. Nat Genet. 2000;24:391. doi: 10.1038/74231. [DOI] [PubMed] [Google Scholar]
- 47.Ishii M. Merrill A.E. Chan Y.S. Gitelman I. Rice D.P. Sucov H.M., et al. Msx2 and Twist cooperatively control the development of the neural crest-derived skeletogenic mesenchyme of the murine skull vault. Development. 2003;130:6131. doi: 10.1242/dev.00793. [DOI] [PubMed] [Google Scholar]
- 48.Aïoub M. Lézot F. Molla M. Castaneda B. Robert B. Goubin G., et al. Msx2−/− transgenic mice develop compound amelogenesis imperfecta, dentinogenesis imperfecta and periodental osteopetrosis. Bone. 2007;41:851. doi: 10.1016/j.bone.2007.07.023. [DOI] [PubMed] [Google Scholar]
- 49.Cheng S.L. Shao J.S. Charlton-Kachigian N. Loewy A.P. Towler D.A. MSX2 promotes osteogenesis and suppresses adipogenic differentiation of multipotent mesenchymal progenitors. J Biol Chem. 2003;278:45969. doi: 10.1074/jbc.M306972200. [DOI] [PubMed] [Google Scholar]
- 50.Dodig M. Tadic T. Kronenberg M.S. Dacic S. Liu Y.H. Maxson R., et al. Ectopic Msx2 overexpression inhibits and Msx2 antisense stimulates calvarial osteoblast differentiation. Dev Biol. 1999;209:298. doi: 10.1006/dbio.1999.9258. [DOI] [PubMed] [Google Scholar]
- 51.Matsubara T. Kida K. Yamaguchi A. Hata K. Ichida F. Meguro H., et al. BMP2 regulates Osterix through Msx2 and Runx2 during osteoblast differentiation. J Biol Chem. 2008;283:29119. doi: 10.1074/jbc.M801774200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng S.L. Shao J.S. Cai J. Sierra O.L. Towler D.A. Msx2 exerts bone anabolism via canonical Wnt signaling. J Biol Chem. 2008;283:20505. doi: 10.1074/jbc.M800851200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen Z. Brand N.J. Chen A. Chen S.J. Tong J.H. Wang Z.Y., et al. Fusion between a novel Krüppel-like zinc finger gene and the retinoic acid receptor-alpha locus due to a variant t(11;17) translocation associated with acute promyelocytic leukaemia. EMBO J. 1993;12:1161. doi: 10.1002/j.1460-2075.1993.tb05757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ikeda R. Yoshida K. Tsukahara S. Sakamoto Y. Tanaka H. Furukawa K., et al. The promyelotic leukemia zinc finger promotes osteoblastic differentiation of human mesenchymal stem cells as an upstream regulator of RUNX2. J Biol Chem. 2005;280:8523. doi: 10.1074/jbc.M409442200. [DOI] [PubMed] [Google Scholar]
- 55.Liu T.M. Guo X.M. Tan H.S. Hui J.H. Lim B. Lee E.H. Zinc-finger protein 145, acting as an upstream regulator of SOX9, improves the differentiation potential of human mesenchymal stem cells for cartilage regeneration and repair. Arthritis Rheum. 2011;63:2711. doi: 10.1002/art.30430. [DOI] [PubMed] [Google Scholar]
- 56.Nakashima K. Zhou X. Kunkel G. Zhang Z. Deng J.M. Behringer R.R., et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 57.Yagi K. Tsuji K. Nifuji A. Shinomiya K. Nakashima K. deCrombrugghe B., et al. Bone morphogenetic protein-2 enhances osterix gene expression in chondrocytes. J Cell Biochem. 2003;88:1077. doi: 10.1002/jcb.10467. [DOI] [PubMed] [Google Scholar]
- 58.Lapunzina P. Aglan M. Temtamy S. Caparrós-Martín J.A. Valencia M. Letón R., et al. Identification of a frameshift mutation in Osterix in a patient with recessive osteogenesis imperfecta. Am J Hum Genet. 2010;87:110. doi: 10.1016/j.ajhg.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nishio Y. Dong Y. Paris M. O'Keefe R.J. Schwarz E.M. Drissi H. Runx2-mediated regulation of the zinc finger Osterix/Sp7 gene. Gene. 2006;372:62. doi: 10.1016/j.gene.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 60.Koga T. Matsui Y. Asagiri M. Kodama T. de Crombrugghe B. Nakashima K., et al. NFAT and Osterix cooperatively regulate bone formation. Nat Med. 2005;11:880. doi: 10.1038/nm1270. [DOI] [PubMed] [Google Scholar]
- 61.Kim S. Koga T. Isobe M. Kern B.E. Yokochi T. Chin Y.E., et al. Stat1 functions as a cytoplasmic attenuator of Runx2 in the transcriptional program of osteoblast differentiation. Genes Dev. 2003;17:1979. doi: 10.1101/gad.1119303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zaidi S.K. Sullivan A.J. Medina R. Ito Y. van Wijnen A.J. Stein J.L., et al. Tyrosine phosphorylation controls Runx2-mediated subnuclear targeting of YAP to repress transcription. EMBO J. 2004;23:790. doi: 10.1038/sj.emboj.7600073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jheon A.H. Ganss B. Cheifetz S. Sodek J. Characterization of a novel KRAB/C2H2 zinc finger transcription factor involved in bone development. J Biol Chem. 2001;276:18282. doi: 10.1074/jbc.M010885200. [DOI] [PubMed] [Google Scholar]
- 64.Kim Y.J. Kim B.G. Lee S.J. Lee H.K. Lee S.H. Ryoo H.M., et al. The suppressive effect of myeloid Elf-1-like factor (MEF) in osteogenic differentiation. J Cell Physiol. 2007;211:253. doi: 10.1002/jcp.20933. [DOI] [PubMed] [Google Scholar]
- 65.Hinoi E. Fujimori S. Wang L. Hojo H. Uno K. Yoneda Y. Nrf2 negatively regulates osteoblast differentiation via interfering with Runx2-dependent transcriptional activation. J Biol Chem. 2006;281:18015. doi: 10.1074/jbc.M600603200. [DOI] [PubMed] [Google Scholar]
- 66.Luan Y. Yu X.P. Xu K. Ding B. Yu J. Huang Y., et al. The retinoblastoma protein is an essential mediator of osteogenesis that links the p204 protein to the RUNX2 transcription factor thereby increasing its activity. J Biol Chem. 2007;282:16860. doi: 10.1074/jbc.M610943200. [DOI] [PubMed] [Google Scholar]
- 67.Jones D.C. Wein M.N. Oukka M. Hofstaetter J.G. Glimcher M.J. Glimcher L.H. Regulation of adult bone mass by the zinc finger adapter protein Schnurri-3. Science. 2006;312:1223. doi: 10.1126/science.1126313. [DOI] [PubMed] [Google Scholar]
- 68.Yoshida C.A. Furuichi T. Fujita T. Fukuyama R. Kanatani N. Kobayashi S., et al. Core-binding factor beta interacts with Runx2 and is required for skeletal development. Nat Genet. 2002;32:633. doi: 10.1038/ng1015. [DOI] [PubMed] [Google Scholar]
- 69.Lee K.S. Hong S.H. Bae S.C. Both the Smad and p38 MAPK pathways play a crucial role in Runx2 expression following induction by transforming growth factor-beta and bone morphogenetic protein. Oncogene. 2002;21:7156. doi: 10.1038/sj.onc.1205937. [DOI] [PubMed] [Google Scholar]
- 70.Hong J.H. Hwang E.S. McManus M.T. Amsterdam A. Tian Y. Kalmukova R., et al. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309:1074. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- 71.Tokuzawa Y. Yagi K. Yamashita Y. Nakachi Y. Nikaido I. Bono H., et al. Id4, a new candidate gene for senile osteoporosis, acts as a molecular switch promoting osteoblast differentiation. PLoS Genet. 2010;6:e1001019. doi: 10.1371/journal.pgen.1001019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shimoyama A. Wada M. Ikeda F. Hata K. Matsubara T. Nifuji A., et al. Ihh/Gli2 signaling promotes osteoblast differentiation by regulating Runx2 expression and function. Mol Biol Cell. 2007;18:2411. doi: 10.1091/mbc.E06-08-0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee K.N. Jang W.G. Kim E.J. Oh S.H. Son H.J. Kim S.H., et al. Orphan nuclear receptor chicken ovalbumin upstream promoter-transcription factor II (COUP-TFII) protein negatively regulates bone morphogenetic protein 2-induced osteoblast differentiation through suppressing runt-related gene 2 (Runx2) activity. J Biol Chem. 2012;287:18888. doi: 10.1074/jbc.M111.311878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang X. Matsuda K. Bialek P. Jacquot S. Masuoka H.C. Schinke T., et al. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry syndrome. Cell. 2004;117:387. doi: 10.1016/s0092-8674(04)00344-7. [DOI] [PubMed] [Google Scholar]
- 75.Poteet-Smith C.E. Smith J.A. Lannigan D.A. Freed T.A. Sturgill T.W. Generation of constitutively active p90 ribosomal S6 kinase in vivo. Implications for the mitogen-activated protein kinase-activated protein kinase family. J Biol Chem. 1999;274:22135. doi: 10.1074/jbc.274.32.22135. [DOI] [PubMed] [Google Scholar]
- 76.Elefteriou F. Ahn J.D. Takeda S. Starbuck M. Yang X. Liu X., et al. Leptin regulation of bone resorption by the sympathetic nervous system and CART. Nature. 2005;434:514. doi: 10.1038/nature03398. [DOI] [PubMed] [Google Scholar]
- 77.Cao H. Yu S. Yao Z. Galson D.L. Jiang Y. Zhang X., et al. Activating transcription factor 4 regulates osteoclast differentiation in mice. J Clin Invest. 2010;120:2755. doi: 10.1172/JCI42106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Teitelbau S.L. Ross F.P. Genetic regulation of osteoclast development and function. Nat Rev Genet. 2003;4:638. doi: 10.1038/nrg1122. [DOI] [PubMed] [Google Scholar]
- 79.Wada T. Nakashima T. Hiroshi N. Penninger J.M. RANKL-RANK signaling in osteoclastogenesis and bone disease. Trends Mol Med. 2006;12:17. doi: 10.1016/j.molmed.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 80.Wu M. Hesse E. Morvan F. Zhang J.P. Correa D. Rowe G.C., et al. Zfp521 antagonizes Runx2, delays osteoblast differentiation in vitro, and promotes bone formation in vivo. Bone. 2009;44:528. doi: 10.1016/j.bone.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hesse E. Kiviranta R. Wu M. Saito H. Yamana K. Correa D., et al. Zinc finger protein 521, a new player in bone formation. Ann N Y Acad Sci. 2010;1192:32. doi: 10.1111/j.1749-6632.2009.05347.x. [DOI] [PubMed] [Google Scholar]
- 82.Correa D. Hesse E. Seriwatanachai D. Kiviranta R. Saito H. Yamana K., et al. Zfp521 is a target gene and key effector of parathyroid hormone-related peptide signaling in growth plate chondrocytes. Dev Cell. 2010;19:533. doi: 10.1016/j.devcel.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hesse E. Saito H. Kiviranta R. Correa D. Yamana K. Neff L., et al. Zfp521 controls bone mass by HDAC3-dependent attenuation of Runx2 activity. J Cell Biol. 2010;191:1271. doi: 10.1083/jcb.201009107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Seriwatanachai D. Densmore M.J. Sato T. Correa D. Neff L. Baron R., et al. Deletion of Zfp521 rescues the growth plate phenotype in a mouse model of Jansen metaphyseal chondrodysplasia. FASEB J. 2011;25:3057. doi: 10.1096/fj.11-183277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jeon E.J. Lee K.Y. Choi N.S. Lee M.H. Kim H.N. Jin Y.H., et al. Bone morphogenetic protein-2 stimulates Runx2 acetylation. J Biol Chem. 2006;281:16502. doi: 10.1074/jbc.M512494200. [DOI] [PubMed] [Google Scholar]
- 86.Vega R.B. Matsuda K. Oh J. Barbosa A.C. Yang X. Meadows E., et al. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119:555. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 87.Schroeder T.M. Kahler R.A. Li X. Westendorf J.J. Histone deacetylase 3 interacts with runx2 to repress the osteocalcin promoter and regulate osteoblast differentiation. J Biol Chem. 2004;279:41998. doi: 10.1074/jbc.M403702200. [DOI] [PubMed] [Google Scholar]
- 88.Westendorf J.J. Zaidi S.K. Cascino J.E. Kahler R. van Wijnen A.J. Lian J.B., et al. Runx2 (Cbfa1, AML-3) interacts with histone deacetylase 6 and represses the p21(CIP1/WAF1) promoter. Mol Cell Biol. 2002;22:7982. doi: 10.1128/MCB.22.22.7982-7992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jensen E.D. Schroeder T.M. Bailey J. Gopalakrishnan R. Westendorf J.J. Histone deacetylase 7 associates with Runx2 and represses its activity during osteoblast maturation in a deacetylation-independent manner. J Bone Miner Res. 2008;23:361. doi: 10.1359/JBMR.071104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Smits P. Bolton A.D. Funari V. Hong M. Boyden E.D. Lu L., et al. Lethal skeletal dysplasia in mice and humans lacking the golgin GMAP-210. N Engl J Med. 2010;362:206. doi: 10.1056/NEJMoa0900158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Byers B.A. Guldberg R.E. García A.J. Synergy between genetic and tissue engineering: Runx2 overexpression and in vitro construct development enhance in vivo mineralization. Tissue Eng. 2004;10:1757. doi: 10.1089/ten.2004.10.1757. [DOI] [PubMed] [Google Scholar]
- 92.Byers B.A. Guldberg R.E. Hutmacher D.W. García A.J. Effects of Runx2 genetic engineering and in vitro maturation of tissue-engineered constructs on the repair of critical size bone defects. J Biomed Mater Res A. 2006;76:646. doi: 10.1002/jbm.a.30549. [DOI] [PubMed] [Google Scholar]
- 93.Wojtowicz A.M. Templeman K.L. Hutmacher D.W. Guldberg R.E. García A.J. Runx2 overexpression in bone marrow stromal cells accelerates bone formation in critical-sized femoral defects. Tissue Eng Part A. 2010;16:2795. doi: 10.1089/ten.tea.2010.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Phillips J.E. Hutmacher D.W. Guldberg R.E. García A.J. Mineralization capacity of Runx2/Cbfa1-genetically engineered fibroblasts is scaffold dependent. Biomaterials. 2006;27:5535. doi: 10.1016/j.biomaterials.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 95.Phillips J.E. Guldberg R.E. García A.J. Dermal fibroblasts genetically modified to express Runx2/Cbfa1 as a mineralizing cell source for bone tissue engineering. Tissue Eng. 2007;13:2029. doi: 10.1089/ten.2006.0041. [DOI] [PubMed] [Google Scholar]
- 96.Phillips J.E. Burns K.L. Le Doux J.M. Guldberg R.E. García A.J. Engineering graded tissue interfaces. Proc Natl Acad Sci USA. 2008;105:12170. doi: 10.1073/pnas.0801988105. [DOI] [PMC free article] [PubMed] [Google Scholar]